Abstract
Spindle cell tumors are clinically heterogeneous but morphologically similar neoplasms. The term refers to the tumor cells' long and slender microscopic appearance. Distinct subgroups of spindle cell tumors are characterized by chromosomal translocations and also fusion genes. Other spindle cell tumors exist that have not yet been found to have characteristic, let alone pathognomonic, genetic or pathogenetic features. Continuous examination of spindle cell tumors is likely to reveal other subgroups that may, in the future, be seen to correspond to meaningful clinical differences and may even be therapeutically decisive. We analyzed genetically a pediatric spindle cell tumor. Karyotyping showed the tumor cells to carry a t(3;17)(p21;q12) chromosomal translocation whereas RNA sequencing identified a SETD2-NF1 fusion gene caused by the translocation. RT-PCR together with Sanger sequencing verified the presence of the above-mentioned fusion transcript. Interphase FISH analysis confirmed the existence of the chimeric gene and showed that there was no reciprocal fusion. The fusion transcript codes for a protein in which the last 114 amino acids of SETD2, i.e., the entire Set2 Rpb1 interacting (SRI) domain of SETD2, are replaced by 30 amino acids encoded by the NF1 sequence. The result would be similar to that seen with truncating SETD2 mutations in leukemias. Absence of the SRI domain would result in inability to recruit SETD2 to its target gene locus through binding to the phosphor-C-terminal repeat domain of elongating RNA polymerase II and may affect H3K36 methylation. Alternatively, loss of one of two functional SETD2 alleles might be the crucial tumorigenic factor.
Keywords: pediatric spindle cell tumor, RNA-sequencing, cytogenetics, fusion gene, SETD2, NF1
Introduction
Spindle cell tumors are clinically heterogeneous but morphologically similar neoplasms that can occur anywhere. The term is descriptive and based on the tumor cells' long and slender microscopic appearance (https://librepathology.org/wiki/Spindle_cell_lesions). The diagnosis of spindle cell tumors relies on histological and morphological features supported by ancillary investigations which include immunohistochemistry, cytogenetics, fluorescence in situ hybridization (FISH), and/or molecular genetics. The diagnosis is prognostically imprecise and even sometimes fails to distinguish benign from low-grade malignant tumors (1,2).
Cytogenetic and molecular genetic analyses of spindle cell tumors have led to the recognition of several distinct karyotypic entities, presumably corresponding to equally distinct pathogenetic subgroups, characterized by chromosomal translocations and also fusion genes that identify specific tumor types (3). For example, congenital fibrosarcomas carry the translocation t(12;15)(p13;q25) which results in the generation of an ETV6-NTRK3 fusion gene (4). Dermatofibrosarcoma protuberans, another subtype of spindle cell sarcoma, is characterized cytogenetically by supernumerary ring chromosomes or the translocation t(17;22)(q22;q13) (5). Either change results in formation of a COL1A1-PDGFB fusion gene in which PDGFB exon 1 is deleted and replaced by a variable segment of the COL1A1 gene (5). A subset of inflammatory myofibroblastic tumor, a neoplasm composed of myofibroblastic spindle cells and infiltrating inflammatory cells, harbor clonal chromosomal rearrangements of chromosome band 2p23 (6). These rearrangements target the ALK gene which may serve as the 3′-partner in fusions with various translocation partners bringing about ALK tyrosine kinase activation (6).
Solitary fibrous tumor, another rare spindle cell tumor, is now defined genetically as carrying a submicroscopic inversion of the long arm of chromosome 12 (12q13) resulting in fusion of the two neighboring genes NAB2 and STAT6 (7–10). This creates a chimeric transcription factor in which the NAB2 repressor domain is substituted by a carboxyl-terminal STAT6 transactivation domain or near-full-length STAT6 (7–10).
In spite of all these genetic-pathologic correlations, other spindle cell tumors exist that have not yet been found to have characteristic, let alone pathognomonic, genetic or pathogenetic features. By way of example, Fruth et al (11) reported a laryngeal spindle cell sarcoma which did not fit into any of the existing spindle cell sarcoma sub-entities: The initially benign-appearing mesenchymal tumor first changed its clinical phenotype without corresponding histological signs of malignancy but later assumed more aggressive histological features. Alaggio et al (12) described two spindle cell tumors with EWSR1-WT1 fusion and favorable prognosis. According to the authors, the tumors could represent ‘an unrecognized subgroup of tumors with spindle cell morphology, bearing the same translocation as desmoplastic small round cell tumor, but characterized by a more favorable clinical course’. In a previous study of ours, we described a spindle cell sarcoma that could not be further sub-classified, but which carried a ring chromosome composed of chromosome 12 material, several fusion genes mapping to 12q, and amplification of MDM2 (13). Nord et al (14) reported a spindle cell sarcoma of the heart with a ring chromosome, amplification of the MDM2 gene, and homozygous deletion of CDKN2A. Finally, Lestou et al (15) reported a case of spindle cell sarcoma in the lower abdominal wall with a complex karyotype, ring chromosomes, amplification of chromosome 18, and co-amplification of 12p11 and 12q13-q22 in the ring chromosomes. The examples above show that continuous examination of tumors with spindle cell morphology is likely to reveal yet other genetic subgroups that may, in the future, be seen to correspond to meaningful clinical and even, when suitable therapeutics are construed against the pathogenetic mechanisms involved, be also therapeutically decisive.
In the present study, we analyzed genetically a pediatric spindle cell tumor. The cytogenetic analysis showed that the tumor cells carried a t(3;17)(p21;q12) chromosomal translocation and RNA sequencing identified a SETD2-NF1 fusion gene caused by the translocation.
Materials and methods
Ethics statement
The study was approved by the Regional Committee for Medical and Health Research Ethics, South-East Norway (REK sør-øst; http://helseforskning.etikkom.no) and written informed consent was obtained from the patient for publication of the case details. The ethics committee's approval included a review of the consent procedure. All patient information has been de-identified.
Case history
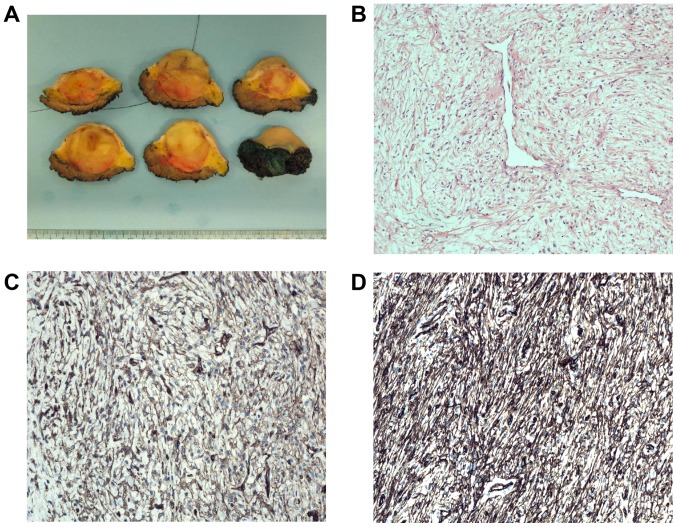
A 16-year-old male presented with a mass in the left deltoid region. After analysis of a needle biopsy, surgical resection was performed. Macroscopic examination disclosed a 4.2 cm large, well demarcated tumor (Fig. 1A). Microscopic examination revealed a moderately cellular tumor with spindle cells without clear atypia intermingled with loose intercellular matrix, partly with myxoid tissue and collagen (Fig. 1B). Dilated vessels were seen. There were some necrotic areas but very few mitotic figures (0–1/10 high power fields). Immunohistochemistry demonstrated positive focal staining for CD34 and CD99 (Fig. 1C and D), but negativity for cytokeratin cocktail (AE1/AE3, EMA, S-100, SMA, and desmin; data not shown). There was no nuclear STAT6 staining and the molecular analysis did not show presence of the NAB2-STAT6 fusion transcript which is pathognomonic for solitary fibrous tumor. FISH analysis was negative for rearrangement of the FUS gene. The histological diagnosis could therefore not be more precise than spindle cell tumor of uncertain malignancy. Three years after treatment, no local recurrence has developed and the patient is in remission.
Figure 1.
Pathologic features of the tumor. (A) Macroscopic picture of the tumor surrounded by subcutaneous fatty tissue. (B) Microscopic examination of a haematoxylin and eosin (H&E)-stained slide showing the moderately cellular tumor with spindle cells in a myxoid and fibrous stroma with dilated vessels. (C) Immunohistochemical analysis demonstrating positivity for CD34. (D) Immunohistochemical analysis demonstrating positivity for CD99.
G-banding and karyotyping
Both a core needle preoperative biopsy and fresh tissue from a representative area of the tumor in the surgical specimen were received and analyzed cytogenetically as part of our diagnostic routine. The samples were disaggregated mechanically and enzymatically with collagenase II (Worthington, Freehold, NJ, USA). The resulting cells were cultured and harvested using standard techniques (16). Chromosome preparations were G-banded with Wright stain and examined. The karyotype was written according to The International System for Human Cytogenetic Nomenclature (ISCN) 2013 guidelines (17).
High-throughput paired-end RNA-sequencing analysis
Tumor tissue adjacent to that used for cytogenetic analysis and histologic examination was frozen and stored at −80°C. Total RNA was extracted using miRNeasy Mini kit according to the manufacturer's instructions (Qiagen Nordic, Oslo, Norway). Tumor tissue was disrupted and homogenized in Qiazol Lysis Reagent (Qiagen Nordic) using a 5 mm stainless steel bead and TissueLyser II (Qiagen Nordic). Subsequently, total RNA was purified using QIAcube (Qiagen Nordic). The RNA quality was evaluated using the Experion Automated Electrophoresis System (Bio-Rad Laboratories, Oslo, Norway). The RNA quality indicator (RQI) was 9.9. Total RNA (3 µg) was sent for high-throughput paired-end RNA-sequencing at the Norwegian Sequencing Centre, Ullevål Hospital (http://www.sequencing.uio.no/). Detailed information about the high-throughput paired-end RNA-sequencing was given elsewhere (18). The software FusionCatcher (19) (https://github.com/ndaniel/fusioncatcher) was used for discovery of fusion transcripts.
Molecular genetic analysis
Total RNA (1 µg) was reverse-transcribed in a 20 µl reaction volume using iScript Advanced cDNA Synthesis kit for RT-qPCR according to the manufacturer's instructions (Bio-Rad Laboratories). The 25 µl PCR volume contained 12.5 µl Premix Ex Taq DNA Polymerase Hot Start version (Takara Bio Europe/SAS, Saint-Germain-en-Laye, France), 2 µl of cDNA, and 0.4 µM of each of the forward primer SETD2-7227F1 (5′-CCTCCCAACTGGAAGACAGCTCGA-3′) and reverse primer NF1-020-452R1 (5′-AGCTTTCCAACCCAGGACTGTGGTC-3′). The PCR was run on a C-1000 Thermal cycler (Bio-Rad Laboratories). The PCR conditions for amplification were: initial denaturation at 94°C for 30 sec followed by 35 cycles of 7 sec at 98°C and 2 min at 68°C, and a final extension for 5 min at 68°C. PCR products (3 µg) were stained with GelRed (Biotium), analyzed by electrophoresis through 1.0% agarose gel, and photographed. The remaining 22 µl PCR products were purified using the MinElute PCR purification kit (Qiagen Nordic) and direct sequenced using the light run sequencing service of GATC Biotech (http://www.gatc-biotech.com/en/sanger-services/lightrun-sequencing.html). The BLAST software (http://www.ncbi.nlm.nih.gov/BLAST/) was used for computer analysis of sequence data.
Fluorescence in situ hybridization (FISH)
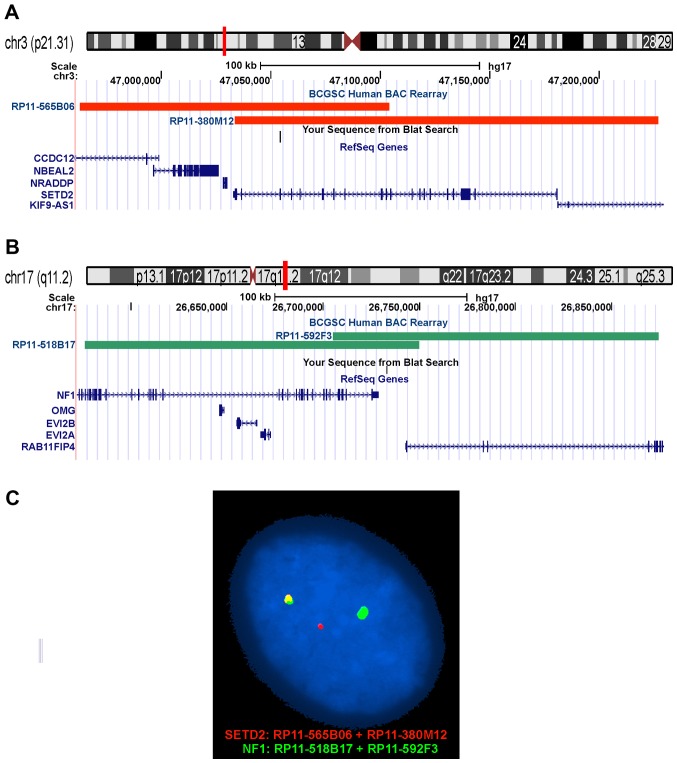
BAC probes were retrieved from the Human ‘32K’ BAC Re-Array library (BACPAC Resources, http://bacpac.chori.org/home.htm). They were selected according to physical and genetic mapping data on chromosomes 3 and 17 (see below) as reported on the Human Genome Browser at the University of California, Santa Cruz website (May 2004, http://genome.ucsc.edu/). FISH mapping of the clones on normal controls was performed to confirm their chromosomal location. The clones were RP11-565B06 (chr3:46962826-47104129) and RP11-380M12 (chr3:47033474-47226748) mapping to chromosome subband 3p21.31 which contains the SETD2 gene (red signal), and RP11-518B17 (chr17:26576215-26749754) and RP11-592F3 (chr17:26705272-26874157) mapping to chromosome subband 17q11.2 which contains the NF1 gene (green signal). FISH was performed as described elsewhere (18). Fluorescent signals were captured and analyzed using the CytoVision system (Leica Biosystems, Newcastle, UK).
Results
G-banding
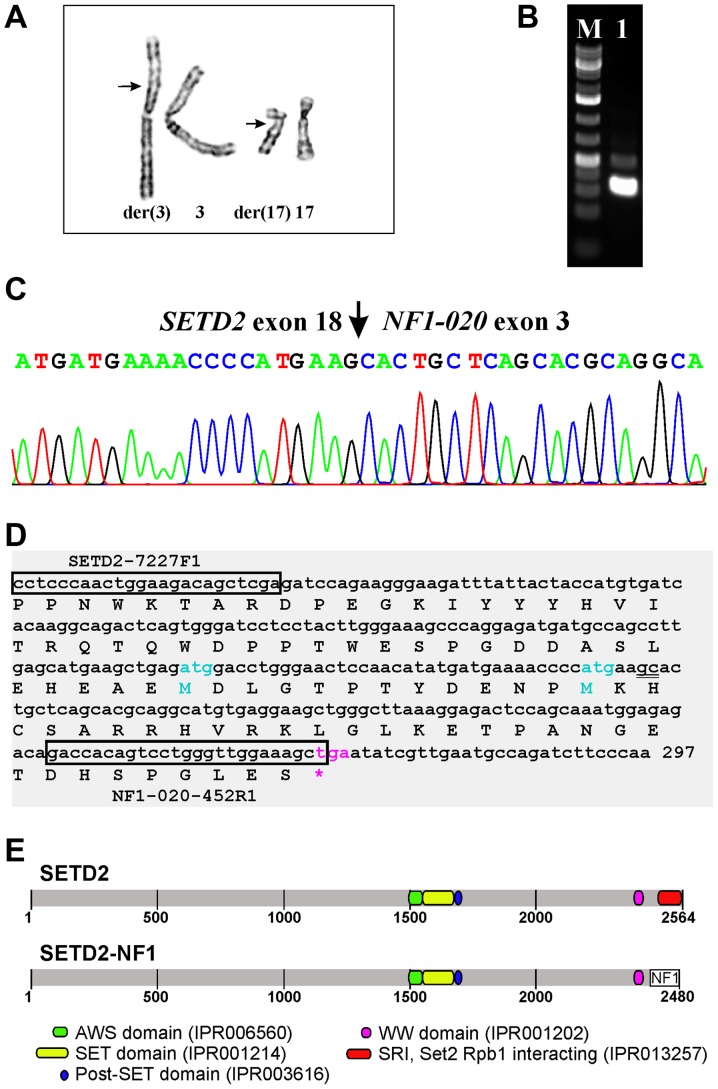
The G-banding analysis of short-term cultured cells from both the core needle biopsy and surgical specimen yielded the karyotype 46,XY,t(3;17)(p21;q12),del(10)(q24)[11] (Fig. 2A).
Figure 2.
Cytogenetic and molecular genetic features of the tumor. (A) Partial karyotype showing the der(3)t(3;17)(p21;q12) and der(17)t(3;17)(p21;q12) together with their normal chromosome homologs; breakpoint positions are indicated by arrows. (B) Amplification of a cDNA fragment (lane 1) using the primers SETD2-7227F1 and NF1-020-452R1. M, 1 kb DNA ladder (GeneRuler, ThermoFisher Scientific). (C) Partial sequence chromatogram of the cDNA fragment showing that exon 18 of SETD2 (sequence with accession number NM_014159 version 6) is fused to exon 3 of NF1-020 (ENST00000422121). (D) Sequence of the amplified cDNA fragment. SETD2-7227F1 and NF1-020-452R1 are shown in boxes. The fusion point ‘gc’ is double underlined. The open reading frame is also shown. (E) Illustration of the SETD2 protein, the putative SETD2-NF1 chimeric protein, and their functional domains.
RNA-sequencing, molecular genetic analysis, and FISH confirmation of SETD2-NF1 fusion
Using the FusionCatcher software with the fastq files obtained from the Norwegian Sequencing Centre, 31 potential fusion transcripts were found (Table I), among them SETD2-NF1. Taking into consideration that SETD2 and NF1 map to chromosome bands 3p21.31 and 17q11.2, respectively (http://genome-euro.ucsc.edu/index.html), the bands identified by G-banding analysis as being recombined by the 3;17-translocation, we decided to investigate further the SETD2-NF1 fusion transcript using molecular techniques. No other fusions were examined.
Table I.
Fusion transcripts detected using FusionCatcher.
| 5′-Chr | 3′-Chr | 5′-Partner gene | 3′-Partner gene | Fusion description | Fusion sequence |
|---|---|---|---|---|---|
| 17 | 3 | COL1A1 | APOD | GCTCCCTCCCACCCAACCAACTTTC*cccccccccataaagacaaaccaat | |
| 3 | 17 | SETD2 | NF1 | AACATATGATGAAAACCCCATGAAG*cactgctcagcacgcaggcatgtga | |
| 7 | 3 | COL1A2 | APOD | CTGGCAACATTGGATTCCCTGGACC*catcggcaccgtactggatcctggc | |
| 19 | 19 | ADCK4 | NUMBL | readthrough | TCCAGCCTCTCAGTGTGTTGGAGAG*acggggcgggcaccatgaacaagtt |
| 1 | 1 | PEAR1 | LRRC71 | readthrough | TCCAGGGCCCTGTGTACATAAACTG*aggagtaccagtgctccggggtcct |
| 2 | 3 | IGFBP5 | APOD | CAGGACTGACCCTCCTTCCTCCAGC*cacccagccccaagatggtgatgct | |
| 19 | 19 | CYP4F12 | CYP4F24P | pseudogene | ACCGCGATCCTAAAGAGATTGAATG*gcattatctgcatcatcaacattat |
| 17 | X | COL1A1 | TIMP1 | CTTTCCACCCTCTCTCCACCTGCCT*ctggcttctggcatcctgttgttgc | |
| 3 | 3 | DVL3 | AP2M1 | readthrough | TTCCGCATGGCCATGGGAAACCCCA*gtctgttctcagagcgatgggccgc |
| 3 | 3 | COL7A1 | UCN2 | readthrough | CGGGTGGTCCAGAGCCAGGGGACAG*cctgacctcacgatgaccaggtgtg |
| 11 | 11 | CTSC | RAB38 | readthrough | TTTGTCAGTCCTGTTCGAAACCAAG*gtcaagaaagatttggaaacatgac |
| 10 | 10 | MTG1 | RP11-108K14.4 | readthrough | CCCTCAACAAACACCAGCGCTTTGG*gtggaccaggtgctctgaggctggc |
| 11 | 11 | LSP1 | TNNT3 | short distance | CCGGCTCCCTAGGCGTCCCATCTCG*aaaccacccaccttcaccatgtctg |
| 1 | 1 | CTBS | GNG5 | readthrough | GCGGGCTCCTTATTATAACTATAAA*gtttcccaggcagctgcagacttga |
| 19 | 19 | GRAMD1A | SCN1B | readthrough | CCTCGGCGGCCACTGCTGGCACGGT*tgtcctcagcctgcgggggctgcgt |
| 10 | 10 | SYNPO2L | MYOZ1 | readthrough | CTAAGCGGCAGAGCCGTGCGGACAG*tgctgcccccacgcctgcccagctc |
| 19 | 19 | XRCC1 | ETHE1 | readthrough | TATGGGGTGGTGCCGCAAGCCTGAA*gcgttggagaccagggccagccctg |
| 11 | 11 | HPX | APBB1 | readthrough | TCTTCTCGGCTCCATATCATGGCAG*gagctgccaaggccatgtctgttcc |
| 16 | 16 | LCAT | PSMB10 | readthrough | TGAATAAAGACCTTCCTTTGCTACC*agtacccagtgagcagcacagaggg |
| 2 | 2 | SOCS5 | LINC01119 | short distance | GAGGCCGCCCCGCGCGCCCCAAACG*atgattccaatgtacagccaatgat |
| 3 | 3 | TBC1D23 | NIT2 | readthrough | GAATAGAAATCTTGGCAATCGAAAG*ctttccgcttggccctcatccagct |
| 2 | 2 | ADCY3 | PTRHD1 | readthrough | CACGGGGGTCATGGGCAACATTCAG*gccccagatgagaccaccctaaagg |
| 19 | 19 | CADM4 | ZNF428 | readthrough | GCGCTCTACGTACTTGTGGTCTACG*catccctctctacctgccaacatcc |
| 20 | 20 | CCM2L | HCK | short distance | CCGACTTCAGCTGCTGCAGCTCCTT*gatggggtgcatgaagtccaagttc |
| 17 | 20 | COL1A1 | CPXM1 | CCACCCAACCAACTTTCCCCCCAAC*catcacctgccattgcccacttact | |
| 16 | 16 | COQ9 | POLR2C | readthrough | TGAGACAAGTGCCTGCTGGACAGAG*gaggccgcagaagacgctcggcagt |
| 1 | 1 | EIF4G3 | HP1BP3 | readthrough | AAAAAGGTGATCATGGAGGAAAAAG*gttgattcttcaccacactgaaacc |
| X | X | MORF4L2-AS1 | TMEM31 | readthrough | AGAAGAGTGCAGGAAAGCAAACCAA*gtgatcactttactgtagaagaaat |
| 7 | 7 | RHBDD2 | POR | readthrough | AGAGGAGGGCAGCCCAGAGCCGGAA*tttcatgatcaacatgggagactcc |
| 14 | 3 | SERPINA3 | APOD | CAACAGGCCCTTCCTGATGATCATT*gtcccttctccagccacccagcccc | |
| 1 | 1 | VPS45 | PLEKHO1 | readthrough | GCACCACAGTGCACAACACGAAAAG*ggacctcaggatggaaaccagcagc |
RT-PCR with the SETD2-7227F1 and NF1-020-452R1 primer combination amplified a 268 bp cDNA fragment (Fig. 2B). Sanger sequencing showed that it was a SETD2-NF1 chimeric cDNA fragment with the fusion point identical to that found using FusionCatcher (Fig. 2C and D; Table I). In this fusion transcript, the sequence of SETD2 coding for the last 114 amino acids of the SETD2 protein are replaced by the NF1 sequence coding for 30 amino acids (Fig. 2D and E).
Interphase FISH analysis confirmed the SETD2-NF1 fusion. All 100 counted nuclei showed a red signal corresponding to the SETD2 (Fig. 3A and C), a green signal corresponding to NF1 (Fig. 3B and C), and a yellow fusion signal corresponding to the SETD2-NF1 (Fig. 3C).
Figure 3.
Interphase FISH for the detection of a SETD2-NF1 fusion gene. (A) The positions of the clones RP11-565B06 (chr3:46962826-47104129) and RP11-380M12 (chr3:47033474-47226748) are indicated in chromosome band 3p21.31. The clones contained the SETD2 gene and were labeled with red. (B) The positions of the clones RP11-518B17 (chr17:26576215-26749754) and RP11-592F3 (chr17:26705272-26874157) are indicated in chromosome band 17q11.2. The clones contained part of the NF1 gene and were labeled with green. (C) Interphase FISH showing nucleus with a green signal, a red signal, and a yellow signal. The yellow signal corresponds to the SETD2-NF1 fusion gene. The fact that there is only one yellow signal indicates that there is no reciprocal fusion.
Discussion
Fusion transcripts of both NF1 and SETD2 with various partners have been described in hematologic malignancies as well as solid tumors (20–22). However, this is the first time that a fusion between SETD2 and NF1 was found.
The SETD2 gene is ubiquitously expressed and codes for a protein which belongs to a class of huntingtin interacting proteins characterized by WW motifs (23,24). SETD2 is also a DNA-binding factor that binds the proximal E1A promoter of adenovirus serotype 12 (24). In addition, SETD2 was shown to possess histone H3 lysine 36 (H3-K36) specific HMTase activity, auto-methylation activity, a novel transcriptional activation domain, and association with hyperphosphorylated RNA polymerase II (25). The SETD2 protein is solely responsible for all H3K36 trimethylation in humans (26). SETD2 interacts with the Ser2/Ser5 hyperphosphorylated RNA polymerase II during transcriptional elongation via its SRI (Set2 Rpb1 interacting) domain, which explains why H3K36 trimethylation is found in the body of actively transcribed genes (27). Deletion of the SRI domain in yeast Set2 abolishes H3K36 methylation, which in turn prevents elongation by RNA polymerase II. This suggests that the SRI domain is responsible for coupling transcription to histone methylation by Set2 (28). It seems that SETD2 serves as a linker between histone H3-K36 methylation and transcriptional regulation in yeast and mammals (25,28).
Involvement of the SETD2 gene has been reported in many types of malignancy (29). Inactivation of SETD is common in clear cell renal carcinoma with loss or decrease of H3K36me3 mark (30), when it is associated with worse prognosis and development of recurrent and/or metastatic disease (31). Downregulation of SETD2 at transcriptional and protein levels was observed in breast cancer (32,33). The expression of SETD2 was lower in malignant samples, decreased with increasing tumor stage, and was lower in samples from patients who developed metastasis, local recurrence, or died from breast cancer compared to those who were disease-free for >10 years (32). SETD2 mutations were also described in high-grade gliomas and in leukemias (34–36). The mutations are either nonsense or frameshift mutations that truncate a portion of the C terminus domain of SETD2. Truncating mutations result in loss of the C terminus SRI domain which is responsible for the recruitment of SETD2 to its target gene locus through binding to the phosphor-C-terminal repeat domain (PCTD) of elongating RNA polymerase II (36). Recently, genomic disruption of SETD2 was reported in chronic lymphocytic leukemia and the data suggested that SETD2 aberrations may be clinically relevant (37). Patients with SETD2 abnormalities and wild-type TP53 and ATM had significantly shorter progression-free and overall survival compared with cases with wild-type for all three genes (37). In malignant mesotheliomas, a combination of the methods array comparative genomic hybridization and targeted next-generation sequencing revealed biallelic SETD2 inactivation in 9 out of 33 examined tumors (38). Gene fusions and splice alterations were also reported to be frequent mechanisms for SETD2 inactivation (39).
SETD2 was found to be the most significantly and recurrently mutated gene in type II enteropathy-associated T-cell lymphoma (EATL-II); 86% (13/15) of EATL-II tumors with 20 distinctive mutations (40). Fourteen of these mutations consisted of premature stop codon, nonsense, frameshift indels or splicing mutations expected to confer critical changes in protein structure. The other six missense mutations occurred in highly conserved residues of functional domains and were predicted to be deleterious with a damaging effect on the protein (40).
The NF1 gene spans approximately 280 kbp, has 58 exons (mRNA transcript variant 1, NM_001042492.2) and codes for the cytoplasmatic and multidomain protein neurofibromin (41). Neurofibromin is a negative regulator of the RAS cellular proliferation pathway (42–45). Several other functions of neurofibromin were also reported, among them positive regulation of adenyl cyclase, regulation of cell adhesion and motility, and suppression of epithelial mesenchymal transition (42–45). The NF1 gene is a classical tumor suppressor gene whose inactivation is responsible for the neurofibromatosis type 1 (NF1) tumor predisposition syndrome (http://omim.org/entry/613113). Mutations of NF1 are also linked to juvenile myelomonocytic leukemia (http://omim.org/entry/607785) and Watson syndrome (http://omim.org/entry/193520). The NF1 syndrome is characterized by the development of multiple neurofibromas, café-au-lait spots, and Lisch nodules (42,45). Patients with NF1 syndrome are at increased risk to develop malignant peripheral nerve sheath tumors, phaeochromocytoma, leukemia, glioma, rhabdomyosarcoma, and breast cancer (42,45). Both alleles of NF1 are inactivated in the tumors in NF1 patients. Mutations in the NF1 gene may also result in cardiovascular, musculoskeletal, and nervous system anomalies (45,46).
Splicing in the NF1 gene is complex and several alternative transcripts were found (47); altogether 23 according to the ensemble genome browser (http://www.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000196712;r=17:31378891-31382106;t=ENST00000422121). The transcript NF1-020 (ENST00000422121) has three exons and is thought to undergo nonsense mediated decay, a process which detects nonsense mutations and prevents the expression of truncated or erroneous proteins (48). Thus, the functional significance, if any, of the NF1-020 transcript is unclear.
In the present case, the t(3;17) resulted in a SETD2-NF1 fusion transcript in which the first 18 exons of SETD2 (sequence with accession number NM_014159 version 6) are fused to exon 3 of the transcript NF1-020 (ENST00000422121) (Fig. 2C and D). The fusion transcript would code for a protein in which the last 114 amino acids of SETD2, in other words the entire SRI domain, are replaced by 30 amino acids encoded by the NF1 sequence (Fig. 2D and E). The result would be similar to that seen with the truncating SETD2 mutations found in leukemias (36). Absence of the SRI domain would result in inability to recruit SETD2 to its target gene locus through binding to the phosphor-C-terminal repeat domain of elongating RNA polymerase II and may affect H3K36 methylation. Alternatively, loss of one of two functional SETD2 alleles might be the crucial factor in tumorigenesis. Whether aberrations of SETD2 are recurrent and define a specific subgroup of spindle cell tumors, remains to be seen.
Acknowledgements
The authors thank Hege Kilen Andersen and Nina Øino for excellent technical assistance. This work was supported by grants from the Norwegian Radium Hospital Foundation.
References
- 1.Collini P, Sorensen PH, Patel S, Blay JY, Issels RD, Maki RG, Eriksson M, del Muro XG. Sarcomas with spindle cell morphology. Semin Oncol. 2009;36:324–337. doi: 10.1053/j.seminoncol.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Fisher C. Spindle cell sarcomas. Surg Pathol Clin. 2011;4:721–744. doi: 10.1016/j.path.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Heim S, Mitelman F, editors. Cancer Cytogenetics: Chromosomal and Molecular Genetic Abberations of Tumor Cells. Fourth. Wiley-Blackwell; 2015. doi: 10.1002/9781118795569. [DOI] [Google Scholar]
- 4.Sandberg AA, Bridge JA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: Congenital (infantile) fibrosarcoma and mesoblastic nephroma. Cancer Genet Cytogenet. 2002;132:1–13. doi: 10.1016/S0165-4608(01)00528-3. [DOI] [PubMed] [Google Scholar]
- 5.Llombart B, Serra-Guillén C, Monteagudo C, López Guerrero JA, Sanmartín O. Dermatofibrosarcoma protuberans: A comprehensive review and update on diagnosis and management. Semin Diagn Pathol. 2013;30:13–28. doi: 10.1053/j.semdp.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F. WHO Classification of Tumours of Soft Tissue and Bone. 4th. IARC; Lyon: 2013. [Google Scholar]
- 7.Chmielecki J, Crago AM, Rosenberg M, O'Connor R, Walker SR, Ambrogio L, Auclair D, McKenna A, Heinrich MC, Frank DA, et al. Whole-exome sequencing identifies a recurrent NAB2-STAT6 fusion in solitary fibrous tumors. Nat Genet. 2013;45:131–132. doi: 10.1038/ng.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohajeri A, Tayebwa J, Collin A, Nilsson J, Magnusson L, von Steyern FV, Brosjö O, Domanski HA, Larsson O, Sciot R, et al. Comprehensive genetic analysis identifies a pathognomonic NAB2/STAT6 fusion gene, nonrandom secondary genomic imbalances, and a characteristic gene expression profile in solitary fibrous tumor. Genes Chromosomes Cancer. 2013;52:873–886. doi: 10.1002/gcc.22083. [DOI] [PubMed] [Google Scholar]
- 9.Robinson DR, Wu YM, Kalyana-Sundaram S, Cao X, Lonigro RJ, Sung YS, Chen CL, Zhang L, Wang R, Su F, et al. Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat Genet. 2013;45:180–185. doi: 10.1038/ng.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schweizer L, Koelsche C, Sahm F, Piro RM, Capper D, Reuss DE, Pusch S, Habel A, Meyer J, Göck T, et al. Meningeal hemangiopericytoma and solitary fibrous tumors carry the NAB2-STAT6 fusion and can be diagnosed by nuclear expression of STAT6 protein. Acta Neuropathol. 2013;125:651–658. doi: 10.1007/s00401-013-1117-6. [DOI] [PubMed] [Google Scholar]
- 11.Fruth K, Hansen T, Katenkamp D, Mann W, Lippert BM. Recurrence of a laryngeal spindle cell sarcoma with a transformation into a higher grade of malignancy. Auris Nasus Larynx. 2009;36:491–495. doi: 10.1016/j.anl.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Alaggio R, Rosolen A, Sartori F, Leszl A, d'Amore ES, Bisogno G, Carli M, Cecchetto G, Coffin CM, Ninfo V. Spindle cell tumor with EWS-WT1 transcript and a favorable clinical course: A variant of DSCT, a variant of leiomyosarcoma, or a new entity? Report of 2 pediatric cases. Am J Surg Pathol. 2007;31:454–459. doi: 10.1097/01.pas.0000213375.02171.43. [DOI] [PubMed] [Google Scholar]
- 13.Panagopoulos I, Bjerkehagen B, Gorunova L, Berner JM, Boye K, Heim S. Several fusion genes identified by whole transcriptome sequencing in a spindle cell sarcoma with rearrangements of chromosome arm 12q and MDM2 amplification. Int J Oncol. 2014;45:1829–1836. doi: 10.3892/ijo.2014.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nord KH, Macchia G, Tayebwa J, Nilsson J, von Steyern F Vult, Brosjö O, Mandahl N, Mertens F. Integrative genome and transcriptome analyses reveal two distinct types of ring chromosome in soft tissue sarcomas. Hum Mol Genet. 2014;23:878–888. doi: 10.1093/hmg/ddt479. [DOI] [PubMed] [Google Scholar]
- 15.Lestou VS, O'Connell JX, Ludkovski O, Gosling H, Lesack D, Horsman DE. Coamplification of 12p11 and 12q13 approximately q22 in multiple ring chromosomes in a spindle cell sarcoma resolved by novel multicolor fluorescence in situ hybridization analysis. Cancer Genet Cytogenet. 2002;139:44–47. doi: 10.1016/S0165-4608(02)00600-3. [DOI] [PubMed] [Google Scholar]
- 16.Mandahl N. Methods in solid tumour cytogenetics. In: Rooney DE, editor. Human cytogenetics: Malignancy and Acquired Abnormalities. 3rd. Oxford University Press; New York: 2001. pp. 165–203. [Google Scholar]
- 17.Schaffer LG, McGowan-Jordan J, Schmid M. ISCN 2013: An International System for Human Cytogenetic Nomenclature Karger, Basel. 2013 doi: 10.1061/9780784412978.045. [DOI] [Google Scholar]
- 18.Panagopoulos I, Gorunova L, Bjerkehagen B, Heim S. The ‘grep’ command but not FusionMap, FusionFinder or ChimeraScan captures the CIC-DUX4 fusion gene from whole transcriptome sequencing data on a small round cell tumor with t(4;19)(q35;q13) PLoS One. 2014;9:e99439. doi: 10.1371/journal.pone.0099439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicorici D, Satalan H, Edgren H, Kangaspeska S, Murumagi A, Kallioniemi O, Virtanen S, Kikku O. FusionCatcher - a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv. 2014 doi: 10.1101/011650. [Google Scholar]
- 20.Andersson AK, Ma J, Wang J, Chen X, Gedman AL, Dang J, Nakitandwe J, Holmfeldt L, Parker M, Easton J, et al. St. Jude Children's Research Hospital-Washington University Pediatric Cancer Genome Project: The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47:330–337. doi: 10.1038/ng.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen K, Navin NE, Wang Y, Schmidt HK, Wallis JW, Niu B, Fan X, Zhao H, McLellan MD, Hoadley KA, et al. BreakTrans: Uncovering the genomic architecture of gene fusions. Genome Biol. 2013;14:R87. doi: 10.1186/gb-2013-14-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshihara K, Wang Q, Torres-Garcia W, Zheng S, Vegesna R, Kim H, Verhaak RG. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34:4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 24.Rega S, Stiewe T, Chang DI, Pollmeier B, Esche H, Bardenheuer W, Marquitan G, Pützer BM. Identification of the full-length huntingtin-interacting protein p231HBP/HYPB as a DNA-binding factor. Mol Cell Neurosci. 2001;18:68–79. doi: 10.1006/mcne.2001.1004. [DOI] [PubMed] [Google Scholar]
- 25.Sun XJ, Wei J, Wu XY, Hu M, Wang L, Wang HH, Zhang QH, Chen SJ, Huang QH, Chen Z. Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J Biol Chem. 2005;280:35261–35271. doi: 10.1074/jbc.M504012200. [DOI] [PubMed] [Google Scholar]
- 26.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kudithipudi S, Jeltsch A. Role of somatic cancer mutations in human protein lysine methyltransferases. Biochim Biophys Acta. 2014;1846:366–379. doi: 10.1016/j.bbcan.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Duns G, van den Berg E, van Duivenbode I, Osinga J, Hollema H, Hofstra RM, Kok K. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 2010;70:4287–4291. doi: 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- 31.Hakimi AA, Ostrovnaya I, Reva B, Schultz N, Chen YB, Gonen M, Liu H, Takeda S, Voss MH, Tickoo SK, et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: A report by MSKCC and the KIRC TCGA research network. Clin Cancer Res. 2013;19:3259–3267. doi: 10.1158/1078-0432.CCR-12-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al Sarakbi W, Sasi W, Jiang WG, Roberts T, Newbold RF, Mokbel K. The mRNA expression of SETD2 in human breast cancer: Correlation with clinico-pathological parameters. BMC Cancer. 2009;9:290. doi: 10.1186/1471-2407-9-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newbold RF, Mokbel K. Evidence for a tumour suppressor function of SETD2 in human breast cancer: A new hypothesis. Anticancer Res. 2010;30:3309–3311. [PubMed] [Google Scholar]
- 34.Fontebasso AM, Schwartzentruber J, Khuong-Quang DA, Liu XY, Sturm D, Korshunov A, Jones DT, Witt H, Kool M, Albrecht S, et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol. 2013;125:659–669. doi: 10.1007/s00401-013-1095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mar BG, Bullinger LB, McLean KM, Grauman PV, Harris MH, Stevenson K, Neuberg DS, Sinha AU, Sallan SE, Silverman LB, et al. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat Commun. 2014;5:3469. doi: 10.1038/ncomms4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu X, He F, Zeng H, Ling S, Chen A, Wang Y, Yan X, Wei W, Pang Y, Cheng H, et al. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat Genet. 2014;46:287–293. doi: 10.1038/ng.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker H, Rose-Zerilli MJ, Larrayoz M, Clifford R, Edelmann J, Blakemore S, Gibson J, Wang J, Ljungström V, Wojdacz TK, et al. Genomic disruption of the histone methyltransferase SETD2 in chronic lymphocytic leukaemia. Leukemia. 2016;30:2179–2186. doi: 10.1038/leu.2016.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshikawa Y, Emi M, Hashimoto-Tamaoki T, Ohmuraya M, Sato A, Tsujimura T, Hasegawa S, Nakano T, Nasu M, Pastorino S, et al. High-density array-CGH with targeted NGS unmask multiple noncontiguous minute deletions on chromosome 3p21 in mesothelioma; Proc Natl Acad Sci USA; 2016; pp. 13432–13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bueno R, Stawiski EW, Goldstein LD, Durinck S, De Rienzo A, Modrusan Z, Gnad F, Nguyen TT, Jaiswal BS, Chirieac LR, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48:407–416. doi: 10.1038/ng.3520. [DOI] [PubMed] [Google Scholar]
- 40.Roberti A, Dobay MP, Bisig B, Vallois D, Boéchat C, Lanitis E, Bouchindhomme B, Parrens MC, Bossard C, Quintanilla-Martinez L, et al. Type II enteropathy-associated T-cell lymphoma features a unique genomic profile with highly recurrent SETD2 alterations. Nat Commun. 2016;7:12602. doi: 10.1038/ncomms12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barron VA, Lou H. Alternative splicing of the neurofibromatosis type I pre-mRNA. Biosci Rep. 2012;32:131–138. doi: 10.1042/BSR20110060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiuru M, Busam KJ. The NF1 gene in tumor syndromes and melanoma. Lab Invest. 2017;97:146–157. doi: 10.1038/labinvest.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahalingam M. NF1 and Neurofibromin: Emerging players in the genetic landscape of desmoplastic melanoma. Adv Anat Pathol. 2017;24:1–14. doi: 10.1097/PAP.0000000000000131. [DOI] [PubMed] [Google Scholar]
- 44.Ratner N, Miller SJ. A RASopathy gene commonly mutated in cancer: The neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer. 2015;15:290–301. doi: 10.1038/nrc3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yap YS, McPherson JR, Ong CK, Rozen SG, Teh BT, Lee AS, Callen DF. The NF1 gene revisited - from bench to bedside. Oncotarget. 2014;5:5873–5892. doi: 10.18632/oncotarget.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larizza L, Gervasini C, Natacci F, Riva P. Developmental abnormalities and cancer predisposition in neurofibromatosis type 1. Curr Mol Med. 2009;9:634–653. doi: 10.2174/156652409788488801. [DOI] [PubMed] [Google Scholar]
- 47.Vandenbroucke I, Callens T, De Paepe A, Messiaen L. Complex splicing pattern generates great diversity in human NF1 transcripts. BMC Genomics. 2002;3:13. doi: 10.1186/1471-2164-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schweingruber C, Rufener SC, Zünd D, Yamashita A, Mühlemann O. Nonsense-mediated mRNA decay - mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta. 2013;1829:612–623. doi: 10.1016/j.bbagrm.2013.02.005. [DOI] [PubMed] [Google Scholar]