Abstract
The growth advantage in stationary phase (GASP) phenotype was shown to be present in two mutants lacking the antifungal phenotype (Af− mutants) of Pseudomonas aureofaciens PA147-2. Complementation demonstrated a correlation between GASP and the antifungal defect in one strain but not in the second. Sequence analysis revealed the Af− GASP strain had a mutation in a gene (finR) encoding a LysR-type regulator. Antifungal-minus mutants arose in starved cultures, and those aged cultures had increased fitness. Taken together, the results show that there are at least two paths to the GASP phenotype in P. aureofaciens, one of which results in a concomitant loss of the antifungal phenotype.
Fluorescent Pseudomonas spp. are ubiquitous inhabitants of bulk soil and the plant rhizosphere and are well adapted to the complex conditions found in these environments. Their success can be at least partly attributed to a range of strategies that enhance their competitive fitness, such as the ability to efficiently scavenge trace materials. An additional characteristic of many soil pseudomonads is the ability to produce antifungal metabolites, enabling them to reduce or eliminate fungal competitors from their shared niche (8, 16, 17, 32). Fungal inhibition by pseudomonads is more than just an ecologically interesting trait; strains capable of producing antifungal metabolites are potential biological alternatives to chemical control of agricultural diseases (35), as demonstrated by experiments with mutants lacking the antifungal phenotype (Af− mutants) (2, 10, 32). However, it has been shown that after inoculation into soils, the population of biocontrol bacteria can decline, probably due to the cost of producing secondary metabolites such as antifungal compounds (15, 34).
Pseudomonas aureofaciens PA147-2 has been shown to inhibit the in vitro growth of phytopathogenic fungi (5) and to suppress Phytophthora rot of asparagus in glasshouse and field trials (6, 15). In a field test P. aureofaciens PA147-2 showed significant protection of asparagus plants from Phytophthora rot; however, this was significantly less effective than treatment with a chemical fungicide. Since it has been demonstrated that colonization is essential for effective biocontrol by a number of different Pseudomonas spp. (PCL1391, WCS365, and SBW25 [9, 12, 27]), it is likely that the lower efficacy of the biological control treatment may have resulted from a decrease in the population size of PA147-2 during the 6-month field trial. Given that PA147-2 forms strong biofilms (26) and adheres strongly to plant roots (unpublished data), we reasoned that the inability to maintain a high population under field conditions might be due to a competitive disadvantage associated with the cost of antifungal metabolite production. Accordingly, mutants of PA147-2 that are defective in antifungal activity should have increased fitness relative to the parental strain. Therefore, we initiated studies to examine the relative fitness of PA147-2 and Af− mutants generated previously (4). The in vitro experiments reported here were designed to assess the fitness cost of antifungal compound production and were undertaken under conditions that gave no measured advantage to any strain in monoculture.
Over the last decade or so, bacteria have been used in experimental systems to study the evolution of “increased fitness” that results from adaptation to defined conditions. In static cultures of P. fluorescens, i.e., under starvation conditions, adaptive radiation of niche specialists has been shown (30). Under both starvation conditions and continuous exponential culture conditions, Escherichia coli strains with higher fitness than the parental strain have evolved (14, 33, 37). Since the conditions encountered in soil over many months are likely to be nutrient deprived, we assessed the relative fitness of PA147-2 Af+ and Af− strains in 10-day “starvation” cultures. During these experiments we observed that two antifungal minus mutants exhibited a GASP-like phenotype in competition with the wild type. The so-called GASP phenomenon (named for growth advantage in stationary phase) refers to bacteria with higher fitness for long-term starvation conditions than their parental strain. GASP has been well-studied in E. coli (14, 37-39) and was recently shown to occur in Enterobacter cloacae, Salmonella enterica serovar Typhimurium, Providencia stuartii, and Shigella dysenteriae (24). However, to our knowledge the genetic basis has only been described in E. coli.
Construction of PA147-2lacZY for competition experiments.
All competition experiments were carried out in Pseudomonas minimal medium (PMM) (19) as follows. For 1:1 competitions, 10 ml of PMM was inoculated with equal numbers of each competing strain (100 μl of each from monocultures grown for the previous 20 h), whereas a dilution of one competitor was used to achieve a minority inoculum for minority experiments. These cultures were grown at 30°C, with shaking at 200 rpm. Samples (100 μl) were taken daily, and 10-fold serial dilutions were plated onto selective media to allow enumeration of each competing strain. To provide a selectable marker for competition experiments, we introduced a lacZY-bearing transposon (Tn7-lac7117) (3) into the PA147-2 genome. The Tn7-lac7117 insertion had no impact on the fitness of PA147-2lacZY relative to PA147-2 or on the production of antifungal compound(s).
Competition between PA147-2lacZY and Af− mutants.
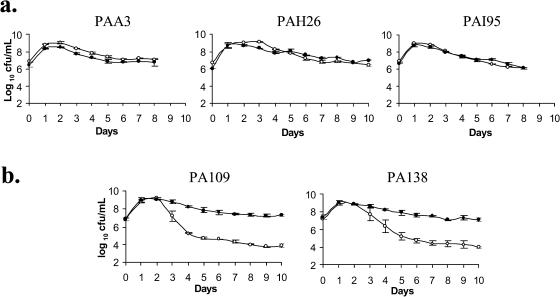
To test whether antifungal metabolite production had an influence on the starvation fitness of PA147-2, five Tn5-generated Af− mutants of PA147-2 (5) were assessed for relative fitness against PA147-2lacZY under defined conditions. These data show that the mutants can be divided into two categories based upon their competitive ability relative to PA147-2lacZY (Fig. 1). Group one mutants (comprising PAI95, PAH26, and PAA3) were of equivalent fitness to PA147-2lacZY, whereas group two mutants (PA109 and PA138) showed an ability to outcompete PA147-2lacZY under these conditions in a manner that strongly resembles the GASP phenomenon. Thus, there are at least two routes by which PA147-2 can become Af−; one results in no loss of fitness, while the second route does, thus supporting our original hypothesis. The possibility that the increased fitness of PA109 and PA138 arose due to additional transposition of Tn5 was ruled out by hybridizing a Tn5-specific probe to total DNA extracted from competition cultures and demonstrating that Tn5 was present as a single copy and in the same-sized DNA fragment in young (overnight) cultures and in those that had grown for 10 days (data not shown). It is worth noting that in competition with PA138 and PA109, PA147-2lacZY was always detectable. The wild type continued to persist as a minority population for the duration of the experiment, in contrast to the findings from Roberto Kolter's group examining prolonged E. coli cultures (for examples, see references 14 and 37).
FIG. 1.
Competition between PA147-2lacZY (○) and Tn5-generated Af− mutants (•). Approximately equal numbers of group 1 mutants (a) or group 2 mutants (b) were mixed in PMM and grown for 8 to 10 days. Strains were enumerated daily. The data shown represent the averages from three independent experiments. Standard errors are shown.
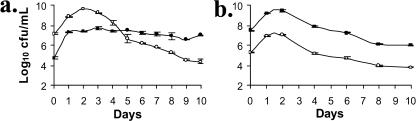
To further examine the competitive nature of the group two mutants relative to PA147-2lacZY, competition experiments were conducted in which the initial ratio of PA138 to wild type (PA147-2lacZY) was 1:250. In these experiments, PA138 was able to establish and eventually dominate the cultures (Fig. 2a). In contrast, when PA147-2lacZY was introduced as a minority in competition with PA147-2, it was unable to increase its relative representation in the population (Fig. 2b). This finding strongly supports the argument that PA138 has increased fitness relative to the wild type under these conditions.
FIG. 2.
(a) Competition between PA138 (•) and PA147-2lacZY (○). PA138 was inoculated as a 1:250 minority population. Despite the low initial inoculum, PA138 was able to outcompete PA147-2lacZY. (b) PA147-2lacZY was inoculated as a 1:150 minority in competition with PA147-2. Unlike the GASP mutant (PA138), PA147-2lacZY is unable to increase its relative representation in the population.
DNA sequences flanking Tn5 insertions.
To understand the molecular basis of the GASP phenotype, we sought to use DNA sequences to identify the genes mutated in the five Af− Tn5 mutants of PA147-2. The sequence adjacent to the Tn5 insertion in PAH26 was reported previously (26). To determine the sequence adjacent to Tn5 in the remaining mutants (PA138, PA109, PAA3, and PA195), a region encompassing 2,684 bp of Tn5 (left of a unique SalI site) and the adjacent P. aureofaciens DNA was ligated into the SalI site of pBluescript KS(−). The resulting clones were sequenced by using a primer that anneals to IS50 (5′-GCACGATGAAGAGCAGAAG -3′). BLAST (1) was used to search GenBank for putative homologues. The group 1 mutant PAH26 has an insertion in pstA, and in PAI95 the transposon has inserted into a gene predicted to encode an efflux protein, whereas the other group one mutant (PAA3) has an insertion in a sequence that has not been described elsewhere. In the group two (GASP) mutants, the transposon insertion was found to be in two different classes of regulatory genes. In PA138, Tn5 interrupted a sequence that we named finR (for fungal inhibition), whose predicted translation product resembles members of the LysR family of transcriptional regulators. The insertion in PA109 interrupted a putative two-component regulator gene (finT) with similarity to sequences specifying predicted hybrid proteins that contain both the sensor histidine kinase and response regulator (receiver) domains in a single protein. This organization is in contrast to the well-known GacA/GacS two-component regulator pair that controls antifungal activity in a number of Pseudomonas sp., which has the sensor (GacS) (11, 18, 20) and response regulator (GacA) (23) domains on different proteins. Thus, the group 2 mutants have insertions in two different classes of putative global regulator genes, which may lead to altered expression of a large number of genes, any of which could influence fitness. The DNA sequence data provide a possible explanation for the results of the initial competition experiments. It appears as if loss of antifungal compound production by mutation of genes that may be directly related to antifungal production (group one mutants) does not usually influence fitness. However, antifungal compound production is tied into regulatory circuits that do affect fitness (group two mutants); thus, changes in these pathways could increase fitness in the field while reducing or inhibiting production of antifungal compounds. Identifying the type of gene interrupted by Tn5 in the Af− mutants led us to propose that regulatory networks involving antifungal activity, rather than antifungal production per se, have an impact on fitness during starvation conditions. Schmidt-Eisenlohr et al. (31) showed a fitness effect from a mutation of the two-component regulator gene gacS in P. chlororaphis. The gacS mutation resulted in reduced numbers of bacteria in a rhizosphere colonization experiment (relative to the wild type) and a reduced lag-phase in culture. Interestingly, these analyses also showed that the gacS mutation did not lead to a competitive defect in the rhizosphere when it was in competition with the parental strain, a finding which is in contrast to our culture-based competition experiments with regulatory mutants that show increased fitness in competition with the wild-type strain.
Complementation of group 2 (GASP) mutants.
To confirm our refined hypothesis that mutations in the putative LysR (PA138) and two-component regulator (PA109) genes (called finR and finT, respectively) were responsible for the observed GASP phenotype, we undertook complementation experiments. Carruthers et al. (5) previously showed that antifungal activity was restored in PA109 by replacing the mutated region with the wild-type sequence by allele exchange (5). We found that the complemented PA109 still exhibited the GASP phenotype, indicating that the finT mutation is responsible for the loss of antifungal activity, but that an additional unknown mutation must cause the GASP effect in this strain. Similarly, the Tn5 insertion in PA138 was replaced by allele exchange with wild-type sequences present in a cosmid that hybridized with a probe flanking Tn5 in PA138 as described previously (5). Southern hybridizations confirmed the allele exchange, and fungal inhibition assays showed that the restored PA138 strain had regained wild-type antifungal activity (data not shown). However, in contrast to PA109, removal of Tn5 in PA138 eliminated the fitness advantage of this strain over PA147-2lacZY, demonstrating that the Tn5 insertion was responsible for the GASP phenotype of this mutant. These data show that our hypothesis was partly correct; a regulator of antifungal activity is also involved in fitness. Furthermore, this experiment demonstrates that only a subset of antifungal activity regulators in PA147-2 are involved in fitness, indicating that there is limited overlap in the genetics underlying these two phenotypes. Given that a finR mutation impacts upon fitness, whereas a finT mutation does not, we can conclude that FinR and FinT are clearly independent regulators controlling aspects of antifungal activity. The finding that finT does not contribute to the increased fitness of PA109 is consistent with the observation that inactivation of the two-component regulator gene gacS has no impact on the competitive fitness of P. chlororaphis (31) and did not result in a competitive advantage for P. aureofaciens 30-84 in rhizosphere colonization experiments conducted in live soil (7). Furthermore, gacA mutations in P. fluorescens CHAO had a negative impact on persistence of viable and culturable cells in bulk soil (25, 28). In contrast, Duffy and Defago (13) observed an accumulation of spontaneous gac mutants (1.3% of cells) after 12 days in a rich medium. Upon stepwise scale-up to increasingly larger cultures (48 h of growth in each culture prior to transfer to the next culture), the gac mutants could accumulate to 7, 23, and 61% of the total viable cells in 20-, 100-, and 500-ml cultures, indicating a fitness advantage. However, whether this is similar to a GASP phenotype is unclear since the cultures were serially transferred rather than grown to starvation. Since the genetic basis for enhanced fitness and lack of antifungal production was known for PA138 (finR::Tn5), it was the strain of choice for the following experiments.
PA138 versus “aged” PA147-2lacZY.
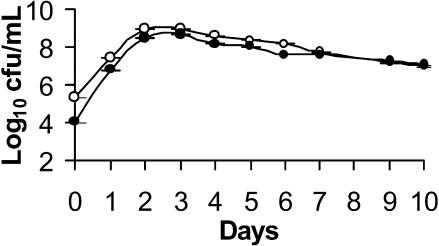
Based upon previous findings in experiments with E. coli (14, 37), we predicted that GASP evolution would occur in long-term cultures of P. aureofaciens, in which case it follows that aged cultures of PA147-2 (and PA147-2lacZY) would have accumulated mutants of greater fitness than the original parental strain. Accordingly, PA138 would be less able to displace populations derived from aged cultures than it can displace “young” PA147-2 (ancestors) in competition experiments. To test this hypothesis, competition experiments were started with PA138 and 10-day-old “evolved” PA147-2lacZY. As predicted, PA138 was not as competitive against the “evolved” PA147-2lacZY (Fig. 3). In competition against PA147-2lacZY progenitors, we have shown that PA138 eventually establishes a population up to 4 orders of magnitude higher than that of PA147-2lacZY. However, in these competitions against evolved PA147-2lacZY, the difference was negligible. This experiment provides an independent line of evidence correlating loss of antifungal activity with increased fitness.
FIG. 3.
Competition between PA138 (•) and 10-day-old PA147-2lacZY (○). The aged PA147-2lacZY is fitter than the parental PA147-2lacZY, as evidenced by the increase in its competitive fitness relative to PA138.
Do Af− mutants arise in PA147-2 monoculture?
Since the loss of ability to produce antifunal compounds has been associated with increased competitive fitness in broth cultures (mutant PA138), and fitter mutants of P. aureofaciens have been shown to accumulate in long-term culture, then Af− mutants should also arise during long-term PA147-2 monoculture. To test this prediction, we grew two independent cultures of PA147-2 in PMM for 20 days. The cultures were serially diluted and plated to obtain single colonies. Individual colonies from both cultures were tested for their ability to inhibit fungal growth. The results (Table 1) show that Af− and antifungal-impaired mutants arise in PA147-2 monocultures, but there is a large variation between replicates. Previous long-term starvation experiments have also shown considerable variation between initially identical lines (14). It is likely that the difference between cultures results from the nature of the GASP mutants that initially take over the population. The higher number of Af− mutants in culture one could result if a finR-type mutation (Af−, GASP) came to dominate the population, whereas an Af+ GASP mutant could have taken over the second culture. Our results with the GASP strains PA138 (finR::Tn5) and PA109 (finT::Tn5) show that Af− and Af+ GASP mutants are possible, and Finkel and Kolter showed that GASP occurred by selection of different mutations in two initially isogenic populations (14).
TABLE 1.
Fungal inhibition by colonies arising in starvation monoculturea
| Replicate group | % Colonies with phenotype:
|
||
|---|---|---|---|
| Antifungal plus | Antifungal reduced | Antifungal minus | |
| 1 | 51 | 2.5 | 46.5 |
| 2 | 75.8 | 21 | 3.2 |
Two cultures of PA147-2 were grown for 20 days in Pseudomonas minimal medium. Upon plating, colonies from both cultures were assessed for the ability to inhibit fungal growth. Data are expressed as percentage of colonies with a particular phenotype.
The ability of PA138 to outcompete PA147-2 bears a striking resemblance to the so-called GASP phenomenon, which is well studied in E. coli (21, 36). During prolonged starvation, the GASP mutants proliferate and take over cultures, displacing the less-fit parental strain. This phenomenon has been associated with a number of different mutations, notably in rpoS (37) and lrp (39). RpoS is a sigma factor involved in the regulation of a cascade of genes in response to the transition to stationary-phase growth (22), and Lrp (the leucine-responsive regulatory protein) is a transcriptional regulator of an extensive regulon of between 35 and 75 genes in E. coli (29). Similarly, PA138 has an insertion in finR, a predicted LysR-type regulator gene, and has the ability to outcompete its parental strain. Thus, we suggest that the mutation in PA138 has produced a GASP phenotype. The correlation of loss of a putative regulator with increased fitness in P. aureofaciens strengthens the relevance of this work to GASP studies, since mutations in global regulator genes have been implicated in the GASP phenotype in E. coli (37, 39). Zinser and Kolter (39) proposed that mutations in regulator genes are highly beneficial during starvation conditions because they result in the simultaneous alteration of multiple cellular activities. PA138 is probably a regulatory mutant and thus supports their proposal. The addition of a mutation in a LysR-type regulator to the spectrum of potential GASP alleles strengthens the hypothesis that fitter strains can emerge as a result of the coordinated repression of a large number of genes (39). It is also conceivable that mutation of a repressor could lead to a fitness increase by derepression of genes whose expression confers an advantage in a given environment.
Our results suggest that under stressful conditions (e.g., starvation) the selection for fitter P. aureofaciens can result in the accumulation of strains in which antifungal production is reduced or eliminated. The fact that there is variation in fitness between replicate starvation cultures of PA147-2 indicates that the loss or reduction of antifungal activity results from mutations arising in culture. It is likely that PA147-2 has a number of different possible solutions to the problem of fitness under starvation conditions. These solutions could result in the same or a similar GASP phenotype, whereas only a subset cause a reduction or loss of antifungal activity. This is supported by PA109, which retains higher fitness even when restored to Af+, whereas PA138 does not. Thus, the increased fitness in PA138 is related to the lack of production of antifungal metabolites, whereas a similar increase of fitness in PA109 is achieved in an unrelated (and unknown) manner. Notably, the loss of Af production does not always result in a fitness advantage (i.e., the group one mutants), demonstrating that there are many routes for the loss of antifungal production, not all of them contributing equally to fitness.
PA147-2 was isolated from the plant rhizosphere, where it retained the ability to inhibit fungal growth. Presumably, the selective force provided by fungal competition in the field was sufficient for maintenance of antifungal production as a successful solution to living in this environment at the time PA147-2 was isolated. However, we have demonstrated that Af− mutants of PA147-2 arise during starvation and that the loss of antifungal production in the case of PA138 led to a strain that could outcompete PA147-2 under starvation conditions. Therefore, if field conditions become unfavorable, such as between growing seasons when plant roots are dormant, starvation may result in the accumulation of starvation-fitter mutants that do not produce antifungal metabolites. Although this could be readily overcome in a biocontrol strategy by reapplication of the bacteria each season, further work to test the field relevance of these data should be performed in order to refine future biocontrol schemes.
Acknowledgments
AGMARDT doctoral scholarships to M.W.S. and S.R.G. are gratefully acknowledged. M.W.S. and S.R.G. were supported in part by a University of Canterbury doctoral scholarship and a University of Canterbury research award, respectively.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anjaiah, V., N. Koedam, B. Nowak-Thompson, J. E. Loper, M. Hofte, J. T. Tambong, and P. Cornelis. 1998. Involvement of phenazines and anthranilate in the antagonism of Pseudomonas aeruginosa PNA1 and Tn5 derivatives toward Fusarium spp. and Pythium. Mol. Plant-Microbe Interact. 11:847-854. [Google Scholar]
- 3.Barry, G. F. 1988. A broad-host-range shuttle system for gene insertion into the chromosomes of gram-negative bacteria. Gene 71:75-84. [DOI] [PubMed] [Google Scholar]
- 4.Carruthers, F. L. 1994. A study of antifungal activity by a potential biological control strain, Pseudomonas aureofaciens strain PA147-2. Ph.D. thesis. University of Canterbury, Christchurch, New Zealand.
- 5.Carruthers, F. L., A. J. Conner, and H. K. Mahanty. 1994. Identification of a genetic locus in Pseudomonas aureofaciens involved in fungal inhibition. Appl. Environ. Microbiol. 60:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carruthers, F. L., T. Shum-Thomas, A. J. Conner, and H. K. Mahanty. 1995. The significance of antibiotic production by Pseudomonas aureofaciens PA147-2 for biological control of Phytophthora megasperma root rot of asparagus. Plant Soil 170:339-344. [Google Scholar]
- 7.Chancey, S. T., D. W. Wood, E. A. Pierson, and L. S. Pierson III. 2002. Survival of GacS/GacA mutants of the biological control bacterium Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. Appl. Environ. Microbiol. 68:3308-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chancey, S. T., D. W. Wood, and L. S. Pierson. 1999. Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 65:2294-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin-A-Woeng, T. F. C., G. V. Bloemberg, I. H. Mulders, L. C. Dekkers, and B. J. J. Lugtenberg. 2000. Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol. Plant-Microbe Interact. 13:1340-1345. [DOI] [PubMed] [Google Scholar]
- 10.Chin-A-Woeng, T. F. C., G. V. Bloemberg, A. J. van der Bij, K. M. G. M. van der Drift, J. Schripsema, B. Kroon, R. J. Scheffer, C. Keel, P. A. H. M. Bakker, H. V. Tichy, F. J. de Bruijn, J. E. Thomas-Oates, and B. J. J. Lugtenberg. 1998. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol. Plant-Microbe Interact. 11:1069-1077. [Google Scholar]
- 11.Corbell, N., and J. E. Loper. 1995. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf-5. J. Bacteriol. 177:6230-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dekkers, L. C., C. C. Phoelich, L. van der Fits, and B. J. J. Lugtenberg. 1998. A site-specific recombinase is required for competitive root colonization by Pseudomonas fluorescens WCS365. Proc. Natl. Acad. Sci. USA 95:7051-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duffy, B. K., and G. Defago. 2000. Controlling instability in gacS-gacA regulatory genes during inoculant production of Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 66:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finkel, S. E., and R. Kolter. 1999. Evolution of microbial diversity during prolonged starvation. Proc. Natl. Acad. Sci. USA 96:4023-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godfrey, S. A. C., M. W. Silby, P. G. Falloon, and H. K. Mahanty. 2000. Biological control of Phytophthora megasperma var. sojae, causal agent of Phytophthora rot of asparagus, by Pseudomonas aureofaciens PA147-2: a preliminary field trial. New Zealand J. Crop Hort. Sci. 28:97-103. [Google Scholar]
- 16.Harrison, L. A., L. Letendre, P. Kovacevich, E. Pierson, and D. Weller. 1993. Purification of an antibiotic effective against Gaeumannomyces graminis var. tritici produced by a biocontrol agent, Pseudomonas aureofaciens. Soil Biol. Biochem. 25:215-221. [Google Scholar]
- 17.Hill, D. S., J. I. Stein, N. R. Torkiewitz, A. M. Morse, C. R. Howell, J. P. Pachlatko, J. O. Becker, and J. M. Ligon. 1994. Cloning of genes involved in the synthesis of pyrrolnitrin from Pseudomonas fluorescens and role of pyrrolnitrin synthesis in biological control of plant disease. Appl. Environ. Microbiol. 60:78-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hrabak, E. M., and D. K. Willis. 1992. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J. Bacteriol. 174:3011-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirner, S., S. Krauss, G. Sury, S. T. Lam, J. M. Ligon, and K. H. vanPee. 1996. The non-haem chloroperoxidase from Pseudomonas fluorescens and its relationship to pyrrolnitrin biosynthesis. Microbiology 142:2129-2135. [DOI] [PubMed] [Google Scholar]
- 20.Kitten, T., T. G. Kinscherf, J. L. McEvoy, and D. K. Willis. 1998. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol. Microbiol. 28:917-929. [DOI] [PubMed] [Google Scholar]
- 21.Kolter, R., D. A. Siegele, and A. Tormo. 1993. The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47:855-874. [DOI] [PubMed] [Google Scholar]
- 22.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5:49-59. [DOI] [PubMed] [Google Scholar]
- 23.Laville, J., C. Voisard, C. Keel, M. Maurhofer, G. Defago, and D. Haas. 1992. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. USA 89:1562-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Garcia, E., A. Tormo, and J. M. Navarro-Llorens. 2003. GASP phenotype: presence in enterobacteria and independence of sigma S in its acquisition. FEMS Microbiol. Lett. 225:201-206. [DOI] [PubMed] [Google Scholar]
- 25.Mascher, F., Y. Moenne-Loccoz, U. Schnider-Keel, C. Keel, D. Haas, and G. Defago. 2002. Inactivation of the regulatory gene algU or gacA can affect the ability of biocontrol Pseudomonas fluorescens CHA0 to persist as culturable cells in nonsterile soil. Appl. Environ. Microbiol. 68:2085-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monds, R. D., M. W. Silby, and H. K. Mahanty. 2001. Expression of the Pho regulon negatively regulates biofilm formation by Pseudomonas aureofaciens PA147-2. Mol. Microbiol. 42:415-426. [DOI] [PubMed] [Google Scholar]
- 27.Naseby, D. C., J. A. Way, N. J. Bainton, and J. M. Lynch. 2001. Biocontrol of Pythium in the pea rhizosphere by antifungal metabolite producing and non-producing Pseudomonas strains. J. Appl. Microbiol. 90:421-429. [DOI] [PubMed] [Google Scholar]
- 28.Natsch, A., C. Keel, H. A. Pfirter, D. Haas, and G. Defago. 1994. Contribution of the global regulator gene gacA to persistence and dissemination of Pseudomonas fluorescens biocontrol strain CHA0 introduced into soil microcosms. Appl. Environ. Microbiol. 60:2553-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman, E. B., R. T. Lin, and R. D'Ari. 1996. The leucine/Lrp regulon, p. 1513-1525. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 30.Rainey, P., and M. Travisano. 1998. Adaptive radiation in a heterogeneous environment. Nature 394:69-72. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt-Eisenlohr, H., A. Gast, and C. Baron. 2003. Inactivation of gacS does not affect the competitiveness of Pseudomonas chlororaphis in the Arabidopsis thaliana rhizosphere. Appl. Environ. Microbiol. 69:1817-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomashow, L. S., and D. M. Weller. 1988. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J. Bacteriol. 170:3499-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Travisano, M., J. A. Mongold, A. F. Bennett, and R. E. Lenski. 1995. Experimental tests of the roles of adaptation, chance, and history in evolution. Science 267:87-90. [DOI] [PubMed] [Google Scholar]
- 34.Viebahn, M., D. C. Glandorf, T. W. M. Ouwens, E. Smit, P. Leeflang, K. Wernars, L. S. Thomashow, L. C. van Loon, and P. A. H. M. Bakker. 2003. Repeated introduction of genetically modified Pseudomonas putida WCS358r without intensified effects on the indigenous microflora of field-grown wheat. Appl. Environ. Microbiol. 69:3110-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weller, D. M., J. M. Raaijmakers, B. B. McSpadden Gardener, and L. S. Thomashow. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40:309-348. [DOI] [PubMed] [Google Scholar]
- 36.Zambrano, M. M., and R. Kolter. 1996. GASPing for life in stationary phase. Cell 86:181-184. [DOI] [PubMed] [Google Scholar]
- 37.Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757-1760. [DOI] [PubMed] [Google Scholar]
- 38.Zinser, E. R., and R. Kolter. 1999. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J. Bacteriol. 181:5800-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zinser, E. R., and R. Kolter. 2000. Prolonged stationary-phase incubation selects for lrp mutations in Escherichia coli K-12. J. Bacteriol. 182:4361-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]