Abstract
The evolutionary processes responsible for the long-term persistence of glycopeptide-resistant Enterococcus faecium (GREF) in nonselective environments were addressed by genetic analyses of E. faecium populations in animals and humans on two Norwegian poultry farms that were previously exposed to avoparcin. A total of 222 fecal GREF (n = 136) and glycopeptide-susceptible (n = 86) E. faecium (GSEF) isolates were obtained from farmers and poultry on three separate occasions in 1998 and 1999. Pulsed-field gel electrophoresis (PFGE) and plasmid DNA analyses discerned 22 GREF and 32 GSEF PFGE types within shifting polyclonal animal and human E. faecium populations and indicated the presence of transferable plasmid-mediated vanA resistance, respectively. Examples of dominant, persistent GREF PFGE types supported the notion that environmentally well-adapted GREF types may counteract the reversal of resistance. PFGE analyses, sequencing of the purK housekeeping gene, and partial typing of vanA-containing Tn1546 suggested a common animal and human reservoir of glycopeptide resistance. Inverse PCR amplification and sequence analyses targeting the right end of the Tn1546-plasmid junction fragment strongly indicated the presence of a common single Tn1546-plasmid-mediated element in 20 of 22 GREF PFGE types. This observation was further strengthened by vanY-vanZ hybridization analyses of plasmid DNAs as well as the finding of a physical linkage between Tn1546 and a putative postsegregation killing system for seven GREF PFGE types. In conclusion, our observations suggest that the molecular unit of persistence of glycopeptide resistance is a common mobile plasmid-mediated vanA-containing element within a polyclonal GREF population that changes over time. In addition, we propose that “plasmid addiction systems” may contribute to the persistence of GREF in nonselective environments.
Community reservoirs of VanA-type glycopeptide-resistant enterococci (GRE) expressing transferable high-level vancomycin resistance were reported in several European countries in the early and mid-1990s (1, 8, 16, 32, 33, 48, 50). Several lines of evidence have supported a link between the use of the glycopeptide antibiotic avoparcin as a growth-promoting agent in livestock and the occurrence of GRE in the community (1, 7, 31, 33, 43, 46, 52, 55). Thus, the use of avoparcin was abandoned in Norway and Denmark in 1995, in Germany in 1996, and in all remaining European Union countries in 1997 in order to reduce human exposure to animal GRE.
The avoparcin ban has given us a unique opportunity for large-scale monitoring of the population dynamics of resistant bacteria and the fate of antibiotic resistance elements when they are not directly selected for. Several reports have demonstrated a decline in the occurrence of GRE in commercial poultry meat as well as in animal and human fecal samples after the discontinued use of avoparcin (30, 38, 49). A comprehensive Danish study revealed that the prevalence of glycopeptide resistance in random isolates of fecal Enterococcus faecium isolated from poultry was reduced from 72.7% in 1995, when the selective pressure was removed, to 5.8% in 2000 (3). Thus, the avoparcin ban seems to have reduced the potential human exposure to GRE through the food chain. Follow-up studies from Norway and Denmark have nevertheless shown that there is a remarkable persistence of GRE on poultry farms that were formerly exposed to avoparcin. In Norway, fecal GRE were isolated by direct selective plating from 99% (72 of 73) of poultry samples and by growth in selective enrichment broth from 18% (13 of 73) of farmers on 73 avoparcin-exposed farms 3 years after the ban (10). Corresponding results were obtained by a similar Danish study 5 years after the ban (24) as well as by a cohort study of 29 Norwegian avoparcin-exposed poultry farms 4 to 7 years after the ban (unpublished data). Accordingly, it appears that the discontinuation of avoparcin use has led to a reduction in the proportion of GRE among enterococci on formerly avoparcin-exposed farms. However, GRE are still present in poultry farm environments and are readily detectable after years without any apparent glycopeptide selection.
The mechanisms of GRE persistence in the absence of selective glycopeptide pressure are unknown. A potential coselection mechanism has been proposed due to the observed genetic linkage of macrolide and copper sulfate resistance determinants to glycopeptide resistance (2, 12, 23). Our previous experimental observations of the persistence of plasmid-mediated VanA resistance in enterococci in vitro and in gnotobiotic mice in the absence of antibiotic selection suggest that environmental adaptation, in vivo gene transfer, and a plasmid maintenance system(s) may counteract the cost-driven reversibility of resistance (28). The present communication is a comprehensive analysis of fecal human and animal glycopeptide-resistant (GREF) and -susceptible (GSEF) E. faecium strains isolated from two Norwegian poultry farms on three occasions 3 to 4 years after the abolishment of avoparcin. The main objective of this study was to investigate the genetic relatedness within and between human and animal GSEF and GREF populations in order to elucidate possible mechanisms for the observed long-term persistence of animal and human GREF in the absence of glycopeptide selection.
MATERIALS AND METHODS
Isolation of GREF and GSEF strains.
GREF and GSEF strains were isolated from fecal samples from two poultry farmers and their poultry on the following three separate occasions over a 1-year period: fall 1998 (F98), spring 1999 (S99), and fall 1999 (F99). The collection of human fecal samples was approved by the Regional Committee for Medical Research Ethics, University of Tromsø, Tromsø, Norway.
The samples were collected, transported, and processed as previously described (10). Briefly, due to low GREF concentrations, human samples were enriched in 10 ml of Enterococcosel broth (Becton Dickinson, Franklin Lakes, N.J.) with 8 mg of vancomycin/liter at 37°C for 24 to 48 h. Ten parallel cultures were made from each human sample in order to enhance the possibility of detecting GREF diversity. Fifty microliters of each parallel culture was inoculated onto cephalexin-, arabinose-, and aztreonam-containing (CAA) agar plates supplemented with 8 mg of vancomycin/liter (CAAV) at 37°C for 24 to 48 h (18). Arabinose-positive colonies were collected for further analysis. Human GSEF strains were isolated by direct plating of 10 μl of fecal solutions onto CAA agar plates. Five to 10 single arabinose-positive colonies were selected from each sample. Animal GREF and GSEF strains were isolated by direct plating of 100-μl fecal dilutions on Slanetz and Bartley's Enterococcus agar plates (Oxoid, Basingstoke, Hampshire, England), with or without 50 mg of vancomycin/liter. The direct plating method for poultry samples had a sensitivity of approximately 1 GREF cell per mg of feces. Five to 10 single enterococcus-like colonies were selected from each sample. The sampling strategy did not allow us to determine whether the isolates were from the same animal or different animals. Strains were named according to the farm from which they were isolated (399 or 64), the time of isolation (F98, S99, or F99), and whether they were of animal (A) or human (H) origin. Glycopeptide-susceptible strains were labeled with an “S” (susceptible). Quality control strains are listed in Table 1.
TABLE 1.
Bacterial control strains used for this study
| Strain | Purpose | Reference or sourcea |
|---|---|---|
| Bacillus subtilis ATCC 6633 | Specificity; Enterococcosel broth | ATCC |
| Escherichia coli ATCC 25922 | Specificity; Enterococcosel broth | ATCC |
| E. faecalis ATCC 29212 | Sensitivity; CAA and CAAV agar, susceptibility testing, and ddl PCR; vancomycin-susceptible control strain. | ATCC |
| E. faecium ATCC 19434 | Sensitivity; CAA agar and ddl PCR | ATCC |
| E. faecium BM4147 | Sensitivity and specificity; CAAV agar, S&Bb agar, and Enterococcosel broth with vancomycin, vanA, and vanY-vanZ hybridizations and PCRs, Tn1546 long PCRs, vanX DdeI digestions (G at position 8,234); VanA-type vancomycin-resistant control strain (vancomycin MIC, >256 mg/liter) | 34 |
| E. faecium BM4147.1 | Sensitivity and specificity; S&B agar and Enterococcosel broth with vancomycin, vanA, tcrB, pRE25 PSK, and Tn1546-P PCRs; plasmid-cured vancomycin-susceptible derivative of BM4147 | 34 |
| E. faecalis ATCC 51299 | Sensitivity; CAA and CAAV agar; VanB-type vancomycin-resistant control strain (vancomycin MIC, 16 to 32 mg/liter) | ATCC |
| E. casseliflavus ATCC 25788 | Sensitivity; CAA and CAAV agar | ATCC |
| E. gallinarum ATCC 49608 | Sensitivity and specificity; CAAV agar, ddl PCR | ATCC |
| E. faecium A17sv1 | Specificity; tcrB PCR | 23 |
| E. faecium 399/F98/H1 | Specificity; Tn1546-P PCR | This study |
| E. faecium 399/F99/A9 | Specificity; pRE25-PSK PCR | This study |
| E. faecium TUH2-13 | Specificity; DdeI digestions (T at position 8,234) | 10 |
ATCC, American Type Culture Collection.
S&B, Slawetz and Bartley’s.
Identification of E. faecium and antimicrobial susceptibility testing.
Gram-positive, catalase-negative, and pyrrolidonyl-β-naphthylamide hydrolysis-positive (PYR disks; Dalynn Biologicals, Calgary, Canada) isolates were further examined by a species-specific enterococcal ddl PCR (17, 43). The glycopeptide susceptibilities of E. faecium isolates were examined by growth on brain heart infusion agar supplemented with 6 mg of vancomycin/liter (Difco Laboratories, Detroit, Mich.), by use of a vancomycin and teicoplanin Etest (AB Biodisk, Solna, Sweden), and genotypically by vanA PCR (44). The PCR primers are presented in Table 2. Extended susceptibility testing was performed according to the manufacturer's instructions (Rosco, Taastrup, Denmark) with tablets containing 250 μg of gentamicin, 500 μg of streptomycin, 33 μg of ampicillin, 78 μg of erythromycin, and 10 μg of tetracycline. The strains were categorized as susceptible (S), intermediately susceptible (I), or resistant (R) according to the criteria of the Norwegian Working Group for Antibiotics (9).
TABLE 2.
PCR primers used for this study
| PCR assay | Primer | Sequence(5′-3′) | Product size (bp) | Reference |
|---|---|---|---|---|
| Tn1546a | ORF1 F9 | AAC CTA AGG GCG ACA TAT GGT G | 10,414 | 21 |
| vanZ R11 | GGT ACG GTA AAC GAG CAA TAA TAC G | |||
| vanAb | vanAF | GTT GCA ATA CTG TTT GGG GG | 1,013 | 44 |
| vanAR | CCC CTT TAA CGC TAA TAC GAT CAA | |||
| vanX | vanX1 | ACT TGG GAT AAT TTC ACC GG | 424 | 44 |
| vanX2 | TGC GAT TTT GCG CTT CAT TG | |||
| vanY-vanZb | vanYF1 | ATG GAT ACG GGT TGC TTG ATA T | 1,375 | 44 |
| vanZR1 | TTT CCC CTC ACT TCA VCAC CTA C | |||
| Tn1546-Pb | vanZF | AGT GCT GAG GAA TTG GTC TCT | 422 | This study |
| P2 R | CCG AGA AAG CTG GTT AAG TCT A | |||
| ORF18-19 (pRE25 PSK)b | ζ-F | GTG GTT TAG GTG GCT GCA AG | 1,044 | This study |
| ɛ-R | TTA ACG AAT TAT CGG CAA GC | |||
| tcrB | 415 | TGA CAA TAA GGC AAC GAT TTC | 871 | 23 |
| 416 | CCA GGC ATG ATG TCC TTG |
Amplicon used as a positive control for Tn1546 hybridizations.
Amplicon used as a probe.
Analyses of chromosomal DNA.
All isolates were examined by pulsed-field gel electrophoresis (PFGE) of SmaI-digested (Promega, Madison, Wis.) total DNA as previously described (44), with the following modifications: mutanolysin was added to the lysis buffer (40 U/ml) and the gels were run at 200 V for 30 h. The criteria presented by Tenover et al. (47) were used for interpretations of genetic relatedness. PFGE data were analyzed with Gel Compare II software (Applied Maths, St. Martens-Latem, Belgium), and a dendrogram was constructed by the use of Dice coefficients and the unweighted pair group method with arithmetic mean, with the bandwidth tolerance set critically at 1.5%. To probe for the chromosomal localization of glycopeptide resistance genes, we separated I-CeuI (New England Biolabs, Beverly, Mass.)-restricted total DNAs from representative strains of all PFGE types by PFGE, transferred them to nylon membranes, and hybridized them separately with 16S rRNA gene and vanA digoxigenin (DIG)-labeled DNA probes as previously described (15). A 10,414-bp Tn1546 PCR amplicon from E. faecium BM4147 was used as a positive control for vanA hybridization (21).
purK allele analysis.
The purK housekeeping gene in representative GREF and GSEF PFGE types was amplified by PCR in order to investigate allele polymorphisms in E. faecium isolates from humans and poultry (29). The purK PCR products were purified by use of the PCR Product Presequencing kit (Amersham Biosciences AB, Uppsala, Sweden) according to the manufacturer's instructions and were sequenced by use of an ABI Prism BigDye cycle sequencing Ready Reaction kit (PE Biosystems, Foster City, Calif.). The products were analyzed in an ABI Prism 3100 DNA sequencer (PE Biosystems). The double-stranded sequences were aligned with purK alleles present at the multilocus sequence type (MLST) website (http://www.mlst.net/links/default.asp).
Analyses of tcrB and vanX DNA sequences.
Strains representing all PFGE types were screened for the presence of tcrB by PCR as previously described in order to investigate possible genetic linkages between CuSO4 resistance and vanA resistance genes (23). DdeI (Promega) digestion of vanX PCR products was performed to differentiate between G and T nucleotides at position 8,234 in Tn1546 (10, 27).
Plasmid DNA analyses.
Plasmid DNAs were isolated by alkaline lysis as previously described (41), with the following modifications: mutanolysin (40 U/ml) was included with lysozyme (20 mg/ml) prior to the lysis step and then incubated at 37°C for 1 h. Plasmid DNA isolated from a representative strain of each GREF PFGE type was digested separately with ClaI, PstI, or BclI according to the instructions of the manufacturer (Promega) and then analyzed by agarose gel electrophoresis. Southern hybridization of ClaI-digested plasmid DNAs with a DIG-labeled vanY-vanZ probe targeting the right end of Tn1546 was performed as previously described (44). The target region was hence designated the ClaI-Tn1546 junction fragment. ClaI-Tn1546 junction fragments from strains 399/F98/H1 and 399/F98/A4 were further characterized by inverse PCR and DNA sequencing (37). Briefly, ClaI-restricted plasmid DNAs were ligated overnight at room temperature by the use of T4 DNA ligase (Promega). Forward and reverse vanY-vanZ PCR primers were inverted, thus priming DNA amplification away from the vanY-vanZ core sequence into the unknown sequence of the adjacent plasmid DNA. Amplicon sequencing was performed as described above. DNA sequence data for strain 399/F98/H1 were used to establish a specific Tn1546-plasmid junction PCR (Tn1546-P PCR) that amplified 372 bp of the ClaI-restricted plasmid junction fragment and 50 bp of ClaI-Tn1546. Tn1546-P PCR products were analyzed by DraI digestion (Promega).
We previously hypothesized that postsegregational killing (PSK) systems can promote the stability of plasmid-mediated vanA resistance in the absence of overt antimicrobial selection (28). Recently, open reading frames (ORFs) 18 and 19 of pRE25 isolated from Enterococcus faecalis (42) were suggested as structural and functional homologues of the ɛ/ζ-PSK system encoded by pSM19035 from Streptococcus pyogenes (36). The presence of pRE25 carrying ORFs 18 and 19 was investigated by a pRE25 PSK PCR. Moreover, a genetic linkage between the putative pRE25 PSK system and Tn1546 was examined by separate hybridizations of PstI-digested plasmid DNAs from pRE25 PSK PCR-positive strains with DIG-labeled vanY-vanZ and pRE25 PSK probes.
Transfer studies.
Representative strains of all GREF PFGE types were examined for transferable vancomycin resistance by filter mating, as previously described (14). E. faecium UW 64/3 (Rifr Fusr) was used as a recipient (53). Donor strains that failed to support transferable vancomycin resistance were retested once. One transconjugant (Vmr Rifr Fusr) from each experiment was examined by SmaI-PFGE analysis and compared to recipient and donor strains. Transconjugants were also examined by ClaI and BclI restriction analyses of plasmid DNAs and were assayed for their susceptibility to streptomycin, ampicillin, gentamicin, erythromycin, and tetracycline.
Nucleotide sequence accession number.
The sequence of the right-end Tn1546-plasmid junction fragment has been given the GenBank accession number AY592978.
RESULTS
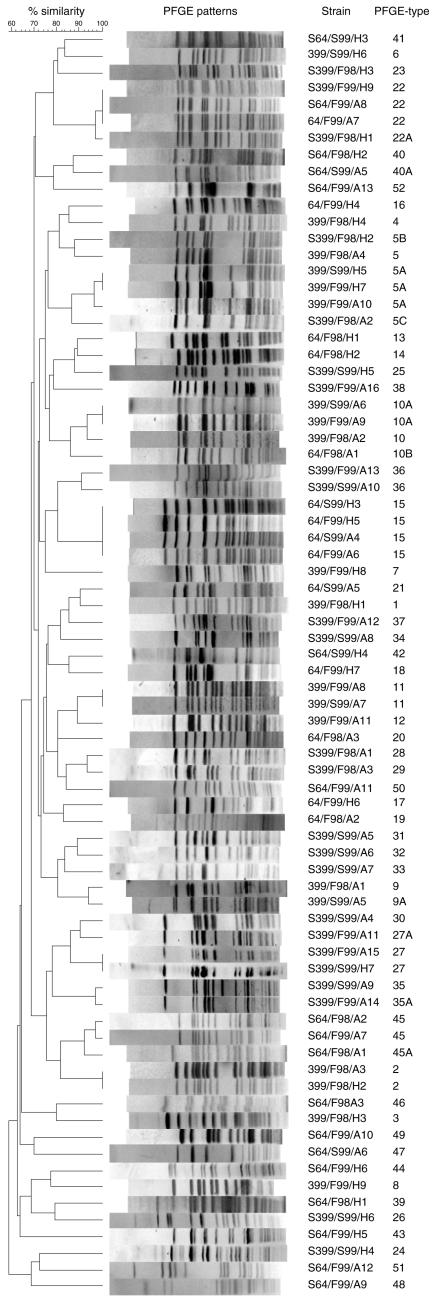
A total of 254 enterococcus-like isolates were collected, 222 of which were confirmed to be E. faecium and were included in this study. Glycopeptide resistance was expressed in 136 isolates (GREF), whereas 86 isolates were susceptible to glycopeptides (GSEF). Totals of 72 (43 samples from farmers and 29 samples from poultry) and 64 (38 samples from farmers and 26 samples from poultry) isolates were confirmed to be GREF for farms 399 and 64, respectively. The GSEF strains numbered 45 (23 samples from farmers and 22 samples from poultry) and 41 (24 samples from farmers and 17 samples from poultry) for farms 399 and 64, respectively. The 222 strains were grouped into 52 different PFGE types, with 22 GREF types and 32 GSEF types. Two PFGE types (5 and 22) were found among both susceptible and resistant isolates. The distributions and relative numbers of the different resistant and susceptible PFGE types are given in Tables 3 and 4, respectively. A dendrogram showing representative strains (n = 76) of all PFGE types and subtypes found among isolates from poultry and farmers at both farms is presented in Fig. 1.
TABLE 3.
Distribution, relative numbers, and characteristics of GREF PFGE types in this study
| Farm | Origin | PFGE type(s) | No. of isolates
|
Transferable glycopeptide resistance | Common Tn1546 junction fragment | pRE25 PSK PCR | Cohybridization Tn1546-pRE25 PSK | ||
|---|---|---|---|---|---|---|---|---|---|
| Fall 1998 | Spring 1999 | Fall 1999 | |||||||
| 399 | Farmer | 1 | 7 | + | + | + | −a | ||
| 2 | 1 | + | + | − | |||||
| 3 | 2 | + | − | + | + | ||||
| 4 | 1 | + | + | + | + | ||||
| 5A | 11 | 15 | + | + | + | + | |||
| 6 | 3 | − | + | − | |||||
| 7 | 1 | + | + | + | + | ||||
| 8 | 2 | − | + | − | |||||
| Poultry | 9, 9A | 4 | 1 | + | + | + | − | ||
| 10, 10A | 4 | 7 | 4 | − | + | + | +b | ||
| 2 | 1 | + | + | − | |||||
| 5, 5A | 1 | 2 | + | + | + | +b | |||
| 11 | 1 | 3 | − | + | + | + | |||
| 12 | 1 | + | + | − | |||||
| 64 | Farmer | 13 | 5 | − | + | − | |||
| 14 | 8 | − | + | − | |||||
| 15 | 12 | 2 | − | + | − | ||||
| 16 | 8 | − | + | + | + | ||||
| 17 | 2 | − | + | − | |||||
| 18 | 1 | + | + | − | |||||
| Poultry | 10B | 8 | − | + | − | ||||
| 19 | 1 | + | + | − | |||||
| 20 | 1 | − | + | − | |||||
| 15 | 6 | 6 | − | + | − | ||||
| 21 | 3 | + | + | − | |||||
| 22 | 1 | − | − | − | |||||
vanY-vanZ and pRE25 PSK probes revealed positive signals for different PstI fragments.
The presence of pRE25 PSK was not a constant feature in PFGE types 5 and 10. The presence of pRE25 PSK and its physical linkage to Tn1546 were demonstrated when they were recovered as PFGE types 5A and 10A in spring and fall 1999.
TABLE 4.
Distribution and relative numbers of GSEF PFGE types
| Farm | Origin | PFGE type | No. of isolates
|
||
|---|---|---|---|---|---|
| Fall 1998 | Spring 1999 | Fall 1999 | |||
| 399 | Farmer | 5B | 2 | ||
| 22, 22A | 2 | 9 | |||
| 23 | 1 | ||||
| 24 | 5 | ||||
| 25 | 1 | ||||
| 26 | 2 | ||||
| 27 | 1 | ||||
| Poultry | 5c | 2 | |||
| 27, 27A | 1 | 2 | |||
| 28 | 2 | ||||
| 29 | 3 | ||||
| 30 | 1 | ||||
| 31 | 1 | ||||
| 32 | 1 | ||||
| 33 | 1 | ||||
| 34 | 1 | ||||
| 35, 35A | 1 | 1 | |||
| 36 | 1 | 2 | |||
| 37 | 1 | ||||
| 38 | 1 | ||||
| 64 | Farmer | 39 | 9 | ||
| 40 | 1 | ||||
| 41 | 9 | ||||
| 42 | 1 | ||||
| 43 | 3 | ||||
| 44 | 1 | ||||
| Poultry | 22 | 2 | |||
| 40A | 1 | ||||
| 45, 45A | 1,a 2 | 4 | |||
| 46 | 1 | ||||
| 47 | 1 | ||||
| 48 | 1 | ||||
| 49 | 1 | ||||
| 50 | 1 | ||||
| 51 | 1 | ||||
| 52 | 1 | ||||
Relative number of PFGE type 45A isolates. PFGE type 45 was isolated in F99 and F98, with the related type 45A only recovered in F98.
FIG. 1.
PFGE dendrogram showing clustering (by UPGMA and the Dice coefficient) of all GREF and GSEF PFGE types recovered from two poultry farmers and their poultry on three different occasions(fall 1998, spring 1999, and fall 1999). The strains were grouped into 52 different PFGE types (including 11 PFGE subtypes, indicated with a letter following the type number). A total of 24 PFGE types were isolated on more than one occasion and/or recovered from both poultry and farmer fecal samples. The strains were named according to the farm from which they were isolated (399 or 64), the time of isolation (F98, S99, or F99), and whether they were of animal (A) or human (H) origin. Glycopeptide-susceptible strains were labeled with an “S” (susceptible). The bandwidth tolerance was critically set at 1.5%.
PFGE analysis of GREF strains.
A total of 22 different PFGE types, with 14 human types and 11 animal types, were found among the 136 GREF strains (Table 3). Three PFGE types were isolated from both poultry and farmer samples. PFGE subtypes 5A, 9A, 10A, and 10B were considered related to the corresponding main PFGE types, differing by four, two, three, and two bands from types 5, 9, and 10, respectively (Fig. 1). Twelve different PFGE types were found on farm 399, with eight human types and six poultry types. Types 2 and 5 were isolated from both poultry and farmers. Four PFGE types were recovered on more than one occasion (types 5, 9, 10, and 11). Only one PFGE type was present throughout the study period, i.e., type 10, which was recovered only from poultry samples. PFGE type 5A strains occurred in farmer specimens from both S99 and F99 in high relative numbers. This observation might reflect a selective advantage during broth enrichment. Thus, the relative distribution of PFGE types is probably better disclosed by direct selective plating. In this regard, it is noteworthy that PFGE type 10, including subtype 10A, was dominant in poultry samples from farm 399 on all three sampling occasions. A total of 11 different PFGE types were recovered from farm 64, with 6 farmer types and 6 poultry types. PFGE type 15, the dominant poultry PFGE type, was found in S99 and F99 in both farmers and poultry. Interestingly, the poultry PFGE type 10B strains, with high relative numbers in F98, were closely related to type 10 strains recovered from poultry on farm 399 throughout the study period (Fig. 1).
PFGE analysis of GSEF strains.
Thirty-two different PFGE types, with 13 human types and 23 animal types, were found among 86 GSEF strains isolated from the two farms (Table 4). Four PFGE types were recovered from both poultry and farmer samples. PFGE subtypes 5B, 5C, 22A, 27A, 35A, 40A, and 45A were considered related or possibly related (5C and 40A) to the corresponding main PFGE types, differing by two, five, two, four, one, five, and two bands, respectively (Fig. 1). Two main GSEF types and their subtypes (5B, 5C, 22, and 22A) showed genetic relatedness to GREF strains. Altogether, 18 different PFGE types were isolated from farm 399, with 7 farmer types and 13 poultry types. Types 5 and 27 were recovered from both poultry and farmer samples in F98. Interestingly, PFGE type 5 was also found within the GREF population from farm 399 and was the dominant type in farmer samples from S99 and F99. Four PFGE types were recovered on two occasions, namely, types 27, 35, and 36, recovered from poultry, and type 22, recovered from farmer specimens. Fifteen PFGE types were found on farm 64, with 6 farmer types and 10 poultry types. PFGE type 40 was recovered from both farmer and poultry samples. Only PFGE type 45 was recovered on two occasions from farm 64, and both times it was recovered from poultry specimens. PFGE type 22 was found within both the GSEF and GREF populations from farm 64 in samples from F99. Surprisingly, an indistinguishable PFGE type, type 22, seemed to dominate within the GSEF population from farm 399 in samples from F99 (Fig. 1).
purK allele.
Five purK alleles were found in the 40 GREF (n = 11) and GSEF (n = 29) PFGE types examined. The purK6 allele was found in all 11 GREF types, including 5 isolates from animals and 6 isolates from humans. The purK pattern within the GSEF population was similar. The purK6 allele appeared in 17 of 18 poultry and 8 of 11 farmer GSEF PFGE types examined. Two new purK types were discerned for an isolate from poultry and an isolate from a farmer, whereas two strains from farmers harbored other types, i.e., purK7 and purK17.
Antimicrobial susceptibility testing of GREF strains.
Representative strains from all 22 PFGE types expressed high-level vancomycin resistance (≥64 mg/liter), as measured by Etest. Teicoplanin resistance (>4 mg/liter) was found in all but one strain, 399/F98/H1 (PFGE type 1), for which the teicoplanin MIC was 0.032 mg/liter. All GREF strains were confirmed as positive for vanA by PCR (data not shown). Seven and four strains were resistant to tetracycline and erythromycin, respectively. Erythromycin and tetracycline resistance was coexpressed in one strain. All GREF PFGE types were susceptible to ampicillin, gentamicin, and streptomycin.
Analyses of tcrB and vanX DNA sequences.
All GREF PFGE types were negative by PCR for the CuSO4 resistance determinant tcrB, indicating that resistance to copper sulfate is not linked to glycopeptide resistance in Norwegian avoparcin-related GREF populations, as opposed to findings for GREF strains isolated from Danish pigs (23). DdeI restriction patterns of vanX amplicons from 21 of 22 GREF PFGE types revealed a G at position 8,234, consistent with the so-called poultry vanX type (27) in GREF strains from both farmers and animals (data not shown). The amplification of vanX failed repeatedly for vanA PCR-positive GREF PFGE type 22 samples, indicating amplification failure due to mutations affecting primer annealing.
Localization of glycopeptide resistance genes and characterization of plasmids.
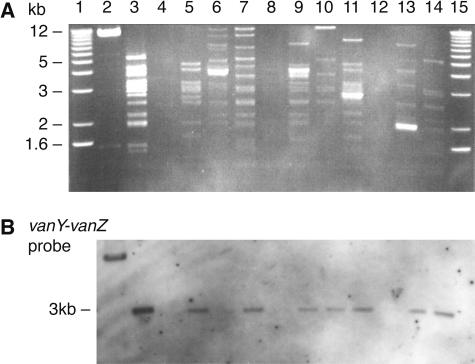
Southern blot hybridization of I-CeuI-digested total DNAs with a 16S rRNA gene probe revealed positive hybridization to four or five DNA fragments for all GREF PFGE types, as expected. Subsequent hybridization with a vanA probe was negative for all GREF PFGE types except for the BM4147-derived Tn1546 long PCR amplicon, which was used as a positive control (data not shown). The results support the plasmid localization of glycopeptide resistance genes for all GREF PFGE types. This was confirmed by plasmid DNA analyses. ClaI restriction fragment length polymorphism (RFLP) analyses of plasmid DNAs from representative strains of all 22 GREF PFGE types showed mainly highly diverse patterns, as illustrated in Fig. 2A, which presents ClaI-RFLP profiles for 9 different GREF PFGE types. Two strains, 399/F98/A4 (PFGE type 5) and 399/S99/A5 (PFGE type 9A), displayed related RFLP patterns (Fig. 2A, lanes 6 and 7, respectively). Indistinguishable ClaI-RFLP patterns were detected for one human-derived strain (399/F99/H8, PFGE type 7) (Fig. 2, lane 5) and one animal-derived strain (399/F99/A9, PFGE type 10A) (data not shown). In contrast to the diverse ClaI-RFLP plasmid profiles, a surprisingly uniform pattern was disclosed when the right-end ClaI-Tn1546-plasmid junction fragments were targeted by vanY-vanZ hybridization. Positive hybridization signals were localized to an approximately 3-kb fragment for 20 of 22 GREF PFGE types, as illustrated in Fig. 2B for 9 of the PFGE types. The hybridization signal for strain 399/F98/A4 is faint (Fig. 2B, lane 6). Sequencing of the inverse PCR amplicons from strains 399/F98/H1 (PFGE type 1) and 399/F98/A4 (PFGE type 5) revealed identical 422-bp Tn1546-plasmid junction fragments. All strains with positive vanY-vanZ hybridization to 3-kb fragments supported the amplification of Tn1546-P PCR products of approximately 420 bp. DraI digestion analyses of Tn1546-P PCR amplicons revealed a common RFLP type for all Tn1546-P PCR-positive strains (data not shown). Thus, molecular evidence for a common right-end Tn1546-plasmid junction fragment was demonstrated for 20 of 22 animal and human GREF PFGE types from both farms.
FIG. 2.
Agarose gel electrophoresis of ClaI-digested plasmid DNAs representing nine different PFGE types (A) and vanY-vanZ hybridization analysis (B). Lanes 1 and 15, 1-kb DNA ladder (Sigma-Aldrich, Oslo, Norway); lane 2, long-PCR product of Tn1546 PCR (positive control); lane 3, 399/F98/H1 (PFGE type 1); lane 5, 399/F99/H8 (PFGE type 7); lane 6, 399/F98/A4 (PFGE type 5); lane 7, 399/S99/A5 (PFGE type 9A); lane 9, 64/F98/H1 (PFGE type 13); lane 10, 64/S99/H3 (PFGE type 15); lane 11, 64/F99/H4 (PFGE type 16); lane 13, 64/F98/A3 (PFGE type 20); lane 14, 64/F99/A5 (PFGE type 21); lanes 4, 8, and 12, failed plasmid isolations.
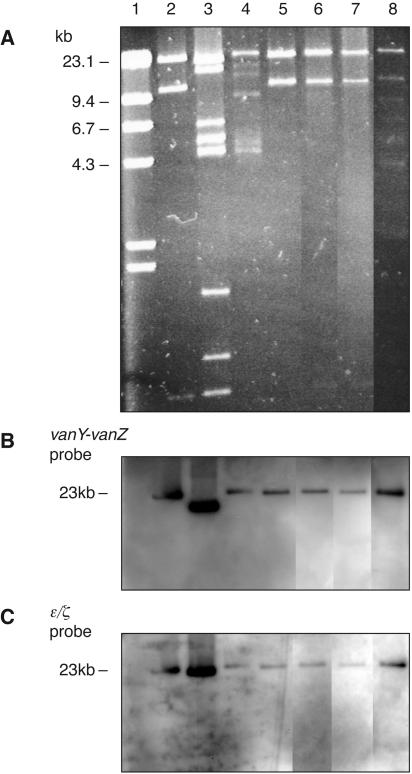
Occurrence of putative PSK system physically linked to Tn1546.
The presence of a putative pRE25 PSK system was demonstrated by PCR for 9 of 22 GREF PFGE types, including 8 from farm 399 and 1 from farm 64 (Table 3). Southern blot hybridizations with DIG-labeled vanY-vanZ and pRE25 PSK probes revealed cohybridization to equally sized PstI plasmid DNA fragments for seven PFGE types (Table 3). Except for cohybridization to a 20-kb fragment in strain 399/S99/A7 (PFGE type 11) (data not shown), the remaining six PFGE types revealed the colocalization of hybridization signals to a 23-kb PstI fragment, as illustrated in Fig. 3. Strain 399/F98/H1 (PFGE type 1) showed vanY-vanZ and pRE25 PSK probe signals on 23- and 20-kb fragments, respectively (Fig. 3A to C, lanes 3). PFGE types 4, 7, and 10 displayed indistinguishable PstI plasmid RFLP profiles (Fig. 3A, lanes 2 and 5 to 7). Thus, PstI-RFLP analysis indicated a more uniform plasmid DNA content for some PFGE types than was illustrated by ClaI restriction analysis (Fig. 2A).
FIG. 3.
Agarose gel electrophoresis of PstI-digested plasmid DNAs from pRE25 PSK PCR-positive strains (A) showing physical linkage between Tn1546 and the putative pRE25 PSK system by vanY-vanZ hybridization (B) and pRE25 PSK hybridization (C). Lane 1, λ-HindIII DNA marker (Promega); lane 2, 399/F99/A9 (PFGE type 10A, positive control); lane 3, 399/F98/H1 (PFGE type 1); lane 4, 399/F98/H3 (PFGE type 3); lane 5, 399/F98/H4 (PFGE type 4); lane 6, 399/F99/H8 (PFGE type 7); lane 7, 399/S99/A6 (PFGE type 10A); lane 8, 64/F99/H4 (PFGE type 16). Strain 399/F98/H1 (PFGE type 1) in lane 3 supported hybridization to separate fragments.
Transfer studies.
A total of 14 GREF strains representing 11 PFGE types were able to support the transfer of glycopeptide resistance (Table 3). Twenty strains representing the 11 PFGE types that were unable to transfer glycopeptide resistance were retested, with negative results. Transfer was confirmed by comparative PFGE analyses of transconjugants, donor strains, and the recipient strain (data not shown). ClaI and BclI analyses of plasmid DNAs from 14 transconjugants demonstrated diverse RFLP patterns (data not shown). All transconjugants were susceptible to tetracycline and erythromycin, supporting the lack of linked resistance genes to vanA resistance determinants. Plasmid DNAs from strains 399/F99/A9 (PFGE type 10A) and 399/F99/H8 (PFGE type 7) had indistinguishable ClaI and PstI patterns (Fig. 3, lanes 2 and 6). Strain 399/F99/A9 did not sustain the transfer of glycopeptide resistance, in contrast to 399/F99/H8, suggesting that plasmid transfer may also be dependent on host factors, as previously described (45).
DISCUSSION
This study addressed the evolutionary processes of the long-term persistence of GREF in apparently nonselective environments by analyses of the genetic relatedness within and between animal and human fecal GREF and GSEF populations on two Norwegian poultry farms that were previously exposed to avoparcin. A collection of 222 E. faecium strains were obtained on three separate occasions during a 1-year period and were examined at the chromosomal level by PFGE analysis of SmaI-restricted DNAs as well as by sequencing of the purK housekeeping genes of selected strains. Moreover, strains representing all 22 PFGE types among the 136 animal and human GREF isolates were examined with regard to the localization of the vanA-containing Tn1546 to a chromosome or plasmid, partial Tn1546 typing, and transferable vanA resistance.
A putative explanation for the observed long-term persistence of GREF after the avoparcin ban might be the survival of specific clones within the farm environment. The presence of GREF in samples from depopulated, cleaned broiler houses and the fecal colonization of broiler chicks soon after their arrival on farms have been demonstrated for formerly avoparcin-exposed Norwegian farms (11). In line with these observations, a Danish study of GREF isolated from consecutive poultry flocks demonstrated the persistence of clonally related strains by PFGE analysis (25). Our PFGE analyses revealed three examples of dominating GREF clones that have persisted over time, specifically PFGE types 5A and 10 on farm 399 and PFGE type 15 on farm 64, consistent with environmentally well-adapted resistant strains counteracting the reversal of resistance. The vanA-carrying Tn1546 supports intracellular mobilization by replicative transposition (6), and the chromosomal insertion of Tn1546 was previously demonstrated (22). A chromosomal position of the inducible vanA gene cluster within fit bacterial clones would confer a stabilization of resistance. However, the negative findings for vanA hybridizations to PFGE-separated I-CeuI-digested DNAs from all GREF types were consistent with the localization of Tn1546 to a plasmid. This observation is also in agreement with the overall picture within the animal and human GREF populations, which are characterized by the presence of multiple clones that change over time on both farms. The finding of 22 different GREF PFGE types on the two farms is most compatible with a readily transferable resistance determinant within a polyclonal E. faecium population.
Studies of VanA-type GREF have mainly demonstrated plasmid localization for Tn1546, as reviewed by Sundsfjord et al. (46). Our hybridization analyses of ClaI-restricted plasmid DNA from each GREF type confirmed the presence of single vanA clusters within diverse RFLPs. In contrast, a surprisingly uniform hybridization fragment pattern was discerned when the right-end Tn1546-plasmid junction was targeted by a vanY-vanZ probe, with a common 3-kb vanY-vanZ-positive plasmid DNA fragment detected for 20 of 22 GREF PFGE types. These findings were confirmed by the sequencing of inverse PCR amplicons targeting the Tn1546-plasmid DNA junction and by subsequent use of the Tn1546-P PCR method designed for this study.
At least two different mechanisms may explain the findings of a common plasmid-mediated vanA-containing element within a polyclonal GREF population. Tn1546 may transpose between different mobile genetic elements within the GREF population (6). Thus, the observed common Tn1546-plasmid junction fragment in 20 GREF PFGE types may represent a “hot spot” for Tn1546 integration. However, experimental data do not support a site-specific integration mechanism for Tn1546 (6). Alternatively, Tn1546 may have spread between strains as part of a larger genetic element, i.e., a composite transposon, or by recombination events between plasmids. The observed diversity in plasmid DNA profiles and the presence of a common Tn1546 right-end plasmid flanking sequence in 20 PFGE types support the latter explanation and suggest that the unit of persistence on both farms was a common mobile plasmid-mediated vanA-containing element within a polyclonal GREF population.
Other lines of evidence also suggest that horizontal gene transfer has played a significant role in the spread and persistence of the vanA cluster on the two farms. A poultry-type G nucleotide at position 8,234 of Tn1546 was found in 21 of 22 GREF PFGE types. Moreover, the vanA donor potential within the GREF population was demonstrated by in vitro transfer studies and was further substantiated by the finding of indistinguishable or highly related vanA-containing plasmid DNA RFLP types in different PFGE types, specifically PFGE types 4, 7, and 10A as well as types 5 and 9A.
Plasmids are considered highly dynamic genetic elements, and sequence analyses of different enterococcal plasmids have revealed mosaic structures of homologous sequences as well as a high prevalence of transposable elements (20, 39, 42). Insertion sequence elements such as IS1216 have previously been suggested as substrates for recombination events and for the creation of composite transposons in enterococci (40). The size of the putative common mobile plasmid-located vanA fragment in the GREF populations on both farms is unknown. DNA sequence analyses are in progress to resolve this issue.
After our observations of a vanA plasmid reservoir within a polyclonal GREF population, we turned to potential mechanisms for the persistence of plasmid-mediated vanA resistance in the absence of glycopeptide selection. The ability of resistance to persist in antimicrobial drug-free environments depends on several key parameters, including (i) the cost of resistance, (ii) the amelioration of fitness costs, (iii) genetic stability, (iv) linked selection, and (v) the horizontal transfer of resistance elements (4, 35). The biological cost of resistance seems to be an important parameter. We previously presented observations suggesting that a newly acquired vanA plasmid reduced the fitness of the E. faecium host strain approximately 4% in vitro. However, the experimental evidence suggested that horizontal gene transfer and environmental adaptation counteract the negative selection of GREF in vitro and in vivo (28), conferring a stabilization of resistance. In the present study, PFGE analysis of GREF revealed three examples of clonal persistence on both farms, suggesting successful environmental adaptation as described previously. Furthermore, PFGE data on GSEF strains from both farms provided indirect evidence for the high stability of plasmid-mediated glycopeptide resistance. If vanA plasmids were unstable in their hosts, then one would expect the isolation of genetically related GREF and GSEF types. Surprisingly, only PFGE types 5 and 22 were common between GREF and GSEF strains. However, we acknowledge the possibility that too few strains were investigated and that the heterogeneity of E. faecium strains within farms may be higher than was detected. The possibility that divergent evolution has separated GSEF and GREF strains that originally were related cannot be ruled out either.
Linked selection is an important mechanism of persistence in environments with changing patterns of antimicrobial selection. In the farm environment, a genetic linkage of Tn1546 to both macrolide and copper sulfate resistance has previously been demonstrated for enterococci (2, 12, 23). A genetic linkage of glycopeptide resistance to other resistance determinants was not observed in this study. However, a physical linkage to genes conferring other selective advantages cannot be ruled out.
Naturally occurring low-copy-number plasmids have developed mechanisms to secure stable maintenance, including partitioning (par), multimer resolution (mrs), and PSKs (19). The recent description of the first functional plasmid stability cassette in E. faecium (20) illustrates that this may be a relevant mechanism for the persistence of plasmid-mediated glycopeptide resistance. The observed genetic linkage between Tn1546 and the putative pRE25 PSK system in seven different PFGE types strengthens this notion. This linkage was recently confirmed by an ongoing plasmid DNA sequencing project with the vanA-containing plasmid p399/F99/A9 showing the presence of three ORFs with 99 to 100% nucleotide sequence homology to ORFs 17 to 19 in pRE25 (42) and strong structural homology to the well-characterized ω/ɛ/ζ-PSK system in pSM19035 (36; unpublished data). Extending these observations, we propose that a genetic linkage between Tn1546 and the putative pRE25 PSK cassette or related systems contributes significantly to vanA-containing plasmid stability by killing off or impeding the growth of plasmid-free segregants. GREF strains that are “addicted” to their glycopeptide resistance plasmids would then constitute a core population in persistent reservoirs. Moreover, 4 of 11 PFGE types that were able to transfer vanA resistance to a recipient in filter mating studies had the putative pRE25 PSK system linked to Tn1546. The horizontal transfer of a Tn1546-linked PSK element would further increase the potential for long-term persistence in the absence of antibiotic selection, when new genetic backgrounds become addicted to the plasmid.
This study supports and further extends our previous observations of a high prevalence of fecal GREF colonization among poultry farmers on avoparcin-exposed farms (10). Several observations support a genetic relationship between poultry and farmer GREF reservoirs. Three examples of indistinguishable GREF PFGE types (2, 5, and 15) isolated from both poultry and farmers were demonstrated in this study, supporting earlier reports of genetically related animal and human GREF strains (43, 50, 51). Indirect evidence for a genetic linkage between the animal and human GREF reservoirs was also provided by plasmid DNA analyses as described above. Moreover, the difference in isolation procedures for poultry and farmer GREF strains demonstrated that there are significantly higher concentrations of GREF in poultry feces than in feces from farmers. Taken together, the data presented in this study and in earlier reports (11, 25) are consistent with a significant persistent GREF reservoir of a poultry or environmental origin that favors resistance transfer to farmers.
The observed long-term persistence of both animal and human GREF strains on previously avoparcin-exposed farms is also interesting from a public health perspective. E. faecium is an important pathogen in hospital-acquired infections, and MLST data suggest that epidemic, virulent lineages of E. faecium have emerged worldwide (26). MLST analyses of clinical strains from several continents as well as recent molecular analyses of Norwegian clinical strains have indicated that purK-1 may be linked to epidemicity (26, 29). Thus, we performed sequencing of the purK genes of selected PFGE types. The purK-6 allele was detected in 36 of the 40 PFGE types examined, representing both poultry and farmer GSEF and GREF populations from both farms. MLST of E. faecium isolates from humans and livestock revealed that purK-6 was the dominant purK allele in poultry strains but that it was also present in strains from several other hosts, including swine, cats, and humans (26). Thus, it seems that the purK-6 allele is not a host-specific marker of E. faecium. However, one should interpret these data cautiously since the E. faecium MLST database is limited and is relatively dominated by antibiotic-resistant strains. More MLST data are needed from non-antibiotic-selected natural populations of E. faecium.
A recent molecular characterization of ampicillin-resistant and -susceptible clinical and fecal E. faecium strains from Norwegian hospitalized patients showed the presence of the purK-1 allele in 80 of 102 strains, while the purK-6 allele was absent (29). Amplified length polymorphism analyses of 218 Norwegian GREF (n = 197) and GSEF (n = 21) strains isolated from poultry feces, carcasses, and farm environments (13) showed that all poultry-associated strains clustered in the previously defined poultry genogroup, supporting the hypothesis of E. faecium host specificity (13, 54). Taken together with data presented by Jureen et al.(29) and Borgen et al. (13), the finding of a dominating purK-6-associated ampicillin-susceptible genetic lineage within GREF populations on Norwegian avoparcin-exposed poultry farms (this study and unpublished data) may partly explain the minor clinical problems with VanA-type GREF in Norway (5, 29) in spite of a prolonged national GREF reservoir in poultry.
In conclusion, a polyclonal but genetically related animal and human GREF population was readily detected 3 to 4 years after the discontinued use of avoparcin on two poultry farms. A few dominant PFGE types persisted throughout the study period, indicating the presence of environmentally well-adapted GREF clones counteracting the reversal of resistance. The common molecular signature within the GREF population was a specific Tn1546-plasmid junction fragment, suggesting that the unit of vanA persistence was a common transferable plasmid-mediated vanA element involved in the spread of Tn1546 into multiple E. faecium genetic backgrounds. In several PFGE types, the vanA-containing Tn1546 was genetically linked to a putative PSK system that might promote Tn1546 stabilization through plasmid addiction. Further studies are needed to address the function of the potential plasmid segregational stability cassette linked to Tn1546.
Acknowledgments
We thank Bettina Aasnes and Aase Mari Kaspersen for technical assistance.
This work was supported by European Commission contract QLK2-CT-2002-00843 “ARTRADI” and the Norwegian Institute for Gene Ecology.
REFERENCES
- 1.Aarestrup, F. M. 1995. Occurrence of glycopeptide resistance among Enterococcus faecium isolates from conventional and ecological poultry farms. Microb. Drug Resist. 1:255-257. [DOI] [PubMed] [Google Scholar]
- 2.Aarestrup, F. M. 2000. Characterization of glycopeptide-resistant Enterococcus faecium (GRE) from broilers and pigs in Denmark: genetic evidence that persistence of GRE in pig herds is associated with coselection by resistance to macrolides. J. Clin. Microbiol. 38:2774-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aarestrup, F. M., A. M. Seyfarth, H. D. Emborg, K. Pedersen, R. S. Hendriksen, and F. Bager. 2001. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 45:2054-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson, D. I. 2003. Persistence of antibiotic resistant bacteria. Curr. Opin. Microbiol. 6:452-456. [DOI] [PubMed] [Google Scholar]
- 5.Anonymous. 2003. NORM NORM-VET 2002. Consumption of antimicrobial agents and occurrence of antimicrobial resistance in Norway. The Norwegian Zoonosis Centre, Oslo, Norway.
- 6.Arthur, M., C. Molinas, F. Depardieu, and P. Courvalin. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bager, F., F. M. Aarestrup, M. Madsen, and H. C. Wegener. 1999. Glycopeptide resistance in Enterococcus faecium from broilers and pigs following discontinued use of avoparcin. Microb. Drug Resist. 5:53-56. [DOI] [PubMed] [Google Scholar]
- 8.Bates, J., Z. Jordens, and J. B. Selkon. 1993. Evidence for an animal origin of vancomycin-resistant enterococci. Lancet 342:490-491. [DOI] [PubMed] [Google Scholar]
- 9.Bergan, T., J. N. Bruun, A. Digranes, E. Lingaas, K. K. Melby, and J. Sander. 1997. Susceptibility testing of bacteria and fungi. Report from the Norwegian Working Group on Antibiotics. Scand. J. Infect. Dis. 103(Suppl.):1-36. [PubMed] [Google Scholar]
- 10.Borgen, K., G. S. Simonsen, A. Sundsfjord, Y. Wasteson, O. Olsvik, and H. Kruse. 2000. Continuing high prevalence of VanA-type vancomycin-resistant enterococci on Norwegian poultry farms three years after avoparcin was banned. J. Appl. Microbiol. 89:478-485. [DOI] [PubMed] [Google Scholar]
- 11.Borgen, K., M. Sorum, H. Kruse, and Y. Wasteson. 2000. Persistence of vancomycin-resistant enterococci (VRE) on Norwegian broiler farms. FEMS Microbiol. Lett. 191:255-258. [DOI] [PubMed] [Google Scholar]
- 12.Borgen, K., M. Sorum, Y. Wasteson, H. Kruse, and H. Oppegaard. 2002. Genetic linkage between erm(B) and vanA in Enterococcus hirae of poultry origin. Microb. Drug Resist. 8:363-368. [DOI] [PubMed] [Google Scholar]
- 13.Borgen, K., Y. Wasteson, H. Kruse, and R. J. Willems. 2002. Vancomycin-resistant Enterococcus faecium (VREF) from Norwegian poultry cluster with VREF from poultry from the United Kingdom and The Netherlands in an amplified fragment length polymorphism genogroup. Appl. Environ. Microbiol. 68:3133-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahl, K. H., E. W. Lundblad, T. P. Rokenes, O. Olsvik, and A. Sundsfjord. 2000. Genetic linkage of the vanB2 gene cluster to Tn5382 in vancomycin-resistant enterococci and characterization of two novel insertion sequences. Microbiology 146:1469-1479. [DOI] [PubMed] [Google Scholar]
- 15.Dahl, K. H., and A. Sundsfjord. 2003. Transferable vanB2 Tn5382-containing elements in fecal streptococcal strains from veal calves. Antimicrob. Agents Chemother. 47:2579-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devriese, L. A., M. Ieven, H. Goossens, P. Vandamme, B. Pot, J. Hommez, and F. Haesebrouck. 1996. Presence of vancomycin-resistant enterococci in farm and pet animals. Antimicrob. Agents Chemother. 40:2285-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ford, M., J. D. Perry, and F. K. Gould. 1994. Use of cephalexin-aztreonam-arabinose agar for selective isolation of Enterococcus faecium. J. Clin. Microbiol. 32:2999-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerdes, K. 2000. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 182:561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grady, R., and F. Hayes. 2003. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 47:1419-1432. [DOI] [PubMed] [Google Scholar]
- 21.Haaheim, H., K. H. Dahl, G. S. Simonsen, O. Olsvik, and A. Sundsfjord. 1998. Long PCRs of transposons in the structural analysis of genes encoding acquired glycopeptide resistance in enterococci. BioTechniques 24:432-437. [DOI] [PubMed] [Google Scholar]
- 22.Handwerger, S., and J. Skoble. 1995. Identification of chromosomal mobile element conferring high-level vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 39:2446-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasman, H., and F. M. Aarestrup. 2002. tcrB, a gene conferring transferable copper resistance in Enterococcus faecium: occurrence, transferability, and linkage to macrolide and glycopeptide resistance. Antimicrob. Agents Chemother. 46:1410-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heuer, O. E., K. Pedersen, J. S. Andersen, and M. Madsen. 2002. Vancomycin-resistant enterococci (VRE) in broiler flocks 5 years after the avoparcin ban. Microb. Drug Resist. 8:133-138. [DOI] [PubMed] [Google Scholar]
- 25.Heuer, O. E., K. Pedersen, L. B. Jensen, M. Madsen, and J. E. Olsen. 2002. Persistence of vancomycin-resistant enterococci (VRE) in broiler houses after the avoparcin ban. Microb. Drug Resist. 8:355-361. [DOI] [PubMed] [Google Scholar]
- 26.Homan, W. L., D. Tribe, S. Poznanski, M. Li, G. Hogg, E. Spalburg, J. D. van Embden, and R. J. Willems. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen, L. B. 1998. Differences in the occurrence of two base pair variants of Tn1546 from vancomycin-resistant enterococci from humans, pigs, and poultry. Antimicrob. Agents Chemother. 42:2463-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnsen, P. J., G. S. Simonsen, O. Olsvik, T. Midtvedt, and A. Sundsfjord. 2002. Stability, persistence, and evolution of plasmid-encoded VanA glycopeptide resistance in enterococci in the absence of antibiotic selection in vitro and in gnotobiotic mice. Microb. Drug Resist. 8:161-170. [DOI] [PubMed] [Google Scholar]
- 29.Jureen, R., J. Top, S. C. Mohn, S. Harthug, N. Langeland, and R. J. Willems. 2003. Molecular characterization of ampicillin-resistant Enterococcus faecium isolates from hospitalized patients in Norway. J. Clin. Microbiol. 41:2330-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klare, I., D. Badstubner, C. Konstabel, G. Bohme, H. Claus, and W. Witte. 1999. Decreased incidence of VanA-type vancomycin-resistant enterococci isolated from poultry meat and from fecal samples of humans in the community after discontinuation of avoparcin usage in animal husbandry. Microb. Drug Resist. 5:45-52. [DOI] [PubMed] [Google Scholar]
- 31.Klare, I., H. Heier, H. Claus, R. Reissbrodt, and W. Witte. 1995. vanA-mediated high-level glycopeptide resistance in Enterococcus faecium from animal husbandry. FEMS Microbiol. Lett. 125:165-171. [DOI] [PubMed] [Google Scholar]
- 32.Klare, I., H. Heier, H. Claus, and W. Witte. 1993. Environmental strains of Enterococcus faecium with inducible high-level resistance to glycopeptides. FEMS Microbiol. Lett. 80:23-29. [DOI] [PubMed] [Google Scholar]
- 33.Kruse, H., B. K. Johansen, L. M. Rorvik, and G. Schaller. 1999. The use of avoparcin as a growth promoter and the occurrence of vancomycin-resistant Enterococcus species in Norwegian poultry and swine production. Microb. Drug Resist. 5:135-139. [DOI] [PubMed] [Google Scholar]
- 34.Leclercq, R., E. Derlot, J. Duval, and P. Courvalin. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157-161. [DOI] [PubMed] [Google Scholar]
- 35.Levin, B. R., M. Lipsitch, V. Perrot, S. Schrag, R. Antia, L. Simonsen, N. M. Walker, and F. M. Stewart. 1997. The population genetics of antibiotic resistance. Clin. Infect. Dis. 24(Suppl. 1):S9-S16. [DOI] [PubMed] [Google Scholar]
- 36.Meinhart, A., J. C. Alonso, N. Strater, and W. Saenger. 2003. Crystal structure of the plasmid maintenance system epsilon/zeta: functional mechanism of toxin zeta and inactivation by epsilon 2 zeta 2 complex formation. Proc. Natl. Acad. Sci. USA 100:1661-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pantosti, A., M. Del Grosso, S. Tagliabue, A. Macri, and A. Caprioli. 1999. Decrease of vancomycin-resistant enterococci in poultry meat after avoparcin ban. Lancet 354:741-742. [DOI] [PubMed] [Google Scholar]
- 39.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 40.Rice, L. B. 2000. Bacterial monopolists: The bundling and dissemination of antimicrobial resistance genes in gram-positive bacteria. Clin. Infect. Dis. 31:762-769. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. W. Russel. 2001. Preparation of plasmid DNA by alkaline lysis with SDS, p. 31-34. In N. Irwin (ed.), Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Schwarz, F. V., V. Perreten, and M. Teuber. 2001. Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid 46:170-187. [DOI] [PubMed] [Google Scholar]
- 43.Simonsen, G. S., H. Haaheim, K. H. Dahl, H. Kruse, A. Lovseth, O. Olsvik, and A. Sundsfjord. 1998. Transmission of VanA-type vancomycin-resistant enterococci and vanA resistance elements between chicken and humans at avoparcin-exposed farms. Microb. Drug Resist. 4:313-318. [DOI] [PubMed] [Google Scholar]
- 44.Simonsen, G. S., M. R. Myhre, K. H. Dahl, O. Olsvik, and A. Sundsfjord. 2000. Typeability of Tn1546-like elements in vancomycin-resistant enterococci using long-range PCRs and specific analysis of polymorphic regions. Microb. Drug Resist. 6:49-57. [DOI] [PubMed] [Google Scholar]
- 45.Sorum, H., T. M. L'Abee-Lund, A. Solberg, and A. Wold. 2003. Integron-containing IncU R plasmids pRAS1 and pAr-32 from the fish pathogen Aeromonas salmonicida. Antimicrob. Agents Chemother. 47:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sundsfjord, A., G. S. Simonsen, and P. Courvalin. 2001. Human infections caused by glycopeptide-resistant Enterococcus spp.: are they a zoonosis? Clin. Microbiol. Infect. 7(Suppl. 4):16-33. [DOI] [PubMed] [Google Scholar]
- 47.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torres, C., J. A. Reguera, M. J. Sanmartin, J. C. Perez-Diaz, and F. Baquero. 1994. vanA-mediated vancomycin-resistant Enterococcus spp. in sewage. J. Antimicrob. Chemother. 33:553-561. [DOI] [PubMed] [Google Scholar]
- 49.van den Bogaard, A. E., N. Bruinsma, and E. E. Stobberingh. 2000. The effect of banning avoparcin on VRE carriage in The Netherlands. J. Antimicrob. Chemother. 46:146-147. [DOI] [PubMed] [Google Scholar]
- 50.van den Bogaard, A. E., L. B. Jensen, and E. E. Stobberingh. 1997. Vancomycin-resistant enterococci in turkeys and farmers. N. Engl. J. Med. 337:1558-1559. [DOI] [PubMed] [Google Scholar]
- 51.van den Bogaard, A. E., R. Willems, N. London, J. Top, and E. E. Stobberingh. 2002. Antibiotic resistance of faecal enterococci in poultry, poultry farmers and poultry slaughterers. J. Antimicrob. Chemother. 49:497-505. [DOI] [PubMed] [Google Scholar]
- 52.Werner, G., I. Klare, and W. Witte. 1997. Arrangement of the vanA gene cluster in enterococci of different ecological origin. FEMS Microbiol. Lett. 155:55-61. [DOI] [PubMed] [Google Scholar]
- 53.Werner, G., R. J. Willems, B. Hildebrandt, I. Klare, and W. Witte. 2003. Influence of transferable genetic determinants on the outcome of typing methods commonly used for Enterococcus faecium. J. Clin. Microbiol. 41:1499-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willems, R. J., J. Top, N. van den Braak, A. van Belkum, H. Endtz, D. Mevius, E. Stobberingh, A. van den Bogaard, and J. D. van Embden. 2000. Host specificity of vancomycin-resistant Enterococcus faecium. J. Infect. Dis. 182:816-823. [DOI] [PubMed] [Google Scholar]
- 55.Willems, R. J., J. Top, N. van den Braak, A. van Belkum, D. J. Mevius, G. Hendriks, M. Santen-Verheuvel, and J. D. van Embden. 1999. Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob. Agents Chemother. 43:483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]