Abstract
Cereulide, a depsipeptide structurally related to valinomycin, is responsible for the emetic type of gastrointestinal disease caused by Bacillus cereus. Due to its chemical structure, (d-O-Leu-d-Ala-l-O-Val-l-Val)3, cereulide might be synthesized nonribosomally. Therefore, degenerate PCR primers targeted to conserved sequence motifs of known nonribosomal peptide synthetase (NRPS) genes were used to amplify gene fragments from a cereulide-producing B. cereus strain. Sequence analysis of one of the amplicons revealed a DNA fragment whose putative gene product showed significant homology to valine activation NRPS modules. The sequences of the flanking regions of this DNA fragment revealed a complete module that is predicted to activate valine, as well as a putative carboxyl-terminal thioesterase domain of the NRPS gene. Disruption of the peptide synthetase gene by insertion of a kanamycin cassette through homologous recombination produced cereulide-deficient mutants. The valine-activating module was highly conserved when sequences from nine emetic B. cereus strains isolated from diverse geographical locations were compared. Primers were designed based on the NRPS sequence, and the resulting PCR assay, targeting the ces gene, was tested by using a panel of 143 B. cereus group strains and 40 strains of other bacterial species showing PCR bands specific for only the cereulide-producing B. cereus strains.
Bacillus cereus is a gram-positive spore-forming food pathogen that causes two types of food poisoning syndromes: emesis and diarrhea. It is an increasing problem, especially in heat-treated food, such as convenience food and food used in catering, because of its resistance to pasteurization and antimicrobial agents. The true incidence of B. cereus food poisoning is unknown for a number of reasons, including misdiagnosis of the illness, which may be symptomatically similar to other types of food poisoning.
The emetic syndrome is mainly characterized by vomiting a few hours after ingestion of the contaminated food. In the diarrheal syndrome, the symptoms appear 8 to 16 h after ingestion and include abdominal pain and diarrhea. Different enterotoxins are the causative agents of the diarrheal syndrome. These toxins are comparatively well characterized at the molecular level (for a review see reference 15), and molecular diagnostic assays are available (14, 19). Far less is known about the emesis-causing molecule cereulide. Cereulide is a small, heat-stable dodecadepsipeptide which, for instance, was shown to be involved in fulminate liver failure in a human case (23). It has been shown to cause cellular damage in animal models and is toxic for mitochondria by acting as a potassium ionophore (2, 27, 36). Recently, it has been reported that cereulide inhibits human natural killer cells and might therefore have an immunomodulating effect (30). Based on its structure, (d-O-Leu-d-Ala-l-O-Val-l-Val)3, one could expect cereulide to be synthesized nonribosomally. Alternating peptide and ester bonds, as well as d-amino acids and a cyclic structure, are often found in nonribosomal peptide synthetase (NRPS) products (24).
NRPSs are large multifunctional proteins with a modular organization. One module contains all catalytic activities which are necessary for incorporation of one amino acid residue into the peptide product. According to the “colinearity rule” (24, 48) the order of modules usually corresponds directly to the order of amino acids in the peptide product. The core of each module is the adenylation domain (A domain) that activates the cognate amino acid as an adenylate. The activated substrate is covalently bound to the thiolation domain (T domain) or peptidyl carrier protein. Chain elongation of the peptide is catalyzed by condensation domains (C domains) that are located at the N-terminal ends of modules accepting acyl groups from the preceding module (40). Following peptide bond formation, the peptidyl moiety is transferred to and condensed with the downstream carrier protein-bound monomer. A carboxyl-terminal thioesterase domain catalyzes the release (and in some cases even oligomerization and/or cyclization) of the mature NRPS-bound peptide product (44). The A and T domains contain highly conserved core motifs that can be used for a universal PCR approach to identify parts of NRPSs (24, 48).
The aim of this work was to identify genes potentially involved in cereulide production by a PCR screening assay by using degenerate primers directed against conserved motifs of known NRPSs (24) in order to gain insight into the synthesis of cereulide and to provide the basis for the development of molecular detection systems for cereulide producers.
MATERIALS AND METHODS
Strains and plasmids.
The cereulide-producing B. cereus strains F4810/72 (18), NC7401 (1), and MHI 87 (35) were used to search for the genetic locus responsible for the production of the emetic toxin cereulide. Cells were grown on plate count plates or in broth at 30°C. Escherichia coli strains were grown at 37°C in Luria-Bertani medium with the appropriate antibiotics. Bacterial strains used to test the specificity of the ces gene PCR assay are listed in Table 1, and additional information on the origins of cereulide-producing strains is provided in Table 2. Plasmids used in this study are listed in Table 3.
TABLE 1.
Bacterial species used to test the specificity of the PCR assay
| Bacterial species | No. of strains tested |
|---|---|
| Bacillus cereus group | |
| Bacillus cereusa | 114 |
| Bacillus anthracis | 7 |
| Bacillus thuringiensis | 6 |
| Bacillus mycoides | 6 |
| Bacillus pseudomycoides | 3 |
| Bacillus weihenstephanensis | 7 |
| Other Bacillus spp. | |
| Bacillus brevis | 3 |
| Bacillus subtilis | 1 |
| Bacillus licheniformis | 3 |
| Bacillus amyloliquefaciens | 1 |
| Non-Bacillus species | |
| Staphylococcus aureus | 10 |
| Staphylococcus equorum | 1 |
| Clostridium perfringens | 3 |
| Listeria monocytogenes | 6 |
| Campylobacter sp. | 3 |
| Escherichia coli (including serovar O157) | 4 |
| Salmonella sp. | 6 |
| Yersinia enterocolitica | 7 |
Including 30 cereulide-producing isolates identified by the HEp-2 cytotoxicity assay and LC-MS analysis.
TABLE 2.
Origins of cereulide-producing B. cereus strains
| Strain(s)a | Origin |
|---|---|
| WSBC 10879, WSBC 10880 | Rice |
| MHI 87, MHI 135, MHI 294,M UHDAM IIFI (1), UHDAM IIFI(2), UHDAM IIFI(3) | Baby food |
| F3080B/87, F3350/87, F3605/73, F3752A/86, F3942/87, F4108/89, F4426/94, | Food remnants associated with emetic outbreaks |
| F47/94, F5881/94, F6921/94, MHI 280, MHI 297, MHI 1305 | |
| F3351/87, F3876/87 | Human feces (patient) |
| F4552/75, F4810/72 (SMR-178), NC7401 | Vomit (patient) |
| UHDAM IH41385 | Dialysis liquid |
| UHDAM NS58, UHDAM NS88, UHDAM NS115 | Spruce tree |
MHI, B. cereus culture collection at the Institute of Hygiene and Technology of Food of Animal Origin, Ludwig-Maximilians-Universität München. Munich, Germany; NC, Nagoya City Public Health Research Institute, Nagoya, Japan; UHDAM, Department of Applied Chemistry and Microbiology, University of Helsinki, Helsinki, Finland; WSBC, Weihenstephan B. cereus culture collection at the Department of Biosciences, Technical University of Munich, Munich, Germany. F strains were obtained from the Public Health Laboratory Service, London, United Kingdom.
TABLE 3.
Plasmids used in this study
| Plasmid | Description | Use | Source |
|---|---|---|---|
| pMK4 | B. subtilis-E. coli shuttle vector; Cmr | Testing of transformation efficiency | Sullivan et al.a |
| pCR2.1-TOPO | Topoisomerase I-activated cloning vector with 3′ overhangs; Kanr Ampr | Subcloning and construction of nonreplicable plasmids for insertion mutagenesis | Invitrogen |
| pME3 | pCR2.1-TOPO containing a 0.7-kb putative NRPS fragment | This study | |
| pME11 | pCR2.1-TOPO containing a 0.7-kb ces PCR fragment | This study | |
| pMEC | pCR2.1-TOPO containing a 1.3-kb ces PCR fragment | Insertion mutagenesis | This study |
See reference 42.
Measurement of toxicity and LC-MS analysis of cereulide-producing strains.
Emetic toxin production by B. cereus strains included in this study was determined by a modification of the cytotoxicity assay described previously (12). In brief, B. cereus isolates were grown in 20 ml of skim milk medium for 18 h, and after autoclaving, an aliquot of the preparation was serially diluted (twofold) in 96-well plates by using Earle’s minimal essential cell culture medium supplemented with 1% fetal calf serum, 1% (vol/vol) sodium pyruvate (100 mmol/liter), 2% (vol/vol) l-glutamine (200 mmol/liter), 0.2% (vol/vol) penicillin-streptomycin (10,000 U/ml), and 2% ethanol as a diluent. Immediately after this, HEp-2 cells (0.15 ml; 105 cells per well) were added, and the plates were incubated for 48 h at 37°C in a 5% CO2 atmosphere. Toxicity titers were determined by using the cell proliferation reagent WST-1 (Roche Diagnostics) essentially as described by Dietrich et al. (9). In addition, all strains were tested by using the boar semen assay as a second bioassay (5). Cereulide production by all strains that were positive in the cytotoxicity assay was confirmed by liquid chromatography (LC)-ion trap mass spectrometry (MS) as described previously (18).
Isolation of DNA.
DNA from gram-positive bacteria was prepared by using an AquaPure genomic DNA isolation kit (Bio-Rad, Munich, Germany). DNA from gram-negative bacteria was prepared by suspending cells from one colony in sterile water. The suspension was heated at 95°C for 3 min and then placed on ice. After centrifugation the supernatant was used as the template for PCR or stored at −20°C. For inverse PCR and Southern analysis, total chromosomal DNA was isolated by phenol-chloroform extraction as described previously (10). Preparation of plasmid DNA was performed by using standard procedures.
Primer design.
Degenerate oligonucleotide primers targeting highly conserved motifs of known nonribosomal peptide synthetases (6, 24) were used to amplify and identify putative NRPS gene fragments in B. cereus. Inverse PCR and module jumping, with a combination of a specific primer located in the known sequence and a degenerate primer targeting conserved motifs in the flanking regions (for details concerning the latter technique see reference 29), were used to obtain sequence information from the flanking regions of the DNA fragments. Based on the sequence information derived from the amplified DNA fragments, specific primers and probes for detection of emetic B. cereus strains were designed. Sequence information for all primers used in this study (except primers used for inverse PCR and control PCR) is shown in Table 4.
TABLE 4.
Oligonucleotide primers used in this studya
| Primer | Sequence (5′-3′) | Use | Source |
|---|---|---|---|
| PSF | GG(AT)C(AGT)AC(ACT)GG(ACT)(AC)A(AGCT)CC(ACT)AA(AG)GG | Degenerate PCR (A domain) | Carnio et al.b |
| PSR2 | GGCA(GT)CCAT(CT)T(CT)GCCA(AG)GTC(AGCT)CC(GT)GT | Degenerate PCR (A domain) | Carnio et al.b |
| F_C3 | GCA(CT)CA(CT)AT(ACT)AT(ACT)TC(AGCT)GA(CT)GG(AGCT)TGG | Module jumping (C-A domain) | This study |
| R_T1 | C(AGT)A(GT)(AGT)A(AG)(AT)GA(AG)TG(ACT)CC(AC)CC | Module jumping (A-T domain) | This study |
| F_Vall | GAACCTTGAACAATTAACAGAAG | Module jumping (A-T domain) | This study |
| CesF1 | GGTGACACATTATCATATAAGGTG | Molecular diversity; cereulide-specific PCR assay | This study |
| CesR1 | GTTTTCTGGTAACAGCGTTTCTAC | Molecular diversity | This study |
| CesR2 | GTAAGCGAACCTGTCTGTAACAACA | Module jumping (C-A domain); cereulide-specific PCR assay | This study |
| 8-26/56 | AGAGTTTGATCCTGGCTCA | 16S rRNA gene amplification (positive control) | Stackebrandt and Liesackc |
| 1511-1493 | CGGCTACCTTGTTACGAC | 16S rRNA gene amplification (positive control) | Stackebrandt and Liesackc |
| CF1 | CCAATTTTCCAAGTGATGATGGG | Probe for hybridization (C domain) | This study |
| CR1 | CAATAATTGTTTCAGGCGAACGCA | Probe for hybridization (C domain) | This study |
| AF1 | TGTTGTTACAGACAGGTTCGCTTAC | Probe for hybridization (A domain) | This study |
| AR1 | GTTCCAATCGGAATGCTGTCTTG | Probe for hybridization (A domain) | This study |
Cloning of PCR amplicons and DNA sequencing.
Amplification products obtained from PCR performed with degenerate primers and amplicons obtained from module jumping were subcloned in the TOPO TA vector (Invitrogen, Karlsruhe, Germany) and sequenced. Primers CesF1 and CesR1 were used to amplify a 2.2-kb DNA fragment from nine cereulide-producing B. cereus strains in order to study the molecular diversity of the ces (cereulide synthetase) gene. PCR amplicons were purified with a QIAquick PCR purification kit (QIAGEN) and were directly sequenced (Sequiserve, Ebersberg, Germany) by using a DNA DyeDeoxy terminator cycle sequencing kit (Applied Biosystems). PCR products obtained by inverse PCR were either subcloned in TOPO TA or directly sequenced.
Sequence analysis.
The resulting sequences were searched against the sequenced genomes of B. cereus strains ATCC 14579T and ATCC 10987, Bacillus anthracis strains A2012, Ames, Kruger B, and Western NA, Bacillus halodurans, and Bacillus subtilis and against the National Center for Biotechnology Information (NCBI) nonredundant protein database by using the Basic Local Alignment Search Tool (BLAST). All sequence similarity searches were performed by using BLASTX and BLASTP at the NCBI website (3, 4). The sequencing analysis software package Vector NTI (Informax Inc.) was used to generate the contig sequence from the sequenced PCR products obtained by inverse PCR and module jumping. The software packages Clustal X and TREECON were used for sequence alignment and cluster analysis (43, 47).
Southern analysis.
Chromosomal DNA was digested with different restriction enzymes. The fragments were separated on a 1% agarose gel and blotted onto nitrocellulose. Southern analysis (37) was performed by using digoxigenin-labeled probes (Roche, Mannheim, Germany) directed against two different domains of ces. The probes were obtained from the emetic reference strain F4810/72 by PCR by using the oligonucleotide primers described in Table 4.
Preparation of electrocompetent cells and insertion mutagenesis.
The electrocompetence of emetic B. cereus strains was tested by using replicative plasmid pMK4 (42). Sixteen cereulide-producing B. cereus strains were cultivated overnight in LB broth at 30°C and 180 rpm. One milliliter of an overnight culture was transferred into 100 ml of LB medium (with 2% [wt/wt] glycine) and incubated at 30°C and 180 rpm until the optical density at 600 nm was 0.4 to 0.7. Cells were harvested by centrifugation, and the pellets were washed with increasing concentrations of ice-cold glycerol (2.5, 5, and 10%), resuspended in precooled electroporation buffer (10% glycerol), and shock frozen in liquid nitrogen. Electroporation was carried out by using a Bio-Rad Gene Pulser with a Pulse Controller (Bio-Rad). Electrocompetent cell suspensions, including the plasmid DNA, were transferred into precooled electroporation cuvettes (Ecu-102; PeqLab Biotechnologie, Erlangen, Germany) and exposed to an electric pulse (voltage, 2.0 kV; initial field strength, 5,000 V/cm; capacitance, 25 μF; resistance, 200 Ω). Cells were overlaid with LB medium and incubated at 30°C and 160 rpm for 2 h. Transformants were selected on LB agar supplemented with chloramphenicol (5 μg/ml). For insertion mutagenesis, amplicons obtained from degenerative PCR and module jumping were subcloned into pCR2.1-TOPO, and the resulting nonreplicative plasmids pME11 and pMEC10 were sequenced. B. cereus strain F4810/72 was transformed with these disruption plasmids as described above, and integration of the plasmids into the chromosome was forced by selection for antibiotic-resistant mutants on LB agar supplemented with kanamycin (50 μg/ml). Successful recombination was checked by PCR by using specific primers for the kan gene and additional primers located in the flanking region of the target DNA.
ces gene-specific PCR.
Each PCR mixture (50 μl) contained each deoxynucleoside triphosphate at a concentration of 0.8 mM, 1.5 mM MgCl2, 50 pM oligonucleotide primer CesF1(Table 4), 50 pM oligonucleotide primer CesR2 (Table 4), 1.25 U of ThermoStart Taq DNA polymerase (ABgene, Epsom, United Kingdom), 5 μl of 10× polymerase buffer (ABgene), and 1 μl of template DNA. The PCR protocol started with a denaturation step consisting of 15 min at 95°C; this was followed by five cycles of 1 min at 95°C, 75 s at 53°C, and 50 s at 72°C, 25 cycles of 1 min at 95°C, 75 s at 58°C, and 50 s at 72°C, and finally an elongation step consisting of 72°C for 5 min. All strains used for evaluation of the assay were checked with the CesF1-CesR2 primer set. Selected strains were amplified in parallel with 16S rRNA primers (41) as a positive control.
Nucleotide sequence accession number.
The nucleotide sequence (5.2 kb) of B. cereus F4810/72 described in this paper has been deposited in the GenBank database under accession no. AY691650.
RESULTS
Identification of putative peptide synthetase genes in B. cereus.
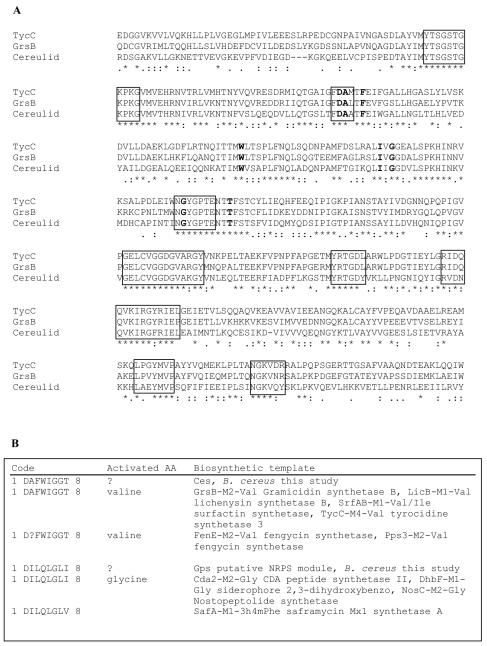
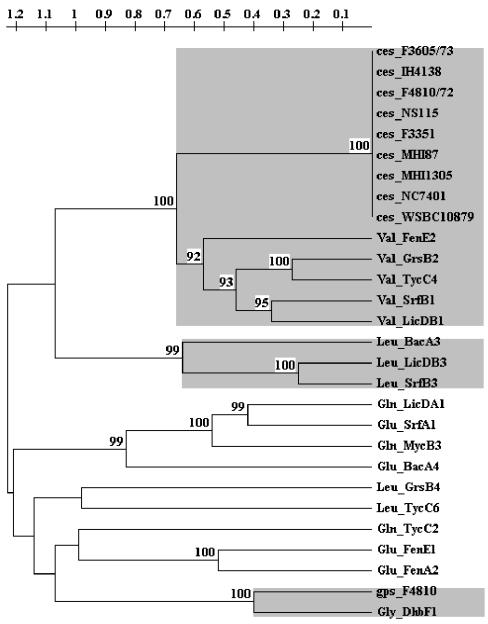
To identify the genetic locus responsible for cereulide production in B. cereus, degenerate PCR primers targeted to conserved sequence motifs of known NRPS genes were used with chromosomal DNA derived from emetic and nonemetic strains. The use of degenerate primers targeting the A3 and A7 core motifs resulted in products of the expected size (700 bp) in all strains tested (data not shown). The 700-bp gene fragments amplified from the cereulide-producing B. cereus strain F4810/72 were subcloned in pCR 2.1 TOPO TA (Invitrogen) and sequenced. The resulting sequences were searched against NCBI's nonredundant database by using the BLASTX or BLASTP algorithms (3). One of the 20 clones analyzed (pME11) showed high homology to the valine activation domain of gramicidin S synthetase (Swissprot database accession no. P14688; 56% identity, 126 of 222 aligned amino acids) and tyrocidine synthetase (Swissprot database accession no. 030409; 57% identity, 127 of 222 aligned amino acids) from Bacillus brevis (28, 46) (Fig. 1). Two other clones (pME3 and pME13) with identical sequences showed significant homology to a glycine-activating module from DhbF (Fig. 2) encoded by a gene belonging to the NRPS complex responsible for catecholic siderophore itoic acid production (EMBL accession no. AF184977; 69% identity, 154 of 223 aligned amino acids) in B. subtilis (25). The deduced amino acid sequences of pME11 and pME3 were used to predict their substrate specificities by the method of Stachelhaus et al. (38) and Challis et al. (7). The primary sequences of the Phe-activating A domain of gramicidin S and the translated sequences of pME3 and pME11 were aligned by using ClustalX (43), and the signature amino acids conferring substrate recognition were compared to a database of 198 known NRPS adenylation domains whose substrate specificity has been determined experimentally (http://raynam.chm.jhu.edu/∼nrps). The eight amino acids lining the putative binding pocket of the A domain from pME11 were identical to the residues of the valine-activating module of peptide synthetases in B. brevis (GrsB and TycC) (28, 46), Bacillus licheniformis (LicB) (21), and B. subtilis (SrfAB) (8), while the eight residues predicted from pME3 were identical to the residues of the glycine-activating module of DhbF of B. subtilis (25) (Fig. 1). A comparison of the sequence of pME11 with the sequenced genome of B. cereus ATCC 14597T yielded no homologous genes, but sequences of several other NRPS modules, including a putative glycine-activating module, were recognized (data not shown). Alignment and cluster analysis of A domains from known Bacillus NRPS genes and translated sequences from pME3 and pME11 derived from B. cereus F4810/72 showed that the A domains cluster according to their predicted substrate specificities (Fig. 2).
FIG. 1.
Comparison of putative NRPS of cereulide-producing B. cereus F4810/72 with known NRPSS. (A) Alignment of a putative valine-activating A domain detected in cereulide-producing B. cereus F4810/72 by degenerative PCR with the A domains of valine-activating modules from GrsB and TycA of B. brevis (28, 46). Conserved core motifs are indicated by boxes, and amino acids that bind specific positions are indicated by boldface type. (B) The selectivity-conferring code for the A domain (38) was used to identify the eight specific signature amino acids of the potential NRPS in cereulide-forming B. cereus. The signature sequences of pME11 (designated Ces) and pME3 (designated Gps) were compared to database sequences of known NRPSs (7). AA, amino acid.
FIG. 2.
Cluster analysis of nonribosomal peptide synthetase modules of Bacillus. Amino acid sequences derived from A domains of known Bacillus NRPSs and putative NRPS modules from F4810/72 obtained by degenerative PCR were aligned, and the regions between the A3 and A7 core motifs, which have been shown to confer substrate specificity (38), were used for cluster analysis. The tree was constructed with TREECON (47) by using the unweighted pair group method with arithmetic averages. All bootstrap values of >80% (100 bootstrap replicates) are shown at the nodes. Abbreviations: Bac, bacitracin synthetase; DhbF, siderophore synthetase; FenE, fengycin synthetase; Grs, gramicidin S synthetase; LicD, lichenysin synthetase; Myc, mycosubtilin synthetase; Srf, surfactin synthetase; Tyc, tyrocidine A synthetase; ces, putative cereulide synthetase; gps, putative NRPS derived from F4810/72 by degenerate PCR.
Sequence analysis of the identified peptide synthetase gene.
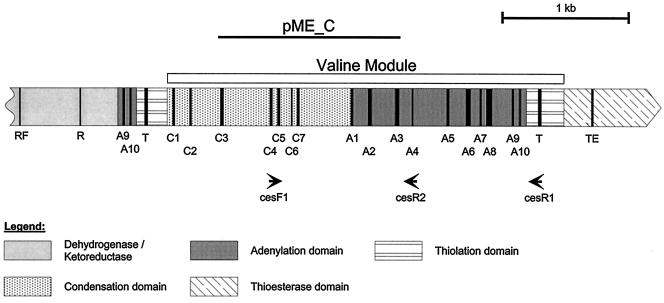
The flanking regions of the DNA fragment from pME11 were sequenced by using a combination of a specific primer located in the known sequence and a degenerate primer located in a conserved domain in the putative flanking regions (module jumping) and by inverse PCR. The resulting 5-kb fragment was sequenced, and the deduced protein sequence was analyzed by using the protein analysis module of the HUSAR Bioinformatics lab at Deutsche Krebsforschungszentrum (http://genome.dfkz-heidelberg.de) and the BLAST conserved motif search tool at NCBI's website (http://www.ncbi.nlm.nih.gov). The corresponding open reading frame that was identified, designated ces (cereulide synthetase), contained a complete putative valine-activating module that had the typical domain organization of NRPS genes (Fig. 3). Strong homology to sequences encoding thioesterases, which are commonly found at the C termini of bacterial NRPS genes, was observed in the C-terminal portion of ces. In the 5′ region of the putative valine activation module the T domain and a predicted A10 core motif of the preceding monomer were found. In addition, sequence motifs typically found in ketoreductase and dehydrogenase genes were detected at the N-terminal end of the ces gene fragment. In addition to the Rossman fold motif (Fig. 3), the typical short-chain dehydrogenase/reductase catalytic Tyr and Ser residues were found (data not shown).
FIG. 3.
Organization and structure of the C-terminal region of cereulide synthetase. The highly conserved core motifs of the peptide synthetase and signature sequences of a short-chain dehydrogenase/reductase (Rossmann fold [RF] motif for cofactor binding and catalytic residues [R] typically found in a short-chain dehydrogenase/reductase [33]) upstream of the valine-activating module are indicated by bars. A1 to A10 and C1 to C5 are highly conserved core sequences in the adenylation domain and the condensation domain, respectively; T indicates the site of 4′-phosphopantetheine binding and substrate acylation located in the thiolation domain; and TE indicates the conserved core motif of thioesterases (for a more detailed description of functional domains see reference 24). pME_C is the DNA fragment used for insertion mutagenesis, and its position is indicated by a bar. The arrows indicate the binding positions of primers used for studying the molecular diversity of the ces gene (CesF1 and CesR1) and in the cereulide-specific PCR assay (CesF1 and CesR2).
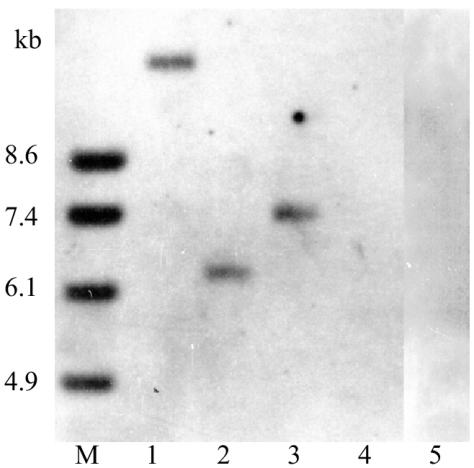
Database searches of sequenced genomes showed that the ces gene fragment is not found in the non-cereulide-producing B. cereus strains ATCC 14579Tand ATCC 10987 and is not present in B. anthracis strains A2012, Ames, Kruger B, and Western NA. Southern analysis of DNA from the cereulide-producing strain F4810/72 that was restricted with different restriction enzymes revealed a single band when a probe that targets the A domain or the C domain of the valine module was used, while no bands were observed for B. cereus ATCC 14579T (Fig. 4) or other non-cereulide-producing B. cereus group strains (data not shown).
FIG. 4.
Southern analysis of chromosomal DNA from the cereulide-producing B. cereus strain F4810/72 and the cereulide-negative strain ATCC 14579T restricted with BamHI (lanes 1 and 4), DraI (lanes 2 and 5), and HpaI (lane 3) and hybridized to a probe from the A domain of the valine module. Lanes 1 to 3, strain F4810/72 (B. cereus emetic reference strain); lanes 4 and 5, strain ATCC 14579T.
Primers CesF1 and CesR1 were designed to amplify a 2.2-kb fragment of the ces peptide synthetase gene of cereulide-producing B. cereus strains (Fig. 3). Direct sequencing of the DNA amplicons of nine different cereulide producers from diverse geographical locations revealed nearly identical sequences of this peptide synthetase for all of the strains sequenced (data not shown). The deduced primary amino acid sequences of all strains were identical, since the only polymorphism detected in the nucleic acid sequence of two strains was located at a wobble position (Fig. 2).
Insertion inactivation of the putative cereulide synthetase gene.
Because not all B. cereus strains are easily transformable, 16 cereulide-producing B. cereus strains were chosen to test their transformability. The food isolate MHI 87 showed the highest transformation efficiency, while the highly toxic food poisoning strains MHI1305, MHI 280, and MHI 297 were not transformable by electroporation, even when different electroporation conditions and different methods for competent cell preparation were used (data not shown). Since the emetic reference strain F4810/72 isolated from a food poisoning case showed a reasonable transformation efficiency, it was chosen for an insertion mutagenesis experiment. A PCR amplicon obtained by module jumping was cloned into TOPO TA, resulting in plasmid pMEC. Since this plasmid is unable to replicate in gram-positive hosts, it was used for insertion mutagenesis (Fig. 3). Mutants were selected on agar plates containing kanamycin, and correct integration of the plasmid in the genome of the antibiotic-resistant cells was demonstrated by PCR (data not shown). Analysis of integrants with the HEp-2 cytotoxicity assay and LC-MS showed that disruption of the C domain yielded a cereulide-deficient phenotype. Wild-type cells were highly toxic (cytotoxicity titer, >160), while the mutant F4810cesCint was nontoxic when it was tested in the HEp-2 cell assay. Quantification of cereulide production by LC-MS revealed a cereulide production level of 0.4 μg mg (wet weight)−1 for wild-type cells, while 0.01 μg of cereulide per mg (wet weight)−1 was measured in the mutant.
Development of a primer system for identification of cereulide-producing B. cereus strains.
The ces gene sequence from F4810/72 was aligned with valine activation modules from B. brevis and predicted NRPS modules from the genomic sequence of B. cereus ATCC 14579T and pME3 in order to design primers specific for the ces gene. These primers were directed to regions of the peptide synthetase gene that did not belong to conserved functional core motifs. The forward primer was located in the C domain, while the reverse primer was located in the A domain. The approximate positions of these primers, which amplified a 1,271-bp DNA product, are shown in Fig. 3.
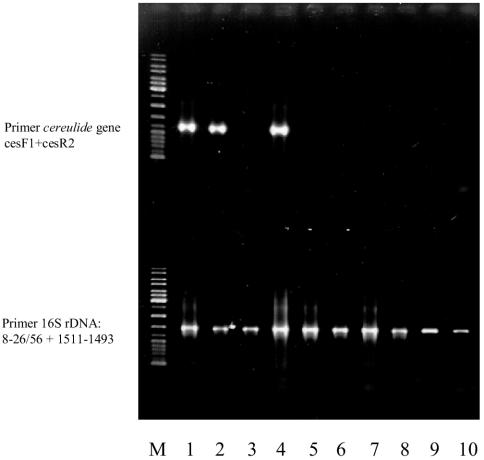
The PCR system developed was tested by using 114 B. cereus isolates from clinical cases, foods, and environments, and cross-reactivity was tested for closely related members of the B. cereus group, as well as for other bacilli and known food pathogens. A total of 143 B. cereus group strains and 40 isolates of other species were included in this survey (Table 1). All 30 cereulide-producing B. cereus isolates (Table 2) were detected by the assay, while no non-cereulide-producing isolates gave any signal in the PCR assay. To check for the absence of PCR-inhibiting substances or any inadequacies of the PCR assay, selected isolates were amplified in parallel with universal primers targeting the 16S rRNA genes. Figure 5 shows the results of PCR amplification of target DNA with the cereulide-specific primers and the results of the control PCR.
FIG. 5.
PCR assay for identification of cereulide-producing B. cereus: gel electrophoresis of PCR products amplified with primers CesF1 and CesR2 (upper portion) and with primers 8-26/56 and 1511-1493 (lower portion) from purified DNA of different food pathogens. Primers CesF1 and CesR2 specifically detect cereulide-producing B. cereus, while primers 8-26/56 and 1511-1493 derived from E. coli (41) target 16S rRNA genes. Lane 1, B. cereus F4810/72 (emetic reference strain derived from vomit of a patient with food poisoning); lane 2, B. cereus MHI 280 (cereulide producer isolated from food remnants connected to food poisoning); lanes 3 and 5, non-cereulide-producing B. cereus food isolates; lane 4, cereulide-producing B. cereus food isolate; lane 6, B. cereus ATCC 14579T; lane 7, B. anthracis Sterne (= CIP7702); lane 8, B. brevis ATCC 9999; lane 9, Staphylococcus aureus WS2604; lane 10, Salmonella enteritidis WS2863; lane M, marker ladder mixture (MBI Fermentas).
DISCUSSION
Identification of the cereulide peptide synthetase (ces) gene.
We identified the genetic locus involved in cereulide synthesis by using a previously described PCR method (45) and degenerate primers derived from conserved core sequences of known NRPSs. One of the subcloned DNA fragments (pME11) obtained by degenerative PCR showed a high level of homology to the valine-activating A domain of gramicidin S. Substrate-binding pocket analysis suggested that the deduced adenylation domain of pME11 indeed activates valine [P = 0.003] (Fig. 1). Southern blot analysis with different restriction enzymes and probes that target the putative valine activation module indicated that only one copy of this module is present in the genome of the cereulide-producing B. cereus strain F4810/72, while this module is not found in the non-cereulide-producing strain ATCC 14579T (Fig. 4) or in other non-cereulide-producing members of the B. cereus group (data not shown). Since the emetic toxin cereulide contains a valine residue, it is tempting to speculate that the genetic locus identified belongs to the gene cluster responsible for cereulide production in B. cereus. In addition, in silico analysis of sequenced genomes of members of the B. cereus group showed that the NRPS gene identified is not found in non-cereulide-producing organisms. Further support for the hypothesis that the NRPS identified contributes to the synthesis of cereulide was provided by insertion mutagenesis experiments. Disruption of the corresponding genetic locus produced cereulide-deficient mutants. Based on these results, we suggest that cereulide is synthesized nonribosomally by the peptide synthetase Ces. Quite recently, Horwood et al. (20) described a small 500-bp gene fragment which was amplified from two emetic B. cereus strains with general NRPS primers and showed sequence homology to the NRPS gene. They suggested that this fragment is associated with cereulide synthesis. However, no substrate specificity was determined for this small DNA fragment, nor have any knockout experiments been performed to provide evidence for their suggestion. A comparison of the ces gene reported here and this 500-bp gene fragment did not support the hypothesis that this DNA fragment has a potential role in cereulide synthesis. In general, many peptide synthetases have been found in the B. cereus genomic sequence (11). In this study, for instance, we identified several putative peptide synthetase gene fragments in emetic B. cereus (e.g., the putative glycine-activating module Gps [Fig. 1B and 2]) which are, based on their predicted substrate specificity, not involved in cereulide synthesis.
Directly downstream of the valine activation module a stretch of 254 amino acids (29 kDa) with sequence similarity to thioesterases was identified, and it is assumed that cereulide synthesis is completed by activity of this domain. In bacteria, release of the completed peptide chain from the peptide synthetase is catalyzed by a thioesterase with a relative molecular mass of 28 to 35 kDa (24, 48), which is in a domain located at the carboxyl terminus of the NRPS. Due to the chemical structure of cereulide ([d-O-Leu-d-Ala-l-O-Val-l-Val-]3) and the colinearity rule (24, 48), an l-O-Val activation module is expected to be located directly upstream of the valine-activating module. In contrast to the valine-activating module that has the typical domain organization of NRPS, the preceding module seems to have a dehydrogenase/ketoreductase domain inserted between the A8 and A9 core motifs. Signature sequences typical of genes encoding short-chain dehydrogenases and ketoreductases (33) were found in the N-terminal region of the ces gene fragment sequenced. Insertions in the A domain between core motifs A8 and A9 have been reported for the EB domain, which is involved in the incorporation of d-2-hydroxyisovaleric acid in enniatin (17), as well as for specialized oxidoreductase domains (for a review see reference 39). Kuse et al. (22) reported that l-leucine and l-valine are the precursors of cereulide. They speculated that O-amino acids are converted into α-amino acids and then reduced to d-O-leucine and l-O-valine or are transaminated to d-alanine. The observed dehydrogenase/ketoreductase catalytic residues in the cereulide synthetase are consistent with this hypothesis. However, sequencing and analysis of the entire NRPS operon(s) are necessary to support this hypothesis and to elucidate the biochemical pathway of cereulide formation in vivo.
ces is highly conserved in cereulide-producing B. cereus strains.
PCR primers derived from the ces gene fragment were designed to study the molecular diversity of the genetic locus responsible for cereulide production in a variety of B. cereus strains. In contrast to B. cereus enterotoxins that show great diversity at the molecular level (16), the partially sequenced cereulide synthetase (approximately 2.2 kb) was highly conserved among 10 emetic strains (Fig. 2). This sequence was not found in the nonemetic strain B. cereus ATCC 14579T (Fig. 4) or in other nonemetic B. cereus group strains tested by Southern blot analysis (data not shown). These results are in accordance with phenotypic and genotypic studies that revealed a close relationship among emetic isolates (10a, 31) while enterotoxin production is commonly found in the different members of the B. cereus group and in other Bacillus species (13, 32, 34). In contrast to many B. cereus strains which possess enterotoxin genes but do not express them (16, 32), all strains tested so far that carry the ces gene, as determined by PCR and Southern analysis, also produce cereulide.
ces-specific PCR assay for identification of cereulide-producing B. cereus strains.
The molecular basis of cereulide synthesis was unknown previously, and the possibilities for fast and reliable detection of cereulide-producing B. cereus strains were therefore limited. Three principal methods for detection of the emetic toxin have been described during the last few years: a cytotoxicity assay, LC-MS analysis, and a sperm-based bioassay (5, 12, 18). These assays are rather difficult to perform on a routine basis and require 1 day to 1 week along with precultivation and laborious sample preparation. Until now, attempts to develop detection systems at an immunological level, such as the systems that are commercially available for B. cereus enterotoxins, failed because of the low antigenic potential of cereulide itself (26). Recently, two PCR systems for detection of emetic strains have been described (10, 20). One assay has been shown to be specific for emetic strains, but the coding potential or the function of the target sequence is unknown, because it did not show any homology to NRPS genes or significant homology to any database sequences (10). The second PCR assay which targets a putative NRPS gene is clearly not specific for cereulide producers, since it amplifies gene fragments from some nonemetic strains too (20). Due to these limitations of the recently described PCR systems, the sequence information for the ces gene was used to develop an improved molecular detection system that (i) directly targets the gene involved in cereulide synthesis and (ii) specifically detects emetic strains.
In conclusion, here we report on a valine module of the peptide synthetase ces gene which is involved in the production of cereulide via the nonribosomal pathway. This finding forms the starting point for a detailed study of the biosynthetic genes responsible for cereulide formation. Based on the unusual structure of the partially characterized l-O-Val activation module, it is expected that it will require quite some biochemical effort to unravel the biochemistry of cereulide synthesis, especially synthesis of the inserted heterocompounds d-O-Leu and l-O-Val.
Acknowledgments
This work was supported by European Commission grant QLK1-CT-2001-00854.
The technical assistance of Agnes Bodor and the help of Natasa Anastasov with the Southern blot analysis are appreciated.
REFERENCES
- 1.Agata, N., M. Mori, M. Ohta, S. Suwan, I. Ohtani, and M. Isobe. 1994. A novel dodecadepsipeptide, cereulide, isolated from Bacillus cereus causes vacuole formation in HEp-2 cells. FEMS Microbiol. Lett. 121:31-34. [DOI] [PubMed] [Google Scholar]
- 2.Agata, N., M. Ohta, M. Mori, and M. Isobe. 1995. A novel dodecadepsipeptide, cereulide, is an emetic toxin of Bacillus cereus. FEMS Microbiol. Lett. 129:17-20. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Andersson, M., R. Mikkola, J. Helin, M. C. Andersson, and M. Salkinoja-Salonen. 1998. A novel sensitive bioassay for detection of Bacillus cereus emetic toxin and related depsipeptide ionophores. Appl. Environ. Microbiol. 64:1338-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carnio, M., S. T. Stachelhaus, K. P. Francis, and S. Scherer. 2001. Pyridinyl polythiazole class peptide antibiotic micrococcin P1, secreted by foodborne Staphylococcus equorum WS2733, is biosynthesized nonribosomally. Eur. J. Biochem. 268:6390-6400. [DOI] [PubMed] [Google Scholar]
- 7.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 8.Cosmina, P., F. Rodriguez, F. de Ferra, G. Grandi, M. Perego, G. Venema, and D. van Sinderen. 1993. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol. Microbiol. 8:821-831. [DOI] [PubMed] [Google Scholar]
- 9.Dietrich, R., C. Fella, S. Strich, and E. Märtlbauer. 1999. Production and characterization of monoclonal antibodies against the hemolysin BL enterotoxin complex produced by Bacillus cereus. Appl. Environ. Microbiol. 65:4470-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehling-Schulz, M., M. Fricker, and S. Scherer. 2004. Identification of emetic toxin producing Bacillus cereus strains by a novel molecular assay. FEMS Microbiol. Lett. 232:189-195. [DOI] [PubMed] [Google Scholar]
- 10a.Ehling-Schulz, M., B. Svensson, M.-H. Guinebretiere, T. Lindbäck, M. Andersson, A. Schulz, M. Fricker, A. Christiansson, P. E. Granum, E. Märtlbauer, C. Nguyen-The, M. Salkinoja-Salonen, and S. Scherer. 2005. Emetic toxin formation of Bacillus cereus is restricted to a single evolutionary lineage of closely related strains. Microbiology 151:183-197. [DOI] [PubMed] [Google Scholar]
- 11.Emmert, E. A. B., A. K. Klimowicz, M. G. Thomas, and J. Handelsman. 2004. Genetics of zwittermicin A production in Bacillus cereus. Appl. Environ. Microbiol. 70:104-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finlay, W. J., N. A. Logan, and A. D. Sutherland. 1999. Semiautomated metabolic staining assay for Bacillus cereus emetic toxin. Appl. Environ. Microbiol. 65:1811-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaviria Rivera, A. M., P. E. Granum, and F. G. Priest. 2000. Common occurrence of enterotoxin genes and enterotoxicity in Bacillus thuringiensis. FEMS Microbiol. Lett. 190:151-155. [DOI] [PubMed] [Google Scholar]
- 14.Granum, P. E., and T. Lund. 1997. Bacillus cereus enterotoxins. FEMS Microbiol. Lett. 157:223-228. [DOI] [PubMed] [Google Scholar]
- 15.Granum, P. E. 2001. Bacillus cereus, p. 373-381. In M. P. Doyle L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers. ASM Press, Washington, D.C.
- 16.Guinebretiere, M. H., V. Broussolle, and C. Nguyen-The. 2002. Enterotoxigenic profiles of food-poisoning and food-borne Bacillus cereus strains. J. Clin. Microbiol. 40:3053-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haese, A., M. Schubert, M. Herrmann, and R. Zocher. 1993. Molecular characterization of the enniatin synthetase gene encoding a multifunctional enzyme catalysing N-methyldepsipeptide formation in Fusarium scirpi. Mol. Microbiol. 7:905-914. [DOI] [PubMed] [Google Scholar]
- 18.Häggblom, M. M., C. Apetroaie, M. A. Andersson, and M. S. Salkinoja-Salonen. 2002. Quantitative analysis of cereulide, the emetic toxin of Bacillus cereus, produced under various conditions. Appl. Environ. Microbiol. 68:2479-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen, B. M., and N. B. Hendriksen. 2001. Detection of enterotoxic Bacillus cereus and Bacillus thuringiensis strains by PCR analysis. Appl. Environ. Microbiol. 67:185-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horwood, P. F., G. W. Burgess, and H. J. Oakey. 2004. Evidence for nonribosomal peptide synthetase production of cereulide (the emetic toxin) in Bacillus cereus. FEMS Microbiol. Lett. 232:319-324. [DOI] [PubMed] [Google Scholar]
- 21.Konz, D., S. Doekel, and M. A. Marahiel. 1999. Molecular and biochemical characterization of the protein template controlling biosynthesis of the lipopeptide lichenysin. J. Bacteriol. 181:133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuse, M., T. Franz, K. Koga, S. Suwan, M. Isobe, N. Agata, and M. Ohta. 2000. High incorporation of l-amino acids to cereulide, an emetic toxin from Bacillus cereus. Bioorg. Med. Chem. Lett. 10:735-739. [DOI] [PubMed] [Google Scholar]
- 23.Mahler, H., A. Pasi, J. M. Kramer, P. Schulte, A. C. Scoging, W. Bär, and S. Krähenbühl. 1997. Fulminant liver failure in association with the emetic toxin of Bacillus cereus. N. Engl. J. Med. 336:1142-1148. [DOI] [PubMed] [Google Scholar]
- 24.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651-2673. [DOI] [PubMed] [Google Scholar]
- 25.May, J. J., T. M. Wendrich, and M. A. Marahiel. 2001. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 276:7209-7217. [DOI] [PubMed] [Google Scholar]
- 26.Melling, J., and B. J. Chapel. 1987. Characteristics of Bacillus cereus emetic toxin. FEMS Microbiol. Lett. 4:133-135. [Google Scholar]
- 27.Mikkola, R., N. E. Saris, P. A. Grigoriev, M. A. Andersson, and M. S. Salkinoja-Salonen. 1999. Ionophoretic properties and mitochondrial effects of cereulide: the emetic toxin of B. cereus. Eur. J. Biochem. 263:112-117. [DOI] [PubMed] [Google Scholar]
- 28.Mootz, H. D., M. A. Marahiel. 1997. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J. Bacteriol. 179:6843-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neilan, B. A., E. Dittmann, L. Rouhiainen, R. A. Bass, V. Schaub, K. Sivonen, and T. Börner. 1999. Nonribosomal peptide synthesis and toxigenicity of cyanobacteria. J. Bacteriol. 181:4089-4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paananen, A., R. Mikkola, T. Sareneva, S. Matikainen, M. Hess, M. Andersson, I. Julkunen, M. S. Salkinoja-Salonen, and T. Timonen. 2002. Inhibition of human natural killer cell activity by cereulide, an emetic toxin from Bacillus cereus. Clin. Exp. Immunol. 129:420-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pirttijärvi, T. S., M. A. Andersson, A. C. Scoging, and M. S. Salkinoja-Salonen. 1999. Evaluation of methods for recognizing strains of the Bacillus cereus group with food poisoning potential among industrial and environmental contaminants. Syst. Appl. Microbiol. 22:133-144. [DOI] [PubMed] [Google Scholar]
- 32.Prüß, B. M., R. Dietrich, B. Nibler, E. Märtlbauer, and S. Scherer. 1999. The hemolytic enterotoxin HBL is broadly distributed among species of the Bacillus cereus group. Appl. Environ. Microbiol. 65:5436-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reid, R., M. Piagentini, E. Rodriguez, G. Ashley, N. Viswanathan, J. Carney, D. V. Santi, C. R. Hutchinson, and R. McDaniel. 2003. A model of structure for ketoreductase domains in modular polyketide synthases. Biochemistry 42:72-79. [DOI] [PubMed] [Google Scholar]
- 34.Rowan, N. J., G. Caldow, C. G. Gemmel, and I. S. Hunter. 2003. Production of diarrheal enterotoxins and other potential virulence factors by veterinary isolates of Bacillus species associated with nongastrointestinal infections. Appl. Environ. Microbiol. 69:2372-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schulz, A. 2004. Zum Nachweis der emetischen Toxine von Bacillus cereus. Ph.D. dissertation. Ludwig-Maximilians-Universität München, Munich, Germany.
- 36.Shinagawa, K., H. Konuma, H. Sekita, and S. Sugii. 1995. Emesis of rhesus monkeys induced by intragastric administration with the HEp-2 vacuolation factor (cereulide) produced by Bacillus cereus. FEMS Microbiol. Lett. 130:87-90. [DOI] [PubMed] [Google Scholar]
- 37.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:742-744. [DOI] [PubMed] [Google Scholar]
- 38.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 39.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 2002. Nonribosomal assembly of peptide antibiotics on modular protein templates, p. 415-434. In A. L. Sonenshein, J. A. Hoch, and R. M. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 40.Stachelhaus, T., H. D. Mootz, V. Bergendahl, and M. A. Marahiel. 1998. Peptide bond formation in nonribosomal peptide biosynthesis. Catalytic role of the condensation domain. J. Biol. Chem. 273:22773-22781. [DOI] [PubMed] [Google Scholar]
- 41.Stackebrandt, E., and W. Liesack. 1992. The potential of rDNA in identification and diagnostics, p. 232-239. In C. Kessler (ed.), Nonradioactive labeling and detection of biomolecules. Springer, Berlin, Germany.
- 42.Sullivan, M., R. Yasbin, and F. Young. 1984. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene 29:21-26. [DOI] [PubMed] [Google Scholar]
- 43.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trauger, J. W., R. M. Kohli, H. D. Mootz, M. A. Marahiel, and C. T. Walsh. 2000. Peptide cyclization catalysed by the thioesterase domain of tyrocidine synthetase. Nature 407:215-218. [DOI] [PubMed] [Google Scholar]
- 45.Turgay, K., and M. A. Marahiel. 1994. A general approach for identifying and cloning peptide synthetase genes. Pept. Res. 7:238-241. [PubMed] [Google Scholar]
- 46.Turgay, K., M. Krause, and M. A. Marahiel. 1992. Four homologous domains in the primary structure of GrsB are related to domains in a superfamily of adenylate-forming enzymes. Mol. Microbiol. 6:529-546. [DOI] [PubMed] [Google Scholar]
- 47.Van de Peer, Y., and R. De Wachter. 1997. Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput. Appl. Biosci. 13:227-230. [DOI] [PubMed] [Google Scholar]
- 48.Von Döhren, H., U. Keller, J. Vater, and R. Zocher. 1997. Multifunctional peptide synthetases. Chem. Rev. 97:2675-2705. [DOI] [PubMed] [Google Scholar]