Abstract
Signal transducers and activators of transcription (STAT) is a family of transcription factors which regulate cell proliferation, differentiation, apoptosis, metastasis, immune and inflammatory responses, and angiogenesis. STAT3 is a latent cytoplasmic transcription factor that belongs to STATs. STAT3 has been reported be regulates genes involved with cellular growth, proliferation and metastasis. Worldwide, colon cancer is one of the leading causes of cancer-related deaths. Cumulative evidence has established that STAT3 is essential for colon cancer progression to advanced malignancy. In our study, we showed that microRNA-1299 (miR-1299) was closely related to the TNM stage of colon cancer, and that the expression of miR-1299 was negatively correlated with the expression of STAT3 in colon cancer which means that miR-1299 can be a negative regulator of STAT3 in colon cancer. A total of 60 cases of different grades of colon samples were used to detect the expression of miR-1299. Results showed that miR-1299 was significantly lower in high-grade colons both in mRNA and protein levels. Furthermore, Overall survival (OS) in patients with low miR-1299 is shorter than 25.6 months, as compared with an OS of 28.4 months in patients with high level of miR-1299. We also confirmed that the overexpression of miR-1299 can not only downregulate the STAT3 pathway, but also inhibited colon cancer cell growth. Our findings could provide new insights into the molecular therapeutic of colon cancer.
Keywords: miR-1299, STAT3, colon cancer, apoptosis, proliferation
Introduction
As the most common digestive malignancy, colon cancer is the most devastating primary tumor (1). Colon cancer is the third most commonly diagnosed cancers worldwide, and it is the third leading cause of cancer-related death worldwide (2). Despite the great progress including surgery and combined radio and chemotherapy, colon cancer remains a ruinous disease with invariable manifestation of tumor recurrence, the 5-year survival rate of colon cancer is still poor (3). Therefore, researchers have intensive interest in the therapeutic strategies aimed at preventing and delaying the disease.
Tumorigenesis is often initiated by mutations activated oncogenes or inhibited by tumor suppressor genes (4). Clinical data and experimental models have shown that signal transducers and activators of transcription (STAT3) played an important role in colon cancer including cell proliferation and survival, invasion, migration, and angiogenesis (5). STAT3 is known to adjust a number of cytokines such as vascular endothelial growth factor (VEGF) and interleukins (ILs). It has been found that STAT3 can be responsible for cellular transformation, and STAT3 may prevent apoptosis by being a downstream protein of Src (6). STAT3 has various effects in malignant transformation, targeting STAT3 can decrease many types of cells and malignant transformation susceptibility (7). In addition to cellular transformation, STAT3 also participates in cellular survival, proliferation, metastasis and angiogenesis.
MicroRNAs (miRNAs) are a group of non-coding RNAs. miRNAs have been reported to be deregulated in a variety of human cancers, and miRNAs were proved to be involved in several physiological and pathological states. Several miRNAs such as miR-9, miR-31, and miR-182 are known to be deregulated in colon cancer, but the expression and function of miR-1299 in colon cancer are unclear. Akao et al found that when SW480 colon cancer cells were overexpression with miR-143 and miR-145, the cell viability was decreased (8). When cells were transfected with hsa-let7a-1 or miR-126 the cell proliferation was decreased. Studies showed that let-7a-1 can inhibit c-MYC and RAS activation, and miR-126 can inhibit the expression of AKT (9,10). In addition, miRNAs have been found to be related to drug resistance, metastasis and prognosis (11–14). All these studies indicate that miRNAs may become therapeutic targets in colon cancer treatment. miR-1299 has been reported that it could affect PIM1 pathway in PI003-induced apoptosis in HeLa cells, however there are no studies on miR-1299 in colon cancer.
In this study, we detected miR-1299 and STAT3 in 60 cases of colon cancer patients with tumor tissues and adjacent tissues to clarify their relationship. We found that miR-1299 as a negative control of STAT3 had a negative correlation with the TNM grade of colon cancer, and that it could affect the growth of colon cancer cells.
Materials and methods
Patients and tissue samples
Sixty cases of tumor tissues and adjacent tissues were obtained from patients undergoing resection of their tumor in Central Hospital Affiliated to Shenyang Medical College. All patients had a clear histologic diagnosis of colon cancer based on the AJCC. All cases were diagnosed with colon cancer and treated between January 2009 and December 2009 at Central Hospital Affiliated to Shenyang Medical College. All cases were classified as shown in Table I. Each patient provided agreement for use of their tissue for research, and the Institutional Research Medical Ethics Committee of Central Hospital Affiliated to Shenyang Medical College granted approval for this study.
Table I.
The relationship between miR-1299 and colon cancer.
| miR-1299 expression | |||||
|---|---|---|---|---|---|
| Parameters | No. of patients | Low | High | χ2 | P-value |
| Sex | 0.028 | 0.866 | |||
| Male | 29 | 20 | 9 | ||
| Female | 31 | 22 | 9 | ||
| Age (years) | 0.082 | 0.775 | |||
| <40 | 25 | 18 | 7 | ||
| ≥40 | 35 | 24 | 11 | ||
| Size (maximal diameter) | 4.898 | 0.027a | |||
| <5 | 18 | 9 | 9 | ||
| ≥5 | 42 | 33 | 9 | ||
| Family history | 0.003 | 0.955 | |||
| No | 27 | 19 | 8 | ||
| Yes | 33 | 23 | 10 | ||
| TNM grade | 10.051 | 0.018a | |||
| I | 13 | 11 | 2 | ||
| II | 9 | 7 | 2 | ||
| III | 29 | 15 | 14 | ||
| IV | 9 | 9 | 0 | ||
Through the Chi-square test we found little correlation between miR-1299 with the sex, age and family history in the tissues we detected; however, there was correlation between the miR-1299 and the tumor size and TNM stage of colon cancer.
Significant difference.
Cell culture
Two human colon cancer cell lines, HCT116 and SW480, were obtained from American Type Culture Collection and cultured according to their recommendations.
MTT assays
3-(4,5-Dimethylthiazolyl) 2,5 diphenyltetrazolium bromide (MTT) assay was used to analyzed the cell viability. Cells (1×103) were seeded to one well of 96-well plates; 12 h later, the transfection was performed. At different time points, 5 mg/ml MTT was added and incubated at 37°C in a CO2 incubator for 4 h, then medium was replaced by 0.1 ml DMSO and the absorbance was measured spectrophotometrically at 490 nm using Microplate Reader (Bio-Rad).
Hochest 33258 staining
After transfected with different miRNAs, cells were fixed with 4% polyoxymethylene, incubated with 10 µg/ml Hochest 33258. Cells were observed with a fluorescence microscope.
Reverse transcription and quantitative real-time PCR
Total RNA was extracted by TRIzol (Invitrogen) through the protocol. The expression of miR-1299 was detected with Stem-Loop RT-PCR assay as reported (15,16). Primer sequences were synthesized as follows: miR-1299, F: 5′-ACACTCCAGCTGGGTTCTGGAAUUCTC-3′, R: 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCCCTCAC-3′; U6, F: 5′-CTCGCTTCGGCAGCACA-3′; R: 5′-AACGCTTCACGAATTTGCGT-3′; STAT3, F: 5′-GGAACAAGCCCCAACCGG-3′, R: 5′-CTAAAATCAGGGGTCCCAACTG-3′; CDK4, F: 5′-CAGAGCTCTTAGCCGAGCGT-3′, R: 5′-GGCACCGACACCAATTTCAG-3′; CDK6, F: 5′-AGTCTGATTACCTGCTCCGC-3′, R: 5′-CCTCGAAGCGAAGTCCTCAA-3′; Bcl-2, F: 5′-GGTGAACTGGGGGAGGATTG-3′, R: 5′-GGCAGGCATGTTGACTTCAC-3′; Bax, F: 5′-AGCTGAGCGAGTGTCTCAAG-3′, R: 5′-GTCCAATGTCCAGCCCATGA-3′; GAPDH, F: 5′-CTCTGCTCCTCCTGTTCGAC-3′, R: 5′-GCGCCCAATACGACCAAATC-3′. All the reactions were carried out as previously described (17).
Western blot analyses
RIPA lysis buffer were used to split cells and tissues. Sample were subjected to 10% SDS-PAGE and transferred onto a PVDF membrane. Target proteins were probed with specific antibodies, STAT3, bcl-2, bax, CDK4, CDK6, MMP2 and Gapdh (Santa Cruz Biotechnology).
Dual luciferase reporter assay
The STAT3 3′-UTR was PCR amplified and cloned into the pMIR-REPORT™ vector (Ambion). The primers were: STAT3-WT, F: 5′-GGAACAAGCCCCAACCGG-3′, R: 5′-CTAAAATCAGGGGTCCCAACTG-3′; STAT3-DEL, F: 5′-CGAATGGGCCTGACAAT-3′, R: 5′-CTAAAATCAGGGGTCCC-3′. Cells were seeded at a 24-well plate and co-transfected with WT or DEL reporter vector and 10 ng pMIR-REPORT βgal control plasmid in miR-1299 mimic and miR-1299 AS. After 36 h, the luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega).
Immunostaining
All the reactions were carried out as previously described (18). All slices were independently assessed by two experienced pathologists who were blinded to the patient clinicopathology and other information. STAT3 expression level was evaluated via positive staining proportion and intensity of tumor cells. Slices with inconsistent results were re-examined by the original two pathologists and a senior pathologist until a consensus was reached. Sections were immunostained with STAT3 antibody (1:200).
Annexin V-FITC
By Annexin V-FITC apoptosis detection kit instructions determination, the specific steps were as follows: cells were washed twice with cold PBS, then re-suspended with binding buffer at a concentration of 1×106 cells/ml. Adding 5 µl of Annexin V-FITC and 10 µl of PI. Finally, adding 400 µl binding buffer to each tube and the apoptosis rate was measured by flow cytometry within 1 h.
Statistical analysis
All data were analyzed with SPSS17.0 (SPSS Inc.). Difference was analyzed by ANOVA test. All experiments were repeated three times. Group comparison and Chi-square test were used to analyze the results.
Results
Expression of STAT3 and miR-1299 in tumor tissues and adjacent tissues
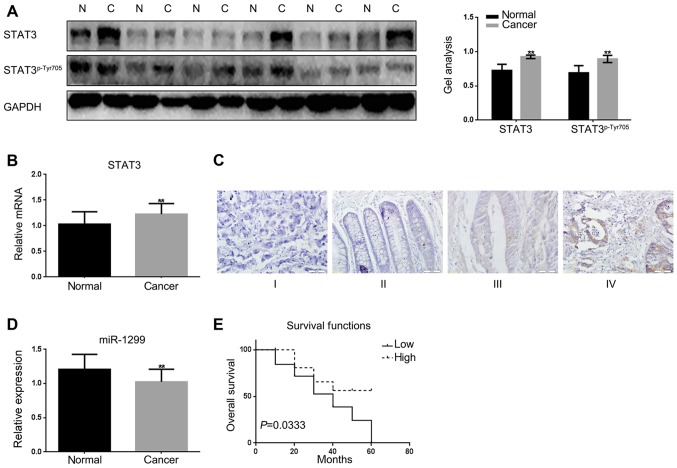
First, the expression levels of STAT3 were detected in colon cancer tissues and adjacent tissues by real-time PCR, western blotting (Fig. 1A and B). The adjacent tissues were used as a control. The results showed that there was a higher expression of STAT3 in colon cancer tissues than the adjacent tissues. Then STAT3 in different stages of colon cancer tissues were detected by immunohistochemical staining. The results showed that STAT3 increased with the increasing of the stage of colon cancer (Fig. 1C). There are few reports on the expression of miR-1299 in cancer; we wondered whether there was a different expression of miR-1299 in colon cancer tissues and adjacent tissues. Real-time PCR was used to test the expression of miR-1299 in colon cancer tissues and adjacent tissues (Fig. 1D). We found that the expression of miR-1299 was significantly downregulated in colon cancer tissues. We analyzed the relationship between miR-1299 and colon cancer in 60 pairs of colon cancer tissues. We found there was little correlation between miR-1299 with the sex, age and family history in the tissues we detected; however there was correlation between the miR-1299 and the tumor size and TNM stage of colon cancer (Table I).
Figure 1.
The expression of STAT3 and miR-1299 in tumor tissues and adjacent tissues. (A) The protein level of STAT3 and STAT3p-Tyr705 in 60 samples of colon cancer tissues and adjacent tissues were detected with western blotting. The expression of STAT3 and STAT3p-Tyr705 were higher in the colon cancer tissues. Data are shown as mean ± SEM. **P<0.01 vs. adjacent tissue group. (B) mRNA expression of STAT3 in 60 samples of colon cancer tissues and adjacent tissues were detected with real-time PCR. The expression of STAT3 was higher in the colon cancer tissues. Data are shown as mean ± SEM. **P<0.01 vs. adjacent tissue group. (C) Immunostaining was used to test the expression of STAT3 in 60 samples of colon cancer tissues and adjacent tissues. The expression of STAT3 was positively correlated with the TNM stage of colon cancer. (D) The levels of miR-1299 were detected by real-time PCR. There was lower expression of miR-1299 in colon cancer tissues. Data are shown as mean ± SEM. **P<0.01 vs. adjacent tissues group. (E) Survival analysis of miR-1299 and colon cancer in 60 cases of colon cancer samples. Results showed that overall survival (OS) in patients with low miR-1299 is 25.6 months, as compared with an OS of 28.4 months in patients with high level of miR-1299. P=0.0333.
miR-1299 inhibits the expression of STAT3 in HCT-116 cells
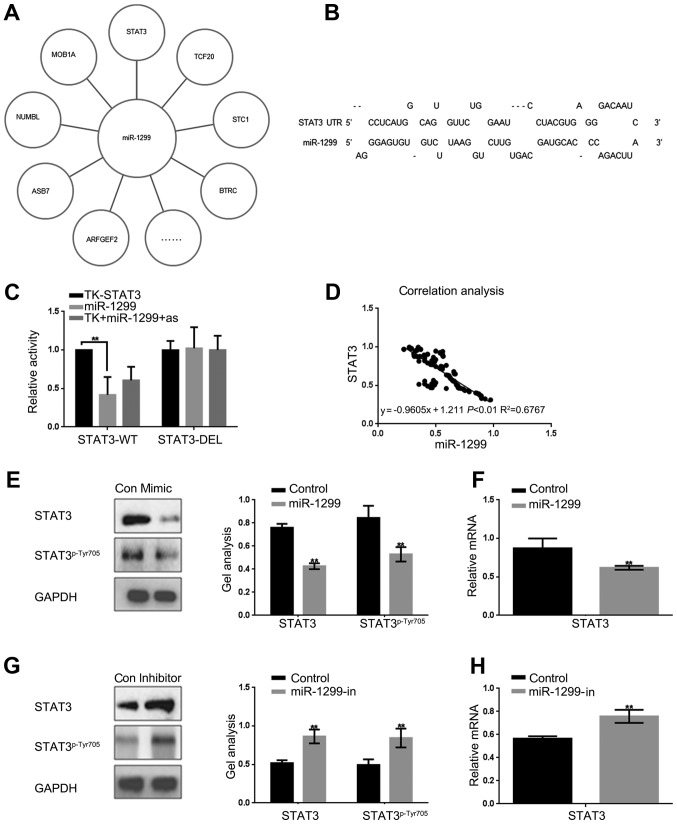
We found there was a variety of proteins could be the target genes of miR-1299 through miRDB tool (Fig. 2A). We evaluated whether there is a correlation between STAT3 and miR-1299 in colon cancer. The miRDB tool showed that miR-1299 could be combined with 3′-UTR of STAT3, which confirmed our conjecture (Fig. 2B). For further confirmation the luciferase reporter assay was carried out to determine whether STAT3 is a potential target gene of miR-1299 (Fig. 2C). The results indicated that miR-1299 inhibited the expression of STAT3 at the gene level. On the other hand, we found that the expression of miR-1299 was negatively correlated with the expression of STAT3 in colon cancer tissue by real-time PCR, which was further evidence that miR-1299 could regulate STAT3 (Fig. 2D). In order to confirm our hypothesis, we overexpressed or inhibited miR-1299 in HCT-116 cells, and then detected the expression of STAT3 by real-time PCR and western blotting. The results show that miR-1299 can negatively regulate the expression of STAT3 in the protein and RNA levels (Fig. 2E-H).
Figure 2.
miR-1299 can inhibit the expression of STAT3 in HCT-116 cells. (A) Site prediction showed that miR-1299 could target at a several of proteins. (B) miRDB predicted that miR-1299 could specifically targeted in STAT3. (C) Luciferase reporter assays of the interaction between miR-1299 and the STAT3 3′-UTR. It was proved that miR-1299 can directly act on STAT3. Data are shown as mean ± SEM. **P<0.01 vs. TK-STAT3 group. (D) The correlation analysis between miR-1299 and STAT3 in colon cancer. The results showed that the expression of mi-1299 was negatively correlated with the expression of STAT3. The formula for that was y= −0.9605x + 1.211 P<0.01 R2=0.6767. (E and F) Western blotting and real-time PCR showed that when the miR-1299 was overexpressed could downregulate the expression of STAT3. Data are shown as mean ± SEM. **P<0.01 vs. control group. (G and H) Western blotting and real-time PCR showed that when the miR-1299 was inhibited the expression of STAT3 was upregulated. Data are shown as mean ± SEM. **P<0.01 vs. control group.
miR-1299 inhibits the proliferation of colon cancer cells
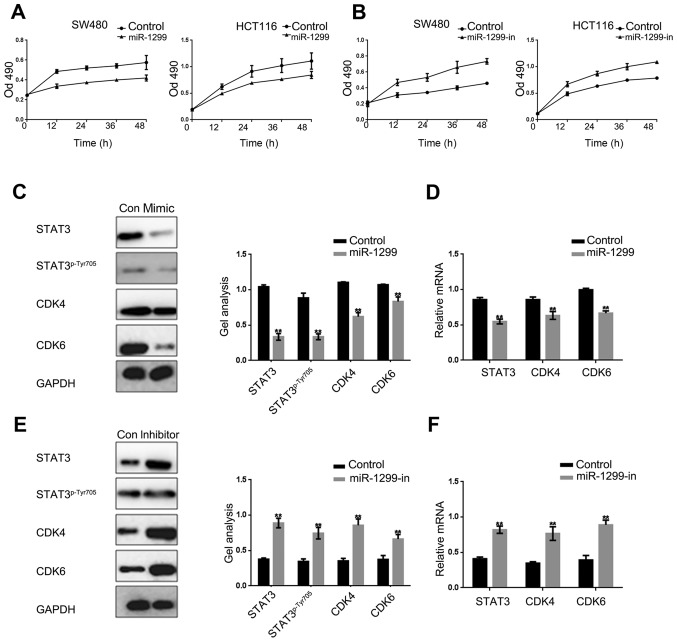
It is well known that STAT3 can regulate cell growth by promoting cell proliferation and inhibiting cell apoptosis. We used MTT assay to examine the effect of miR-1299 to HCT-116 and SW480 cells. With the overexpression or inhibition of miR-1299, we found that miR-1299 can suppress the proliferation of HCT-116 cells and SW480 cells (Fig. 3A and B). Further, we examined the CDK4 and CDK6 downstream proteins of the STAT3 pathway in HCT-116 cells. Results showed that the expression of STAT3, STAT3p-Tyr705, CDK4 and CDK6 were negatively correlated with the expression of miR-1299 (Fig. 3C-F). These results were sufficient to explain that miR-1299 can inhibit cells proliferation by negatively regulating phosphorylation of STAT3, CDK4 and CDK6.
Figure 3.
miR-1299 inhibits the proliferation of colon cancer cells by suppressing the STAT3 pathway. (A) HCT-116 cells and SW480 cells were overexpressed with miR-1299, cell proliferation was detected by MTT assay. Data are shown as mean ± SEM. (B) HCT-116 cells and SW480 cells were downregulated with miR-1299, cell growth was detected by MTT assay. Data are shown as mean ± SEM. (C and D) Western blotting and real-time PCR showed that when the miR-1299 was overexpressed the expression of STAT3, CDK4 and CDK6 were downregulated. Data are shown as mean ± SEM. **P<0.01 vs. control group. (E and F) Western blotting and real-time PCR showed that when the miR-1299 was inhibited the expression of STAT3, CDK4 and CDK6 were upregulated. Data are shown as mean ± SEM. **P<0.01 vs. control group.
miR-1299 promotes apoptosis of colon cancer cells by suppressing the STAT3 pathway
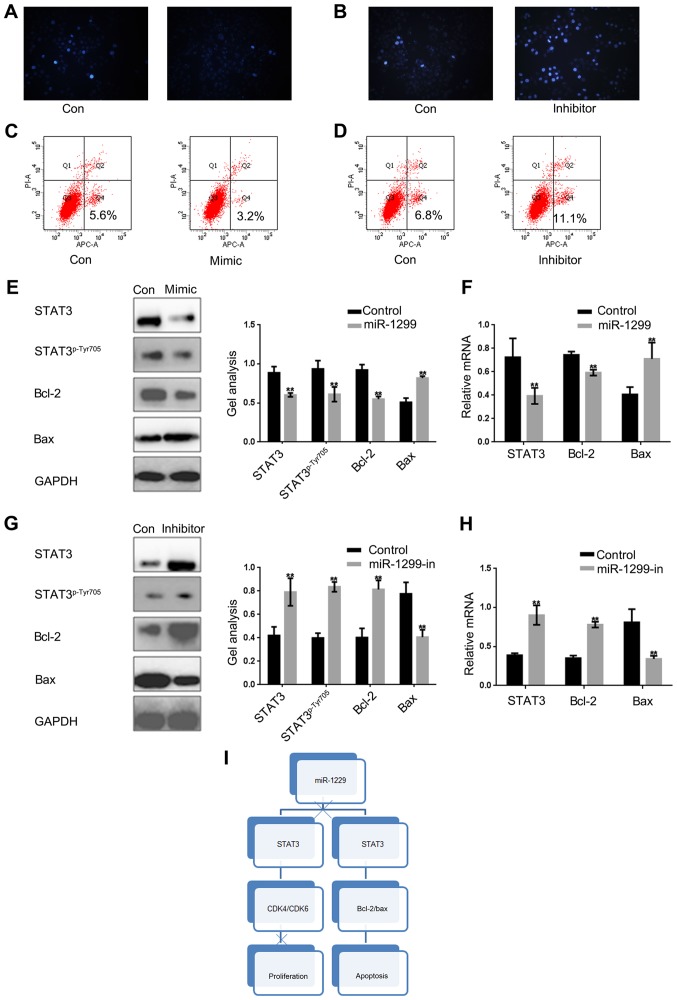
STAT3 has been proved to be able to promote the growth of tumor cells by inhibiting apoptosis. Since miR-1299 can inhibit cell proliferation through negative regulation of STAT3, we investigated whether miR-1299 can affect cell apoptosis by regulating the expression of STAT3. Hochest 33258 staining was used to detect whether mi-1299 can affect cell apoptosis by regulating the STAT3 in HCT-116 cells (Fig. 4A and B). Then by Annexin V-FITC apoptosis detection we obtained similar results (Fig. 4C and D). We found that apoptosis of HCT-116 cells was stimulated when miR-1299 was overexpressed. On the contrary, apoptosis of HCT-116 was obviously decreased when the expression of miR-1299 was inhibited. Moreover, we found that regardless of the protein level or the RNA level, miR-1299 can promote apoptosis of HCT-116 cells by inhibiting the phosphorylation of STAT3 (Fig. 4E-H).
Figure 4.
miR-1299 promotes apoptosis of colon cancer cells by suppressing the STAT3 pathway. (A and B) miR-1299 significantly promoted apoptosis of cells. When HCT-116 cells were overexpressed or inhibited with miR-1299, apoptosis of HCT-116 cells were detected by Hoechst 33258. (C and D) By Annexin V-FITC apoptosis detection, we found that miR-1299 significantly promoted apoptosis of the cells. (E and F) Western blotting and real-time PCR showed that when the miR-1299 was overexpressed the expression of STAT3 and bcl-2 was downregulated, however the expression of bax was upregulated. Data are shown as mean ± SEM. **P<0.01 vs. control group. (G and H) Western blotting and real-time PCR showed that when the miR-1299 was inhibited the expression of STAT3 and bcl-2 were upregulated, however the expression of bax was downregulated. Data are shown as mean ± SEM. **P<0.01 vs. control group. (I) The diagram of this study.
These above results confirmed that miR-1299 could inhibit the growth of colon cancer cells by regulating the expression of STAT3, the phosphorylation level of STAT3 and the expression of its downstream protein.
Discussion
Signal transducers and activators of transcription (STAT) family proteins were found in 1994, they are latent cytoplasmic transcription factors (19). STAT3 as a very important protein in the STAT family is activated by phosphorylation of a tyrosine residue located at position 705. There is convincing evidence to show that STAT3 activation plays a critical role in the occurrence and development of tumors. Cancer cell growth, survival, metastasis, and angiogenesis are closely related to the activation of STAT3. Researchers have shown that STAT3 could be responsible for cellular transformation in NIH3T3 cells and in mouse skin epithelial cells (20). In a number of cell types when STAT3 was inhibited the malignant transformation was decreased. There is also research showing that STAT3 can participate in cellular growth. Sufficient evidence shows that the upregulation of cyclin D1, CDK4, CDK6 and cMyc are associated with the STAT3 pathway (21). STAT3 has also been shown to upregulate the expression of PIM1 (22).
In addition to promoting proliferation, STAT3 can also achieve the function of promoting cell growth by inhibiting apoptosis. The effect of STAT3 on cell apoptosis is through regulating the expression of bcl-2, bax, p53, surviving and clAP2 (23). Evidence has been provided that STAT3 plays a crucial role in cancer metastasis. Several lines of evidence strongly implicate that STAT3 can regulate the MMPs to play a crucial role in cell metastasis (24–27). There are many studies showing that STAT3 can regulate angiogenesis of tumors by adjusting the expression of VEGF and HIF-1α (28–30).
MicroRNAs are proved to play crucial roles in cancer development. There are two main categories of microRNAs that are involved in cancer progression, one can enhance cell growth, survival, and metastasis, and the other suppressed these activities. Several RNA therapeutics, with diverse modes of action, are being evaluated in large late-stage clinical trials, and many more are in early clinical development (31). For example, studies identified that miR-143 and miR-145 as the tumor suppressors were downregulated in colon cancer. miR-1299 was reported downregulated in many cancers and reported to negatively adjust the expression of PIM1 (32). Studies showed that miR-1299 may be involved in the cellular proliferation and apoptosis. However, there are no previous research on the miR-1299 in colon cancer. We were the first to find that miR-1299 can target STAT3 and suppress the expression of STAT3 then inhibiting the STAT3 pathway.
In our study, we found that miR-1299 is closely related to the occurrence and development of colon cancer. It is negatively correlated with the TNM staging of colon cancer, and it is closely related to the prognosis of colon cancer. Furthermore, the expression of miR-1299 and STAT3 in colon carcinoma was negatively correlated. We hypothesized that miR-1299 could affect the occurrence and development of colon cancer by regulating the expression of STAT3. After a series of studies we found that miR-1299 can regulate STAT3. miR-1299 inhibited the proliferation of colon cancer cells by inhibiting the expression of STAT3, and promoted apoptosis of colon cancer cells.
Our findings suggest that overexpression of miR-1299 may be considered as a promising strategy for targeted therapies in colon cancer.
Acknowledgements
This work was supported by grants from the Technological innovation fund of Shenyang Technology Division (no. F15-139-9-07), National Natural Science Foundation of China (no. 81502333) and China Postdoctoral Science Foundation (no. 2016M591437).
References
- 1.Sun KX, Xia HW, Xia RL. Anticancer effect of salidroside on colon cancer through inhibiting JAK2/STAT3 signaling pathway. Int J Clin Exp Pathol. 2015;8:615–621. [PMC free article] [PubMed] [Google Scholar]
- 2.Cekaite L, Rantala JK, Bruun J, Guriby M, Agesen TH, Danielsen SA, Lind GE, Nesbakken A, Kallioniemi O, Lothe RA, et al. MiR-9, −31, and −182 deregulation promote proliferation and tumor cell survival in colon cancer. Neoplasia. 2012;14:868–879. doi: 10.1593/neo.121094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terzić J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114 e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen AV, Wu YY, Liu Q, Wang D, Nguyen S, Loh R, Pang J, Friedman K, Orlofsky A, Augenlicht L, et al. STAT3 in epithelial cells regulates inflammation and tumor progression to malignant state in colon. Neoplasia. 2013;15:998–1008. doi: 10.1593/neo.13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamran MZ, Patil P, Gude RP. Role of STAT3 in cancer metastasis and translational advances. Biomed Res Int. 2013;2013:421821. doi: 10.1155/2013/421821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/MCB.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251:199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 8.Akao Y, Nakagawa Y, Naoe T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep. 2006;16:845–850. [PubMed] [Google Scholar]
- 9.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 10.Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47:939–946. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, et al. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene. 2009;28:4065–4074. doi: 10.1038/onc.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 13.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schimanski CC, Frerichs K, Rahman F, Berger M, Lang H, Galle PR, Moehler M, Gockel I. High miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol. 2009;15:2089–2096. doi: 10.3748/wjg.15.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng J, Wang K, Liu X, Chen S, Chen J. The quantification of tomato microRNAs response to viral infection by stem-loop real-time RT-PCR. Gene. 2009;437:14–21. doi: 10.1016/j.gene.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 16.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel D, Chaudhary J. Increased expression of bHLH transcription factor E2A (TCF3) in prostate cancer promotes proliferation and confers resistance to doxorubicin induced apoptosis. Biochem Biophys Res Commun. 2012;422:146–151. doi: 10.1016/j.bbrc.2012.04.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akira S, Nishio Y, Inoue M, Wang XJ, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 20.Yu CY, Wang L, Khaletskiy A, Farrar WL, Larner A, Colburn NH, Li JJ. STAT3 activation is required for interleukin-6 induced transformation in tumor-promotion sensitive mouse skin epithelial cells. Oncogene. 2002;21:3949–3960. doi: 10.1038/sj.onc.1205499. [DOI] [PubMed] [Google Scholar]
- 21.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/MCB.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirogane T, Fukada T, Muller JM, Shima DT, Hibi M, Hirano T. Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity. 1999;11:709–719. doi: 10.1016/S1074-7613(00)80145-4. [DOI] [PubMed] [Google Scholar]
- 23.Bhattacharya S, Ray RM, Johnson LR. STAT3-mediated transcription of Bcl-2, Mcl-1 and c-IAP2 prevents apoptosis in polyamine-depleted cells. Biochem J. 2005;392:335–344. doi: 10.1042/BJ20050465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Huang C, Huang K, Wu W, Jiang T, Cao J, Feng Z, Qiu Z. STAT3 knockdown reduces pancreatic cancer cell invasiveness and matrix metalloproteinase-7 expression in nude mice. PLoS One. 2011;6:e25941. doi: 10.1371/journal.pone.0025941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie TX, Wei D, Liu M, Gao AC, Ali-Osman F, Sawaya R, Huang S. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene. 2004;23:3550–3560. doi: 10.1038/sj.onc.1207383. [DOI] [PubMed] [Google Scholar]
- 26.Dechow TN, Pedranzini L, Leitch A, Leslie K, Gerald WL, Linkov I, Bromberg JF. Requirement of matrix metalloproteinase-9 for the transformation of human mammary epithelial cells by Stat3-C; Proc Natl Acad Sci USA; 2004; pp. 10602–10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itoh M, Murata T, Suzuki T, Shindoh M, Nakajima K, Imai K, Yoshida K. Requirement of STAT3 activation for maximal collagenase-1 (MMP-1) induction by epidermal growth factor and malignant characteristics in T24 bladder cancer cells. Oncogene. 2006;25:1195–1204. doi: 10.1038/sj.onc.1209149. [DOI] [PubMed] [Google Scholar]
- 28.Chen Z, Han ZC. STAT3: A critical transcription activator in angiogenesis. Med Res Rev. 2008;28:185–200. doi: 10.1002/med.20101. [DOI] [PubMed] [Google Scholar]
- 29.Xu Q, Briggs J, Park S, Niu G, Kortylewski M, Zhang S, Gritsko T, Turkson J, Kay H, Semenza GL, et al. Targeting Stat3 blocks both HIF-1 and VEGF expression induced by multiple oncogenic growth signaling pathways. Oncogene. 2005;24:5552–5560. doi: 10.1038/sj.onc.1208719. [DOI] [PubMed] [Google Scholar]
- 30.Oh MK, Park HJ, Kim NH, Park SJ, Park IY, Kim IS. Hypoxia-inducible factor-1alpha enhances haptoglobin gene expression by improving binding of STAT3 to the promoter. J Biol Chem. 2011;286:8857–8865. doi: 10.1074/jbc.M110.150557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullenger BA, Nair S. From the RNA world to the clinic. Science. 2016;352:1417–1420. doi: 10.1126/science.aad8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z, He W, Gao J, Luo J, Huang X, Gao C. Computational prediction and experimental validation of a novel synthesized pan-PIM inhibitor PI003 and its apoptosis-inducing mechanisms in cervical cancer. Oncotarget. 2015;6:8019–8035. doi: 10.18632/oncotarget.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]