Abstract
Byssochlamys species are responsible for spoilage and degradation of fruits and silages and can also produce the mycotoxin patulin. We analyzed secondary metabolite production by Byssochlamys nivea. Mycophenolic acid and its precursors, 5-methylorsellinic acid and 5,7-dihydroxy-4-methylphthalide, were identified in all of the B. nivea strains that we examined.
Mycotoxins are secondary metabolites of fungi that may contaminate animal and human feeds and that constitute a risk factor for human and animal health. Maize and grass silage are frequently contaminated by fungal toxins, such as patulin (7), roquefortine C (14, 19), and mycophenolic acid (MPA) (20). In addition to its antimicrobial and antitumoral activities (1, 21), MPA also has immunosuppressive properties due to its inhibitory effect on IMP dehydrogenase (2, 3, 15). Although its acute toxicity appears to be low (the oral 50% lethal dose in rats is 700 mg/kg of body weight) (5), MPA may be a predisposing factor for infectious diseases.
One of the most common molds isolated from silage, Penicillium roqueforti, can produce MPA. However, in a recent study of the natural occurrence of MPA contamination in silage, only 42% of the MPA-positive samples were contaminated by P. roqueforti (18), suggesting that one or more other fungal species could also synthesize this mycotoxin. Monascus purpureus, Trichoderma viride, Geotrichum candidum, Paecilomyces variotii, and Byssochlamys nivea are fungal species commonly recovered from ensiled maize (8, 16). B. nivea can synthesize two other toxins, patulin (17) and byssochlamic acid (8).
The objectives of this study were to determine if B. nivea produced mycophenolic acid and whether this production could explain discrepancies observed in previous analyses of silage samples.
Fungal strains and culture conditions.
We used B. nivea strain NRRL 2615 (USDA-ARS National Center for Agricultural Utilization Research, Peoria, Ill.). Ascospore suspensions were used to inoculate three 1-liter Roux flasks with 100 ml of sterile Czapek-dextrose broth (7). These cultures were incubated at 25°C, in the dark, without shaking, for 1 to 42 days. Other B. nivea strains used for these studies were NRRL 5253, NRRL 1678, and NRRL 5254 from Northern Region Research Laboratory, Peoria, Ill.; MUCL 39714 from Mycothèque de l'Université Catholique de Louvain, Louvain, Belgium; and IHEM 3076 from Collection mycologique de l'Institut d'Hygiene et d'Epidémiologie, Brussels, Belgium.
Contamination control.
Purity of the B. nivea NRRL 2615 culture on the eighth day of incubation was verified by macroscopic and microscopic examination and by internal transcribed spacer (ITS) rRNA and 5.8S rRNA gene amplification and sequencing. Genomic DNA was isolated as previously described (13); primer pairs (ITS-5 and ITS-4) and PCR conditions were those described by White et al. (20). Amplified fragments were cloned into the pCR 2.1-TOPO cloning vector. Amplification products were sequenced once, and cloned fragments were sequenced twice. The amplified ITS DNA coding sequences from these cultures differed from those already deposited in GenBank (accession number U18361) at three sites, two of which were gaps.
Dry weight determinations.
Culture samples were filtered through tared Whatman no. 1 filter paper. The washed cells were dried overnight in an oven at 70°C and cooled to room temperature before weighing.
Chemical standards used for mycotoxin analysis.
Patulin and mycophenolic acid were purchased from Sigma (St. Louis, Mo.) and used without further purification. The sources of the other secondary metabolites were P. M. Shoolingin-Jordan, University of Southampton, Southampton, United Kingdom, and Y. Ebizuka, Graduate School of Pharmaceutical Sciences, University of Tokyo, Tokyo, Japan (6-methylsalicylic acid); H. Fujimoto, Graduate School of Pharmaceutical Sciences, Chiba University, Chiba, Japan (5,7-dihydroxy-4-methylphthalide); J. D. White, Oregon State University, Corvallis (byssochlamic acid); and T. Anke, University of Kaiserslautern, Kaiserslautern, Germany (orsellinic acid).
Extraction procedures.
Filtered medium (100 ml) was adjusted to pH 2.0 with 2 M H2SO4 and extracted with 70 ml of ethyl acetate. The organic phase was evaporated in vacuo at 50°C. The residue was dissolved in 200 μl of methanol, filtered through a 0.45-μm-pore-size filter, and injected (20 μl) into a chromatographic column.
HPLC and LC-MS analysis.
Mycotoxin analysis was performed by high-performance liquid chromatography (HPLC) with diode array detection by using a 150- by 4-mm Zorbax octyldecyl silane 5-μm column. For the detection and identification of secondary metabolites, HPLC analysis was performed with a linear elution gradient by using 33 mM acetic acid (solvent A) and acetonitrile (solvent B) according to the method described by Frisvad (10). For MPA quantification, the gradient program was: at time zero, 100% solvent A; linear gradient to 10% solvent B within 20 min, to 20% solvent B in 10 min, and to 90% solvent B in 4 min; a plateau for 4 min; and, finally, a decrease within 7 min to 10% solvent B. Liquid chromatography-mass spectrometry (LC-MS) analysis was performed with a Thermo Finnigan HPLC-LCQ DUO Ion Trap (San Jose, Calif.) equipped with an electrospray ionization (ESI) source. HPLC was performed with a 150- by 2.1-mm Kromasil 5C18 column. Twenty microliters of the methanol suspension was injected directly into the LC system. Gradient chromatography (33-min run) was performed with 17 mM acetic acid acetonitrile as the mobile phase at a flow rate of 0.2 ml/min. The gradient started with 90% solvent A (17 mM acetic acid) for 2 min, and then solvent B (acetonitrile) was increased to 75% within 25 min; that percentage remained constant for 3 min before a return to 90% solvent A within 1 min and stabilization for 5 min. ESI was performed at room temperature in a negative mode, and the spray voltage was maintained at 4.5 kV with the capillary temperature at 250°C by using a nitrogen sheath (70 ml/min) and gas flows (20 ml/min). MS and MS2 tuning of the MS instrument were made with authentic 20 μM MPA at 2.5 μl/min in a 50/50 ratio of 17 mM acetic acid to acetonitrile. The collision energy was set at 30%.
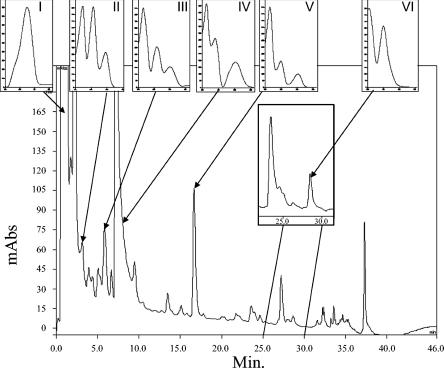
Six metabolites were detected and identified by HPLC and LC-MS analysis from the culture supernatants (Fig. 1), and the bracketed retention indices (RI) were calculated for all identified metabolites as described by Frisvad (11, 12). Major metabolite I (RI = 660) had the retention index and UV spectrum of patulin. Other metabolites were identified on the basis of their UV, MS, and MS2 spectra after comparison with reference standards. Metabolites II (RI = 710), IV (RI = 783), and V (RI = 957) showed, respectively, a base peak at m/z 179, 151, and 319, and were identified as 5-7-dihydroxy-4-methylphthalide, 6-methylsalicylic acid, and MPA. The MS2 fragments of these three acidic compounds had an important M-44 peak resulting from carboxyl group cleavage. The MS2 peaks of metabolite V were observed at m/z 287 (100%; M − CH3OH), 275 (77%; M − CO2), 269 (14%; M − H2O, − CH3OH) and 243 (20%; M − CO2, − CH3OH), 191 (49%), 179 (45%; metabolite II). Metabolite III (RI = 763) had a base peak at m/z 181, and its collision fragmentation led to fragments at m/z 137 and 121. These ions correspond to a decarboxylation and to the loss of a fragment of 60 atomic mass units and are consistent with the decarboxylated fragments observed in the tandem MS spectra of the orsellinic acid and the 6-methylsalicylic acid standards, respectively. Metabolite III has one more methyl group (Δ MS = 14) than orsellinic acid. On the basis of these data, metabolite III was identified as 5-methylorsellinic acid. Metabolite VI (RI = 1103) was identified as byssochlamic acid after comparisons with UV and MS data of a reference compound. Indeed, this metabolite displayed a pseudomolecular weight at m/z 331. The MS2 peaks were observed at m/z 287 (100%; M − CO2), 243 (3%; M − 2 CO2), 259 (2%; M − CO2, − C2H5) and 215 (4%; M − 2 CO2, − C2H5). These six metabolites can be placed into three biosynthetic pathways: patulin (patulin, 6-methylsalicylic acid), mycophenolic acid (MPA and its two precursors, 5-methylorsellinic acid and 5,7-dihydroxy-4-methylphthalide), and byssochlamic acid (byssochlamic acid).
FIG. 1.
HPLC traces of CHCl3 extracts of B. nivea culture filtrate (day 5) and UV spectra of metabolites detected in these cultures. UV absorption readings (in nm) were 276 (100%) for patulin (metabolite I); 216 (100%), 257, and 296 for 5,7-dihydroxy-4-methylphthalide (II); 216 (100%), 260, and 298 for 5-methylorsellinic acid (III); 205 (100%), 240, and 302 for 6-methylsalicylic acid (IV); 214 (100%), 252, and 303 for mycophenolic acid (V); and 204 (100%) and 250 for byssochlamic acid (VI). Inset, sample of chromatogram of B. nivea culture filtrate (day 13); metabolite VI (byssochlamic acid) is first detected on day 11. mAbs, milli-absorbance units.
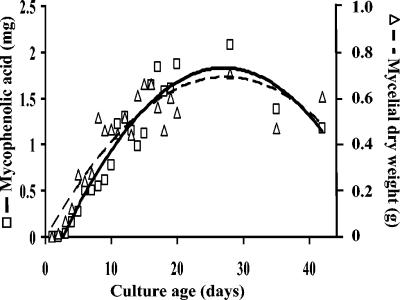
The first stable precursor of mycophenolic acid, 5-methylorsellinic acid, was detected from days 3 to 14, and 5,7-dihydroxy-4-methylphthalide was identified from days 2 to 42. MPA was first detected on day 2 and increased gradually until day 42. MPA concentration increases with fungal mass (Fig. 2). A Pearson correlation coefficient was calculated between MPA and the biomass (ρ = 0.763) and was found significant with a two-sided Gaussian test (P < 0.001) as implemented by S-plus software (Insightful, Seattle, Wash.). Unlike most secondary metabolites, which are formed in the idiophase after primary growth is complete, MPA was produced continuously by B. nivea and was proportional to the fungal biomass. MPA is produced in a similar manner by both batch and continuous-flow cultures of Penicillium brevicompactum (6).
FIG. 2.
Amount of mycophenolic and growth of B. nivea on Czapek-dextrose broth during production kinetics. □ and solid line, mycophenolic acid (mg/100 ml of culture medium); ▵ and dashed line, dry weight (g/Roux flask).
We tested several strains of B. nivea for MPA production. All of the B. nivea strains produced MPA and its two precursors.
This report is the first of mycophenolic acid production by a Byssochlamys species. Until now, this compound was known only from cultures of a few Penicillium species. Thus, B. nivea should be added to the list of fungi, including P. brevicompactum, P. roqueforti, Penicillium carneum, and Penicillium raciborskii, that are known to synthesize this toxin (4, 12). The origin of MPA in silage has previously been attributed exclusively to P. roqueforti, the most common fungal contaminant of silage. Like P. roqueforti, B. nivea can grow at very low oxygen concentration and is often isolated from 3- to 4-month-old silage (9), but further work is needed to document the production of MPA by B. nivea in naturally contaminated silage.
The consumption of patulin or mycophenolic acid or a combination of both by domesticated animals may substantially increase their susceptibility to infectious diseases. The simultaneous exposure to additional toxins, e.g., roquefortine C, PR toxin, and byssochlamic acid, would increase the animal health risk even further, and the concomitant isolation of B. nivea and P. roqueforti from a silage sample should serve as an indicator of potentially increased toxin exposure.
Nucleotide sequence accession number.
Sequences of the B. nivea stain NRRL 2615 were deposited in GenBank under accession number AF486189.
Acknowledgments
This work was supported in part by the National Institute of Agronomic Research Program on Mycotoxins (contract 00263), by a Midi-Pyrénées Regional grant (01002728), and by an Environment and Health 2002 grant (RD2003-009).
We thank P. M. Shoolingin-Jordan, Y. Ebizuka, H. Fujimoto, J. D. White, and T. Anke for chemical standards; N. Ledger, T. Pineau, and P. Martin for critical comments on the manuscript; and S. Peterson, curator of the ARS Culture Collection, for providing cultures of some of the strains examined.
REFERENCES
- 1.Abraham, E. P. 1945. The effect of mycophenolic acid on the growth of Staphylococcus aureus in heart broth. Biochem. J. 39:398-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, A. C., and E. M. Eugui. 2000. Mycophenolate mofetil and its mechanisms of action. Immunopharmacology 47:85-118. [DOI] [PubMed] [Google Scholar]
- 3.Bentley, R. 2000. Mycophenolic acid: a one hundred year odyssey from antibiotic to immunosuppressant. Chem. Rev. 100:3801-3826. [DOI] [PubMed] [Google Scholar]
- 4.Boysen, M., P. Skouboe, J. Frisvad, and L. Rossen. 1996. Reclassification of the Penicillium roqueforti group into three species on the basis of molecular genetic and biochemical profiles. Microbiology 142:541-549. [DOI] [PubMed] [Google Scholar]
- 5.Cole, R. J., and R. H. Cox. 1981. Handbook of toxic fungal metabolites, p. 866. Academic Press, New York, N.Y.
- 6.Doerfler, D. L., C. D. Bartman, and I. M. Campbell. 1979. Mycophenolic acid production by Penicillium brevicompactum in two media. Can. J. Microbiol. 25:940-943. [DOI] [PubMed] [Google Scholar]
- 7.Escoula, L. 1975. Toxinogenic moulds in silage. II. In vitro kinetics of patulin and byssochlamic acid biosynthesis by Byssochlamys nivea Westling in liquid medium. Ann. Rech. Vet. 6:155-163. [PubMed] [Google Scholar]
- 8.Escoula, L. 1975. Toxinogenic moulds in silage. V. Production of byssochlamic acid in liquid medium with by Byssochlamys nivea Westling, Byssochlamys fulva Olliver and Smith and Paecilomyces varioti Bainier isolated in forages. Ann. Rech. Vet. 6:311-314. [PubMed] [Google Scholar]
- 9.Escoula, L. 1977. Moisissures des ensilages et conséquences toxicologiques. Fourrages 69:97-114. [Google Scholar]
- 10.Frisvad, J. C. 1987. High-performance liquid chromatography determination profiles of mycotoxins and other secondary metabolites. J. Chromatogr. 392:333-347. [DOI] [PubMed] [Google Scholar]
- 11.Frisvad, J. C., and U. Thrane. 1987. Standardized high-performance liquid chromatography of 182 mycotoxins and other fungal metabolites based on alkylphenone retention indices and UV-VIS spectra (diode array detection). J. Chromatogr. 404:195-214. [DOI] [PubMed] [Google Scholar]
- 12.Frisvad, J. C., and U. Thrane. 1993. Liquid column chromatography of mycotoxins, p. 253-372. In V. Betina (ed.), Chromatography of mycotoxins: techniques and applications. Elsevier, Amsterdam, The Netherlands.
- 13.Girardin, H., J. P. Latgé, T. Srikantha, B. Morrow, and D. R. Soll. 1993. Development of DNA probes for fingerprinting Aspergillus fumigatus. J. Clin. Microbiol. 31:1547-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Häggblom, P. 1990. Isolation of roquefortine C from feed grain. Appl. Environ. Microbiol. 56:2924-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonsson, C. A., and H. Carlsten. 2003. Mycophenolic acid inhibits inosine 5′-monophosphate dehydrogenase and suppresses immunoglobulin and cytokine production of B cells. Int. Immunopharmacol. 3:31-37. [DOI] [PubMed] [Google Scholar]
- 16.Pelhate, J. 1975. Mycoflore des mais-fourrages ensilés. Déterminisme de son évolution. Rev. Mycol. 39:65-95. [Google Scholar]
- 17.Rice, S. L., L. R. Beuchat, and R. E. Worthington. 1977. Patulin production by Byssochlamys spp. in fruit juices. Appl. Environ. Microbiol. 34:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneweis, I., K. Meyer, S. Hormansdorfer, and J. Bauer. 2000. Mycophenolic acid in silage. Appl. Environ. Microbiol. 66:3639-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tüller, G., G. Armbruster, S. Wiedenmann, T. Hänichen, D. Schams, and J. Bauer. 1998. Occurrence of roquefortine in silage—toxicological relevance to sheep. J. Anim. Physiol. Anim. Nutr. 80:246-249. [Google Scholar]
- 20.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, Calif.
- 21.Williams, R. H., D. H. Lively, D. C. DeLong, J. C. Cline, and M. J. Sweeny. 1968. Mycophenolic acid: antiviral and antitumor properties. J. Antibiot. (Tokyo) 7:463-464. [DOI] [PubMed] [Google Scholar]