Abstract
Given our vast methane reserves and the difficulty in transporting methane without substantial leaks, the conversion of methane directly into electricity would be beneficial. Microbial fuel cells harness electrical power from a wide variety of substrates through biological means; however, the greenhouse gas methane has not been used with much success previously as a substrate in microbial fuel cells to generate electrical current. Here we construct a synthetic consortium consisting of: (i) an engineered archaeal strain to produce methyl-coenzyme M reductase from unculturable anaerobic methanotrophs for capturing methane and secreting acetate; (ii) micro-organisms from methane-acclimated sludge (including Paracoccus denitrificans) to facilitate electron transfer by providing electron shuttles (confirmed by replacing the sludge with humic acids), and (iii) Geobacter sulfurreducens to produce electrons from acetate, to create a microbial fuel cell that converts methane directly into significant electrical current. Notably, this methane microbial fuel cell operates at high Coulombic efficiency.
Microbial fuel cells generate electricity from a variety of sources, however from methane only negligible electrical power has been reported so far. Here the authors convert methane into electricity using a synthetic consortium consisting of an engineered archaeal strain, microorganisms from methane-acclimated sludge, and Geobacter sulfurreducens.
Electrical current may be generated through biological means from organic substrates in microbial fuel cells (MFCs) in which microbes transfer electrons extracellularly (exoelectrogens)1,2. In MFCs, exoelectrogens deposit electrons on an anode, from which the electrons migrate to the cathode to create an electrical current; at the cathode, the electrons are passed to another electron acceptor. In a typical dual-chamber set-up, a proton-conducting membrane is used between the anode and cathode chambers to complete the circuit by allowing excess positive charge in the anode (in the form of protons) to migrate to the cathode3 (Supplementary Fig. 1). Use of MFCs has led to the successful generation of electrical current from a wide variety of substrates, from both natural and artificial sources (including acetate4, glucose5, and natural6 and artificial7 wastewaters)8. However, with methane as a substrate, only negligible electrical power has been reported using uncultured anaerobic methane-oxidizing consortia isolated from oceanic sediment9,10.
Most studies related to MFCs involve natural exoelectrogens that make electrically conductive pili, such as Geobacter sulfurreducens11,12, or that secrete their own redox-active electron shuttles, such as Shewanella oneidensis13. Even organisms not naturally capable of external electron transfer may generate electrical current through the use of externally supplied electron shuttling molecules, such as thionine14, neutral red14 or methylene blue15. However, these exogenously supplied mediators may be toxic and expensive16.
The potential substrate for MFCs, methane, is also a potent greenhouse gas that can trap heat 38 times more efficiently on a molar basis than carbon dioxide over a span of 20 years17,18. Methane extraction from shale deposits has gained considerable attention as methane is an energy dense fuel with decreased carbon dioxide emissions per unit energy19. However, procedures of extraction, supply and storage lead to leaks of methane into the atmosphere, with ∼8% of the methane produced from a well lost to leaks; up to half of this leaking is due to downstream processes of distribution, transportation, and storage18. Conversion of methane to other biofuels, such as through the Fischer-Tropsch process that yields higher molecular weight hydrocarbons, is well established in industry, but large initial capital investments (on the order of $20 billion) are needed for these chemical processing plants20. Therefore, processes to convert methane biologically to electricity at the source, if developed, could reduce leaks (from distribution, transportation and storage) and capital cost. Furthermore, harnessing methane has been recognized as one of the most important near-term goals for biochemical engineering21.
Critically, anaerobic methanotrophs (ANME) participate in anaerobic oxidation of methane (AOM) in nature to play an important role in curbing methane emissions, with roughly 75 Tg of methane each year (88% of methane from natural sources of leaks) in the ocean consumed22. AOM in nature typically involves consortia of archaeal ANME paired with sulfate reducing bacteria23,24 where surplus electrons from methane catabolism are likely transferred between syntrophs directly via multi-haem cytochromes25 with the terminal electron acceptor (including sulfate24, nitrate26, ferric iron27 and manganic manganese27) being replaceable by others such as humic acids and anthraquinone-2,6-disulfonate (AQDS)28. However, since no individual species has been isolated that participates in AOM, little is known about it28,29.
We30 recently reversed methanogenesis to allow for growth on methane as the main carbon source by engineering the archaeal strain Methanosarcina acetivorans to produce methyl-coenzyme M reductase (Mcr) from ANME in an unculturable microbial mat from the Black Sea31; the engineered archaeal strain passes electrons to external Fe3+ (in the form of FeCl3) and converts methane to acetate30. In a follow-up study, we further metabolically engineered the methane-consuming M. acetivorans strain to synthesize lactate by adding the gene for the production of a 3-hydroxybutyryl-CoA dehydrogenase from Clostridium acetobutylicum32. Furthermore, the archaeal host, M. acetivorans, was adapted to small pulses of oxygen (named air-adapted M. acetivorans (AA)33), making it more robust in terms of biofilm production for potential use in a MFC. Here, we combine the metabolically engineered M. acetivorans that produces Mcr and grows on methane30 with G. sulfurreducens and methane-consuming sludge to generate substantial electrical current from methane.
Results
Acclimation of sludge samples to methane
Given our goal of creating an MFC that utilizes methane, we desired a consortium that would oxidize methane to provide electrons for the anode via pili11,12 or multi-haem complexes (possibly associated with other electron shuttles)34. We focused on using a consortium since most environmental processes occur within consortia35, and natural anaerobic methane oxidation involves consortia23.
To begin formulating our consortium for oxidizing methane, sludge samples from a local anaerobic digester for treating wastewater were acclimated over 567 days to methane as the main carbon source via three successive culturing cycles in high salts (HS) liquid medium with various terminal electron acceptors (Supplementary Table 1). This led to consortia that could be inoculated into methane MFCs for possibly aiding electricity production with electron transfer to the anodes. These cultures were visualized via transmission electron microscopy to reveal cells (both rod-shaped and irregular cocci) attached to each other with pili and subcellular structures (diameter <50 nm) to form biofilm networks (Supplementary Fig. 2); hence, the consortium held promise for conducting electrons in a MFC.
Electricity from methane
MFCs were first inoculated with the air-adapted M. acetivorans strain33 that produces ANME Mcr via pES1-MATmcr3 (Supplementary Table 2) so that methane could be converted to acetate30. The M. acetivorans strain hosting pES1-MATmcr3 can transfer electrons to ferric iron30; here, the electrons were consumed by converting ferricyanide to ferrocyanide at the cathode to complete the circuit for the generation of electricity3. Although the air-adapted strain can tolerate oxygen and oxidize methane to produce acetate (Supplementary Fig. 3), the whole apparatus was operated anaerobically to eliminate oxygen as a terminal electron acceptor that competes with the generation of an electrical current. G. sulfurreducens was added to potentially catabolize acetate to produce electricity36. Several days after adding G. sulfurreducens, methane-acclimated sludge was added to the MFCs to provide other potential consortial members to produce electricity from methane and other by-products of methane catabolism.
Voltages between the anode and cathode across a fixed 1 kΩ resistance increased to significant levels (ca., 0.6 V with current density 273 mA m−2) a few days after sludge addition for MFCs containing both the air-adapted M. acetivorans strain producing Mcr and G. sulfurreducens (Fig. 1a, Table 1) while the absence of sludge led to no appreciable current (Fig. 1b); these results show that sludge is necessary to generate electricity from methane. G. sulfurreducens is also required for electricity, since there is little current in its absence (Fig. 1c, Table 1). Compared to wild-type M. acetivorans, the air-adapted strain was also a better host (Fig. 1d, Table 1). Furthermore, there was no current without the air-adapted M. acetivorans strain producing Mcr (Fig. 1e) or with only the engineered strain (Fig. 1f). Sludge by itself but with methane did not yield electricity (Fig. 1g, Table 1), no current was found without any of the micro-organisms but with methane (Fig. 1g), and without methane, the air-adapted M. acetivorans strain producing Mcr, G. sulfurreducens, and sludge did not produce current (Fig. 1h). Therefore, electricity generation from methane depends on the presence of the air-adapted M. acetivorans strain producing Mcr, G. sulfurreducens and sludge.
Figure 1. Voltages generated in the MFCs with combinations of micro-organisms.
Micro-organisms in the MFCs include combinations of the following: the air-adapted M. acetivorans host containing pES1-MATmcr3 (‘AA/Mcr3'), the air-adapted M. acetivorans host containing the empty plasmid pES1(Pmat) (‘AA/Empty'), M. acetivorans C2A containing pES1-MATmcr3 (‘C2A/Mcr3'), G. sulfurreducens and sludge. The absence of the air-adapted M. acetivorans strain is indicated as ‘no AA/no Mcr3,' the absence of G. sulfurreducens is indicated as ‘no G. sulfurreducens', the absence of sludge is indicated as ‘no sludge' and the replacement of the methane headspace with a nitrogen headspace is indicated as ‘no methane'. Sludge was added to the indicated MFCs once the voltage of each MFC decreased to a threshold value of 150 mV or below, at time 0. For MFCs not including sludge, time 0 here is indicated as 132 h after inoculation and set-up. All values (voltages between the anode and cathode across a 1 kΩ fixed resistance) are represented as means±s.e.m. from at least three replicate MFCs. (a–e) The red voltage values for ‘AA/Mcr3/G. sulfurreducens/sludge' are repeated to aid in comparing results to values for (a) ‘AA/Empty/G. sulfurreducens/sludge', (b) ‘AA/Mcr3/G. sulfurreducens/no sludge', (c) ‘AA/Mcr3/no G. sulfurreducens/sludge', (d) ‘C2A/Mcr3/G. sulfurreducens/sludge' and (e) ‘no AA/no Mcr3/G. sulfurreducens/sludge'. (f) Values from ‘AA/Mcr3/no G. sulfurreducens/no sludge' is compared to values from ‘AA/Empty/no G. sulfurreducens/no sludge', (g) values from ‘no AA/no Mcr3/no G. sulfurreducens/no sludge' are compared to values from ‘no AA/no Mcr3/no G. sulfurreducens/sludge', and (h) values from ‘AA/Mcr3/G. sulfurreducens/sludge/no methane' are displayed.
Table 1. Maximum power generation and current densities in the MFCs.
| MFC conditions | Maximum power (mW m−2) | Maximum current density (mA m−2) |
|---|---|---|
| AA/Mcr3/G. sulfurreducens/sludge | 168±9 | 273±7 |
| AA/Empty/G. sulfurreducens/sludge | 80±70 | 200±100 |
| C2A/Mcr3/G. sulfurreducens/sludge | 26±9 | 110±20 |
| AA/Mcr3/no G. sulfurreducens/sludge | 20±10 | 90±30 |
| AA/Mcr3/G. sulfurreducens/sludge/no methane | 3.0±0.2 | 36±1 |
| no AA/no Mcr3/no G. sulfurreducens/no sludge | 4.5±0.4 | 45±2 |
| no AA/no Mcr3/no G. sulfurreducens/sludge | 0.2±0.1 | 9±3 |
| AA/Mcr3/no G. sulfurreducens/no sludge | 20±20 | 80±40 |
| AA/Empty/no G. sulfurreducens/no sludge | 7±5 | 90±50 |
Maximum power and current densities are normalized by the cathode surface area of 0.00227, m2. All MFCs included methane in the headspace unless otherwise indicated. MFCs included at time 0 as indicated the air-adapted M. acetivorans host containing pES1-MATmcr3 (‘AA/Mcr3'), the air-adapted M. acetivorans host containing pES1(Pmat) (‘AA/Empty'), M. acetivorans C2A containing pES1-MATmcr3 (‘C2A/Mcr3'), no M. acetivorans strains (‘no AA/no Mcr3') and G. sulfurreducens. Sludge was added to the MFCs as indicated once the voltage of each MFC decreased to below a threshold value of 150 mV. Averages and s.d.'s between at least three replicates are shown.
Methane consumption followed the voltage generation trends in that MFCs with the air-adapted M. acetivorans producing Mcr paired with G. sulfurreducens and sludge having the highest methane consumption (Supplementary Table 3). Methane consumption in MFCs with the air-adapted M. acetivorans strain producing Mcr but without both G. sulfurreducens and sludge had little methane consumption; hence, electricity is generated as a means to remove the excess electrons from the process for methane oxidation to occur, and the sludge and G. sulfurreducens provide a means to conduct the electrons to the anode. Methane losses were not due to leaks, since no oxygen was detected. In MFCs including G. sulfurreducens and sludge, production of Mcr from ANME allowed for greater consumption of the methane substrate.
The re-filling of MFCs with more methane (Supplementary Fig. 4) caused an increase in voltage only for MFCs with air-adapted M. acetivorans producing Mcr from ANME with G. sulfurreducens and methane-acclimated sludge. Corroborating the methane consumption results, production of Mcr from ANME is required for sustained production of electrical current (with a threefold increase in voltage compared to no production of Mcr from ANME, Supplementary Fig. 4). Production of Mcr also increases maximum power output (Table 1). Acetate was confirmed to be used as an intermediate in the methane-to-electricity process, since acetate was not detected in MFCs with air-adapted M. acetivorans producing Mcr from ANME with G. sulfurreducens and methane-acclimated sludge with a methane headspace after 40 days of incubation (results not shown), while acetate is the main product of methane consumption by air-adapted M. acetivorans producing Mcr from ANME (Supplementary Fig. 3).
Visualization of the MFC anodes
To determine the presence of potential biofilm structures, MFC anode fibres were visualized by scanning electron microscopy (SEM). Only individual cells could be seen attached to the anode (Supplementary Fig. 5), suggesting that electricity was generated through indirect contact (via electron shuttle molecules) rather than direct contact. The absence of such cell-like structures in the MFCs not inoculated with cells indicates that these structures are cells. The absence of distinctly rod-shaped bacteria attached to the anodes indicated that G. sulfurreducens cells, where included, did not attach to the anode. M. acetivorans producing Mcr from ANME develops biofilms on FeCl3 precipitates when grown with methane as the substrate30; however, the carbon fibre anode surfaces (Supplementary Fig. 5) are much smoother and more structured than the FeCl3 precipitates (Supplementary Fig. 6), so it is likely more difficult for M. acetivorans strains to attach to the anodes.
Sludge replacement by electron shuttles
Due to the absence of a robust biofilm, we postulated that the bulk of the electron transfer was unlikely due to direct electron transfer but instead occurred via electron shuttles provided by the sludge. To test this hypothesis, we replaced the sludge with five sources of electron shuttles (0.5 mM riboflavin 5′-monophosphate (FMN)37, 0.5 mM flavin adenine dinucleotide37, 5 mM AQDS28, 0.5% humic acids28 and supernatants from methane-acclimated sludge) and found that of these tested electron shuttles, only humic acids (a complex mix of acids containing carboxyl and phenolate groups) could substitute for the sludge component in MFCs containing the air-adapted M. acetivorans strain with pES1-MATmcr3 and G. sulfurreducens (Supplementary Fig. 7).
Characterization of the methane-acclimated sludge
To determine which micro-organisms were likely active in the methane-acclimated sludge, 16S rDNA analyses were made for the original sludge samples, for the sludge after methane acclimation, and for the sludge used both with MFCs producing and not producing electricity (that is, MFCs with no methane). Critically, the prevalent micro-organism belonged to the genus Paracoccus (Fig. 2), and Paracoccus denitrificans can significantly substitute for the sludge component in the methane MFCs (Supplementary Fig. 7). As internal controls, both M. acetivorans and G. sulfurreducens were detected as substantial community members in the MFCs where they were added as part of the synthetic consortium (Fig. 2). The omission of methane (and lack of significant electrical current) did not lead to notable changes in community composition (Fig. 2) which indicates that not many members proliferated substantially under the MFC conditions; however, the relative metabolic activity of each genus member remains to be shown. As expected, the acclimation of the sludge samples to methane decreased the overall diversity (Fig. 2).
Figure 2. Genera identified in sludge and in the MFCs.
Relative compositions are displayed based on the number of identified 16S rDNA reads compared to the total reads and are characterized to the genus taxonomic level. Genera consisting of at least 1% of the relative compositions of each sample are displayed, and DNA reads belonging to these genera are considered for the total reads in each sample. ‘Original sludge' is the initial sludge 811 days after isolation, and the ‘Acclimated sludge' is the pooled seven Round 3 methane-acclimated sludge cultures (Supplementary Table 1) after 567 days of incubation, with both of these sludge samples used to inoculate MFCs. MFCs included the air-adapted M. acetivorans host containing pES1-MATmcr3 (‘AA/Mcr3') and G. sulfurreducens. Sludge was added to the indicated MFCs once the voltage of each MFC decreased to a threshold value of 150 mV or below. All MFCs included methane in the headspace except the ‘AA/Mcr3/G. sulfurreducens/sludge/no methane' sample. Samples from MFCs were taken after 40 or 65 days of incubation for the two ‘AA/Mcr3/G. sulfurreducens/sludge' samples, or after 16 days of incubation for the ‘AA/Mcr3/G. sulfurreducens/sludge/no methane' sample.
Discussion
We demonstrate here the generation of significant electrical current using methane as a substrate in MFCs. We also show that our breakthrough technology for extracting electrical power from methane yields a Coulombic efficiency of 90±10% (in MFCs with the air-adapted M. acetivorans producing Mcr, G. sulfurreducens, and methane-acclimated sludge). Although this Coulombic efficiency is high, it is still reasonable, as other Coulombic efficiencies from MFCs at 90% (ref. 38) and 85% (ref. 39) have been reported. The high Coulombic efficiency of this system, compared to other systems where oxygen from the cathode can be used, or other electron acceptors may be present in the solution, reflects the lack of other suitable electron acceptors in the medium other than the anode.
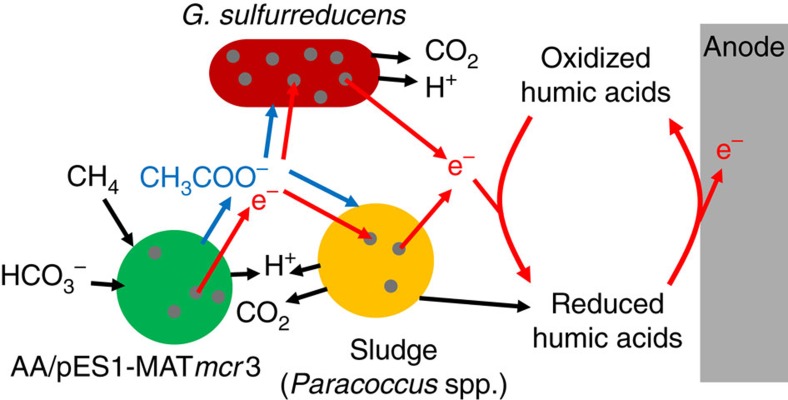
The electricity was generated by a synthetic consortium that includes known species (the air-adapted M. acetivorans strain engineered to produce ANME Mcr and G. sulfurreducens) combined with either partially characterized species from methane-acclimated sludge isolates or exogenous electron shuttles in the form of humic acids. We also could produce electricity by using only known species, that is, with the sludge replaced by P. denitrificans. Hence, the primary role of the sludge was to provide soluble electron shuttles that function like humic acids which are known to transport electrons28. The most prevalent genus in the methane-acclimated sludge, Paracoccus sp., has demonstrated enhancement of electrical power generation40, and many Paracoccus spp. are found in soil41,42,43. Although G. sulfurreducens is capable of forming electrically conductive biofilm structures11,12 and the sludge samples acclimated to methane consumption here could form biofilm structures, the bulk of the electron transfer appears to be via electron shuttles. Geobacter species are well known to use humic acids and other quinone analogues for electron transfer44,45,46,47,48, and G. sulfurreducens readily transfers electrons to humic acids (and other similar electron acceptors) using multi-haem cytochromes and consumes acetate49,50, so G. sulfurreducens plays a role in transferring electrons to the electron shuttles. Hence, our results corroborate the identification of Geobacter in MFCs in a previous study that attempted to use methane as a substrate in MFCs10. We postulate then that in the MFC with the synthetic consortium, the ANME Mcr-producing M. acetivorans consumes methane to produce oxidized intermediates (including acetate) that are consumed by the G. sulfurreducens and sludge components and that the Mcr-producing M. acetivorans generates electrons; G. sulfurreducens uses multi-haem cytochromes to rapidly transfer electrons to shuttles produced by the sludge or by P. denitrificans (Fig. 3).
Figure 3. Proposed model of biological production of electricity from methane in MFCs.
The air-adapted strain producing Mcr from ANME (‘AA/pES1-MATmcr3') consumes methane to produce acetate and excess electrons. Acetate is further oxidized completely to carbon dioxide, producing more excess electrons by G. sulfurreducens or sludge that can be substituted to a significant extent by P. denitrificans (‘Sludge (Paracoccus spp.)'). The sludge has a second role in providing humic acid-like electron shuttles, and G. sulfurreducens has a second role in providing optimized electron transfer to electron shuttles via multi-haem cytochromes (grey circles) that are located in the outer membrane or S-layer in bacteria and archaea, respectively.
Methods
Bacterial/archaeal strains and cultivation conditions
The M. acetivorans strains (Supplementary Table 1) were routinely grown anaerobically as pre-cultures at 37 °C in an 80% N2/19% CO2/1% H2 atmosphere with mild shaking in 10 ml HS medium51 or HSYE (HS medium with 2.5 g l−1 yeast extract) with 125 mM methanol as the carbon source, unless otherwise indicated. All 28-ml culture tubes (18 × 150 mm, Bellco Glass, Vineland, NJ, USA) were sealed by aluminium crimp seals. pES1-based plasmids were maintained in M. acetivorans with 2 μg ml−1 puromycin, and methane served to induce ANME Mcr production30 in the MFCs. Mcr. G. sulfurreducens PCA was routinely grown anaerobically in Geobacter Basal medium52 with 10 mM sodium acetate as the electron donor and 40 mM sodium fumarate as the electron acceptor53. Before addition to the MFCs, P. denitrificans DSM413 was cultivated aerobically in nutrient broth at 30 °C in a shaker with constant agitation (250 r.p.m.) for 48 h (ref. 54).
For testing oxygen tolerant (ca., 5 days) consumption of methane for the M. acetivorans air-adapted strain with high cell-density inocula30, 2 ml of the strain was pre-grown in 200 ml of HS medium with 125 mM methanol (and 2 μg ml−1 puromycin when plasmids were present) at 37 °C for 5 days (turbidity at 600 nm ∼1.0). Cells were collected by centrifugation (5,000 r.p.m. for 20 min), then were washed three times with HS medium and puromycin alone to remove residual methanol. The final cell pellet was resuspended using 5 ml of HS medium supplemented with 10 mM FeCl3 and 2 μg ml−1 puromycin to yield a density of 4 × 1010 CFU ml−1. After filling the headspace of each tube with methane, oxygen was added, where indicated, by replacing a portion of the headspace with air to a final concentration of 1% oxygen. The tubes were incubated at 37 °C with shaking at 250 r.p.m. for 5 days.
Anaerobic sludge from treated wastewater was collected (∼10 ml) just above the sedimentation layer of an anaerobic digester (maintained at 35 °C) from the Office of Physical Plant at the Pennsylvania State University, then kept under a headspace of methane at room temperature. The sample was inoculated (1:100) into HS medium under a methane headspace (HS-methane) with varying concentrations of 0.001–100 mM FeSO4 or FeCl3 for 56 days, then subcultures (1:100 inoculation) were made in HS-methane with 0.001 mM FeSO4 or 1 mM FeCl3, respectively, for 176 days, then the subcultures were grown (1:100 inoculation) in HS-methane with 40 mM ferrihydrite, 1 mM FeCl3, 0.001 mM FeSO4 (with or without 5% H2 in the headspace), or 0.001 mM FeCl3. All incubation steps were conducted at 37 °C.
Transformation of M. acetivorans
All plasmids were transformed into M. acetivorans using a liposome-mediated procedure55 with slight modifications. Cells (2 ml) grown in HS medium or HSYE with 125 mM methanol to a turbidity at 600 nm of 0.2–0.5 were centrifuged and resuspended into 1 ml transformation buffer (850 mM sucrose and 80 mM sodium bicarbonate, pH 7.4). Plasmid DNA (4 μg) was mixed with 15–25 μl of DOTAP (n-(1-(2,3-dioleoyloxy)propyl)-n,n,n-trimethylammonium methyl-sulfate, Sigma-Aldrich, St Louis, MO, USA) prepared in transformation buffer in a final reaction volume of 50 μl and incubated at 37 °C for at least 15 min to make DNA:liposome complexes. The 1 ml cell resuspension was mixed with the 50 μl DNA:liposome complex and incubated at 37 °C for 4 h. The cells were then inoculated into 10 ml HS or HSYE medium with 125 mM methanol, then after 48 h of incubation, 1 ml of the culture was added to 10 ml selective HSYE medium with 125 mM methanol and 2 μg ml−1 puromycin.
Electron microscopy
Cell morphology was examined via transmission electron microscopy (FEI Tecnai G2 Spirit BioTwin, Hillsboro, OR, USA) using uranyl acetate-stained cells56. Cellular attachments to anodes were visualized by SEM (Sigma VP-FESEM, Zeiss, Oberkochen, Germany). Samples for SEM were fixed by placing carbon fibres from the anode brush in 2.5% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.2), and incubating at room temperature for 1 h. The samples were then washed with increasing concentrations of ethanol: 25, 50, 70, 85, 95 and 100% (three times). Samples were dried via critical point drying before visualizing by SEM.
Microbial fuel cells
Two-bottle MFC reactors (total volume of each bottle is 155 ml) were constructed3 as in Supplementary Fig. 1. All MFC-related power and voltage generation results were performed with three replicates. The two bottles with sideports (2.4 cm inner diameter) were clamped together with a treated Nafion 117 proton exchange membrane (Dupont, Wilmington, DE) and one rubber gasket (3.5 cm outer diameter). The Nafion 117 membranes were cut into 4 × 4 cm squares, then pre-treated by heating for 1 h in solutions of 30% hydrogen peroxide, water, 0.5 M sulfuric acid and water. The top of each MFC chamber consisted of a rubber septum (42 cm diameter, ¼-inch thick) and plastic cap with a hole (Corning, Corning, NY, USA). All rubber septa had holes drilled in the centre to allow an electrode to pass. For each anode chamber top, a carbon fibre brush electrode (Mill-Rose, Mentor, OH, USA) from carbon fibres (PANEX 35 50 K, Zoltek, St Louis, MO, USA), twisted into two titanium wires57 12.7 cm long and heat-treated at 450 °C for 30 min58, was passed through the hole in the septum so that 2.8 cm of the top protruded, then the hole was closed with epoxy (Loctite, Dusseldorf, Germany). For each cathode chamber top, a carbon cloth circle (38 mm diameter, Fuel Cell Store, Boulder, CO, USA) was attached to a 10-cm-long titanium wire (1.0 mm diameter, Alfa Aesar, Haverhill, MA, USA), then the wire was passed through the hole in the septum so that 3 cm of the wire protruded, and the hole was closed with epoxy.
To inoculate each MFC anode chamber, cells (M. acetivorans and G. sulfurreducens) from 200 ml cultures each were collected by centrifugation (5,000 r.p.m. for 20 min) and washed three times with HS medium lacking resazurin (HSNR, redox-active resazurin would short-circuit the MFC system). The final cell pellet was resuspended in 100 ml HSNR with puromycin and placed in the anode chamber. 100 ml of catholyte solution (100 mM ferricyanide in 100 mM sodium phosphate, 5.8 mM ammonium chloride and 1.7 mM potassium chloride pH 7.0) was placed into each cathode chamber, and the caps were closed tightly before removing the MFC from the anaerobic atmosphere. The headspace of each anode chamber was filled with methane. The MFCs were incubated at 30 °C and measurements of the voltage differential across the cathode and anode of each MFC was taken over 1 kΩ resistance at a frequency of 0.05 s−1 (with data displayed daily) using a 16-channel differential analogue input module (NI 9205, National Instruments, Austin, TX, USA). P. denitrificans DSM413 was added to the MFCs by collecting cells from 200 ml cultures by centrifugation (5,000 r.p.m. for 20 min) and washing three times with HSNR anaerobically. The pellet was left inside the anaerobic chamber to de-aerate completely for 1 h, then resuspended in 5 ml HSNR with puromycin and placed in the anode chamber. To inoculate with sludge, 2 ml of each of the seven Round 3 methane-acclimated sludge cultures (Supplementary Table 1) were combined with 4 ml of the initial sludge isolate. This 18 ml of sludge was centrifuged and resuspended in a total of 8 ml HSNR with puromycin, then 2 ml of the resuspension was injected into each anode chamber.
The addition of sludge supernatants was done similarly by combining 2 ml of each of the seven Round 3 methane-acclimated sludge cultures (Supplementary Table 1) with 4 ml of the initial sludge isolate. This 18 ml of sludge was centrifuged, then 4 ml of filter-sterilized supernatant was added to each MFC. Electron shuttles (humic acids and FMN (Sigma-Aldrich), flavin adenine dinucleotide (Alfa Aesar), AQDS (Carbosynth, Berkshire, UK) and supernatants from methane-acclimated sludge) were included where specified by adding the sterile solutions into HSNR with puromycin to the final concentrations specified.
Gas chromatography and high-pressure liquid chromatography
For gas chromatography30, 50 μl of the gas phase was passed through a 60/80 Carboxen-1000 column (4600 × 2.1 mm, Supelco catalogue no. 12390-U) and a thermal conductivity detector on a 6890 N Agilent gas chromatograph. The injector, column and detector temperatures were maintained at 150, 180 and 280 °C, respectively. Carrier gas flow and reference gas flow (nitrogen used for both) were each maintained at 20 ml per min. Gases were identified based on their retention times, and their concentrations were calculated per comparisons with standards.
High-pressure liquid chromatography30 was conducted for the quantification of acetic acid. Samples were filtered through a 0.22 μm polyvinylidene fluoride membrane before fractionating 60 μl of a 1:6 dilution (in running buffer, 0.0025 M sulfuric acid in water) with a reversed-phase column [Phenomenex Rezex ROA-Organic Acid H + (8%) (300 × 7.8 mm)], employing a Waters 717 autosampler, a model 515 pump, and a 2996 photodiode array detector (absorbance at 210 nm was used). An isocratic flowrate of 0.4 ml per min 0.0025 M sulfuric acid in water was used. Glacial acetic acid (EMD Millipore, catalogue no. AX0073-6) was used as a standard for comparisons. The peak corresponding to acetic acid was identified based on retention time, and concentrations were calculated per comparisons with standards.
Characterization of sludge consortia
Genomic DNA was extracted directly from anode chambers using the UltraClean Microbial DNA Isolation Kit (MoBio, Carlsbad, CA, USA). The v3–v4 hypervariable region of 16S rDNA was amplified using primers PRO341-f and PRO801-r (Supplementary Table 4) universal for prokaryotes (both Archaea and Bacteria domains)59 with Ilumina (San Diego, CA, USA) adaptors attached for downstream processing. Amplicon libraries were further processed and analysed by the Genome Sciences Facility at the Penn State Hershey College of Medicine using the MiSeq (Illumina) platform according to manufacturer specifications.
Calculations
The Coulombic efficiency CE, defined as the fraction of electrons recovered from the substrate, was calculated38 with modifications using equation (1):
 |
where I is average current (current from abiotic reactions with methane from MFCs not inoculated with cells and current not from methane from MFCs inoculated with air-adapted M. acetivorans producing Mcr from ANME, G. sulfurreducens and sludge, without a methane in the headspace subtracted), t is time, e is the moles of electrons from each mole of methane consumed (8 for methane, from the half reaction equations (2) and (3) below), n is the total mol methane consumed, and F is Faraday's constant.
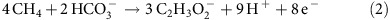 |
 |
Data availability
The raw 16S rDNA sequencing reads for the methane-acclimated sludge, original sludge, MFC with methane replicate 1, MFC with methane replicate 2 and MFC without methane replicate 1 have been submitted to the NCBI SRA database ( http://www.ncbi.nlm.nih.gov) under biosample accessions SAMN06562530, SAMN06562531, SAMN06562532, SAMN06562533 and SAMN06562534, respectively. The other data that support the main findings in this study are available from the authors upon reasonable request.
Additional information
How to cite this article: McAnulty, M. J. et al. Electricity from methane by reversing methanogenesis. Nat. Commun. 8, 15419 doi: 10.1038/ncomms15419 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Supplementary Figures, Supplementary Tables, Supplementary Note and Supplementary References
Acknowledgments
This work was supported by the Department of Energy Advanced Research Projects Agency—Energy. We thank Dr. Yuka Imamura Kawasawa and Anna C. Salzburg at The Pennsylvania State University College of Medicine for work with 16S rDNA sequencing and its analysis.
Footnotes
The authors declare no competing financial interests.
Author contributions T.K.W. conceived the project. M.J.M. and V.G.P. designed experiments and interpreted the data. R.J.-C. provided the air-adapted strain of M. acetivorans. K.-Y.K and B.E.L. provided insights into the MFC set-up. M.J.M, V.G.P. and T.K.W. wrote the manuscript.
References
- Berk R. S. & Canfield J. H. Bioelectrochemical energy conversion. Appl. Microbiol. 12, 10–12 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. B. & Yarbrough H. F. Preliminary experiments on a microbial fuel cell. Science 137, 615–616 (1962). [DOI] [PubMed] [Google Scholar]
- Oh S., Min B. & Logan B. E. Cathode performance as a factor in electricity generation in microbial fuel cells. Environ. Sci. Technol. 38, 4900–4904 (2004). [DOI] [PubMed] [Google Scholar]
- Logan B., Cheng S., Watson V. & Estadt G. Graphite fiber brush anodes for increased power production in air-cathode microbial fuel cells. Environ. Sci. Technol. 41, 3341–3346 (2007). [DOI] [PubMed] [Google Scholar]
- Catal T., Li K., Bermek H. & Liu H. Electricity production from twelve monosaccharides using microbial fuel cells. J. Power Sources 175, 196–200 (2008). [Google Scholar]
- Oh S. E. & Logan B. E. Hydrogen and electricity production from a food processing wastewater using fermentation and microbial fuel cell technologies. Water Res. 39, 4673–4682 (2005). [DOI] [PubMed] [Google Scholar]
- Jang J. K. et al. Construction and operation of a novel mediator- and membrane-less microbial fuel cell. Process Biochem. 39, 1007–1012 (2004). [Google Scholar]
- Pant D., Van Bogaert G., Diels L. & Vanbroekhoven K. A review of the substrates used in microbial fuel cells (MFCs) for sustainable energy production. Bioresource Technol. 101, 1533–1543 (2010). [DOI] [PubMed] [Google Scholar]
- Girguis P. & Reimers C. E. Methane-powered microbial fuel cells. US 2011/0123835 A1 (2011).
- Ding J. et al. Decoupling of DAMO archaea from DAMO bacteria in a methane-driven microbial fuel cell. Water Res. 110, 112–119 (2016). [DOI] [PubMed] [Google Scholar]
- Richter H. et al. Electricity generation by Geobacter sulfurreducens attached to gold electrodes. Langmuir 24, 4376–4379 (2008). [DOI] [PubMed] [Google Scholar]
- Reguera G. et al. Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl. Environ. Microbiol. 72, 7345–7348 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloski N. J. & Gralnick J. A. Flavin electron shuttles dominate extracellular electron transfer by Shewanella oneidensis. mBio 4, e00553-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D. H. & Zeikus J. G. Electricity generation in microbial fuel cells using neutral red as an electronophore. Appl. Environ. Microbiol. 66, 1292–1297 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimnejad M. et al. Methylene blue as electron promoters in microbial fuel cell. Int. J. Hydrogen Energy 36, 13335–13341 (2011). [Google Scholar]
- Das S. & Mangwani N. Recent developments in microbial fuel cells: a review. J. Sci. Ind. Res. 69, 727–731 (2010). [Google Scholar]
- Shindell D. T. et al. Improved attribution of climate forcing to emissions. Science 326, 716–718 (2009). [DOI] [PubMed] [Google Scholar]
- Howarth R. W., Santoro R. & Ingraffea A. Methane and the greenhouse-gas footprint of natural gas from shale formations. Clim. Change 106, 679–690 (2011). [Google Scholar]
- Howarth R. W. Methane emissions and climatic warming risk from hydraulic fracturing and shale gas development: implications for policy. Energy Emiss. Control Technol. 3, 45–54 (2015). [Google Scholar]
- Haynes C. A. & Gonzalez R. Rethinking biological activation of methane and conversion to liquid fuels. Nat. Chem. Biol. 10, 331–339 (2014). [DOI] [PubMed] [Google Scholar]
- Lee S. Y. & Kim H. U. Systems strategies for developing industrial microbial strains. Nat. Biotechnol. 33, 1061–1072 (2015). [DOI] [PubMed] [Google Scholar]
- Reeburgh W. S. Oceanic methane biogeochemistry. Chem. Rev. 107, 486–513 (2007). [DOI] [PubMed] [Google Scholar]
- Valentine D. L. Biogeochemistry and microbial ecology of methane oxidation in anoxic environments: a review. Antonie Van Leeuwenhoek 81, 271–282 (2002). [DOI] [PubMed] [Google Scholar]
- Timmers P. H. et al. Anaerobic oxidation of methane associated with sulfate reduction in a natural freshwater gas source. ISME J. 10, 1400–1412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn S. E., Chadwick G. L., Kempes C. P. & Orphan V. J. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526, 531–535 (2015). [DOI] [PubMed] [Google Scholar]
- Haroon M. F. et al. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500, 567–570 (2013). [DOI] [PubMed] [Google Scholar]
- Ettwig K. F. et al. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc. Natl Acad. Sci. USA 113, 12792–12796 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller S. et al. Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 351, 703–707 (2016). [DOI] [PubMed] [Google Scholar]
- Knittel K. & Boetius A. Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334 (2009). [DOI] [PubMed] [Google Scholar]
- Soo V. W. et al. Reversing methanogenesis to capture methane for liquid biofuel precursors. Microb. Cell Fact. 15, 11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima S. et al. Structure of a methyl-coenzyme M reductase from Black Sea mats that oxidize methane anaerobically. Nature 481, 98–101 (2012). [DOI] [PubMed] [Google Scholar]
- McAnulty M. J. et al. Metabolic engineering of Methanosarcina acetivorans for lactate production from methane. Biotechnol. Bioeng. 114, 852–861 (2017). [DOI] [PubMed] [Google Scholar]
- Jasso-Chavez R. et al. Air-adapted Methanosarcina acetivorans shows high methane production and develops resistance against oxygen stress. PLoS ONE 10, e0117331 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto A. et al. Cell-secreted flavins bound to membrane cytochromes dictate electron transfer reactions to surfaces with diverse charge and pH. Sci. Rep. 4, 5628 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice S. A., Wuertz S. & Kjelleberg S. Next-generation studies of microbial biofilm communities. Microb. Biotechnol. 9, 677–680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond D. R. & Lovley D. R. Electricity production by Geobacter sulfurreducens attached to electrodes. Appl. Environ. Microbiol. 69, 1548–1555 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez-Orta S. B. et al. The effect of flavin electron shuttles in microbial fuel cells current production. Appl. Microbiol. Biotechnol. 85, 1373–1381 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. COD removal characteristics in air-cathode microbial fuel cells. Bioresource Technol. 176, 23–31 (2015). [DOI] [PubMed] [Google Scholar]
- Devasahayam M. & Masih S. A. Microbial fuel cells demonstrate high coulombic efficiency applicable for water remediation. Indian J. Exp. Biol. 50, 430–438 (2012). [PubMed] [Google Scholar]
- Kiely P. D. et al. Anodic biofilms in microbial fuel cells harbor low numbers of higher-power-producing bacteria than abundant genera. Appl. Microbiol. Biotechnol. 88, 371–380 (2010). [DOI] [PubMed] [Google Scholar]
- Sun X., Luo P. & Li M. Paracoccus angustae sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 65, 3469–3475 (2015). [DOI] [PubMed] [Google Scholar]
- Nguyen N. L. et al. Paracoccus panacisoli sp. nov., isolated from a forest soil cultivated with Vietnamese ginseng. Int. J. Syst. Evol. Microbiol. 65, 1491–1497 (2015). [DOI] [PubMed] [Google Scholar]
- Dastager S. G. et al. Paracoccus niistensis sp. nov., isolated from forest soil, India. Antonie Van Leeuwenhoek 99, 501–506 (2011). [DOI] [PubMed] [Google Scholar]
- Lovley D. R. & Blunt-Harris E. L. Role of humic-bound iron as an electron transfer agent in dissimilatory Fe(III) reduction. Appl. Environ. Microbiol. 65, 4252–4254 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. A., Nevin K. P. & Lovley D. R. Syntrophic growth via quinone-mediated interspecies electron transfer. Front. Microbiol. 6, 121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub K. L. & Schink B. Evaluation of electron-shuttling compounds in microbial ferric iron reduction. FEMS Microbiol. Lett. 220, 229–233 (2003). [DOI] [PubMed] [Google Scholar]
- Scott D. T. et al. Quinone moieties act as electron acceptors in the reduction of humic substances by humics-reducing microorganisms. Environ. Sci. Technol. 32, 2984–2989 (1998). [Google Scholar]
- Lovley D. R. et al. Humic substances as electron acceptors for microbial respiration. Nature 382, 445–448 (1996). [Google Scholar]
- Voordeckers J. W., Kim B. C., Izallalen M. & Lovley D. R. Role of Geobacter sulfurreducens outer surface c-type cytochromes in reduction of soil humic acid and anthraquinone-2,6-disulfonate. Appl. Environ. Microbiol. 76, 2371–2375 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K., Schicklberger M. & Gescher J. Dissimilatory reduction of extracellular electron acceptors in anaerobic respiration. Appl. Environ. Microbiol. 78, 913–921 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf W. W., Zhang J.-K., Shi X. & Wolfe R. S. Molecular, genetic, and biochemical characterization of the serC gene of Methanosarcina barkeri Fusaro. J. Bacteriol. 178, 5797–5802 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccavo F. et al. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl. Environ. Microbiol. 60, 3752–3759 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotaru A. E., Woodard T. L., Nevin K. P. & Lovley D. R. Link between capacity for current production and syntrophic growth in Geobacter species. Front. Microbiol. 6, 744 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y. et al. Alteration of intracellular protein expressions as a key mechanism of the deterioration of bacterial denitrification caused by copper oxide nanoparticles. Sci. Rep. 5, 15824 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf W. W. et al. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl Acad. Sci. USA 94, 2626–2631 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer E. R. et al. Scanning electron microscopy. Curr. Protoc. Microbiol. Chapter 2, Unit 2B 2 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanas V. & Logan B. E. Evaluation of multi-brush anode systems in microbial fuel cells. Bioresource Technol. 148, 379–385 (2013). [DOI] [PubMed] [Google Scholar]
- Feng Y. J., Yang Q., Wang X. & Logan B. E. Treatment of carbon fiber brush anodes for improving power generation in air-cathode microbial fuel cells. J. Power Sources 195, 1841–1844 (2010). [Google Scholar]
- Takahashi S. et al. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 9, e105592 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures, Supplementary Tables, Supplementary Note and Supplementary References
Data Availability Statement
The raw 16S rDNA sequencing reads for the methane-acclimated sludge, original sludge, MFC with methane replicate 1, MFC with methane replicate 2 and MFC without methane replicate 1 have been submitted to the NCBI SRA database ( http://www.ncbi.nlm.nih.gov) under biosample accessions SAMN06562530, SAMN06562531, SAMN06562532, SAMN06562533 and SAMN06562534, respectively. The other data that support the main findings in this study are available from the authors upon reasonable request.