Abstract
Primers were designed to amplify a 592-bp region within a conserved structural gene (g20) found in some cyanophages. The goal was to use this gene as a proxy to infer genetic richness in natural cyanophage communities and to determine if sequences were more similar in similar environments. Gene products were amplified from samples from the Gulf of Mexico, the Arctic, Southern, and Northeast and Southeast Pacific Oceans, an Arctic cyanobacterial mat, a catfish production pond, lakes in Canada and Germany, and a depth of ca. 3,246 m in the Chuckchi Sea. Amplicons were separated by denaturing gradient gel electrophoresis, and selected bands were sequenced. Phylogenetic analysis revealed four previously unknown groups of g20 clusters, two of which were entirely found in freshwater. Also, sequences with >99% identities were recovered from environments that differed greatly in temperature and salinity. For example, nearly identical sequences were recovered from the Gulf of Mexico, the Southern Pacific Ocean, an Arctic freshwater cyanobacterial mat, and Lake Constance, Germany. These results imply that closely related hosts and the viruses infecting them are distributed widely across environments or that horizontal gene exchange occurs among phage communities from very different environments. Moreover, the amplification of g20 products from deep in the cyanobacterium-sparse Chuckchi Sea suggests that this primer set targets bacteriophages other than those infecting cyanobacteria.
Unicellular cyanobacteria are a major component of the prokaryotic biomass in the oceans (6, 15), account for more than half of the fixed carbon in some regions, and play a key role in the transfer of energy through the microbial loop (6, 15, 21). Consequently, knowledge of the regulation of cyanobacterial communities is required to understand global nutrient and energy cycles.
Viruses are the most abundant biological entities in fresh and marine waters and are typically 5- to 10-fold more abundant than prokaryotes (1, 19, 29). As mortality agents of heterotrophic and photosynthetic microbes (8, 25, 37), they affect the abundance and diversity of host cell communities (13, 18) as well as the cycling of carbon and nutrients (37).
The proportion of primary producers lost to viral lysis is uncertain; however, viruses infecting a single strain of the marine cyanobacterium Synechococcus are widespread and can reach levels of abundance of >105 ml−1 (16, 27, 36). Estimates from the proportions of visibly infected cells and viral decay rates suggest that approximately 3 to 10% of Synechococcus spp. are destroyed daily by viral lysis (19, 26, 37).
The impact of viruses on host communities can be inferred from viral community diversity, as viral taxonomy can provide insights into the hosts that they infect. For example, phylogenetic analyses of algal virus DNA polymerase genes revealed that phycoviruses are monophyletic relative to other viral families and that genetically distinct groups are clearly resolved and correspond to the host taxon infected (3, 4). Subsequently, denaturing gradient gel electrophoresis (DGGE) was combined with PCR (22) to “fingerprint” and compare spatial (23) and temporal (24) differences among algal virus communities.
The power of combining PCR with DGGE persuaded us to adopt this approach to examine the diversity of cyanophages in a wide range of natural environments. Previously, primers specific for conserved regions of g20 (a structural gene) in three cyanophages (S-PM2, S-BnM1, and S-WHM1) were developed to amplify a 165-bp fragment (9, 40). DGGE fingerprints of PCR-amplified g20 gene fragments showed that cyanophage communities differed over spatial scales from meters (7) to thousands of kilometers (39). Because the size of the fragment was small for phylogenetic analysis, other primers were developed to amplify a 592-bp region of cyanophage g20 genes (41). With these primers, cyanophage gene fragments were amplified from estuarine and coastal locations in the Atlantic Ocean (17, 35, 41) and from one freshwater lake (5) and were compared phylogenetically.
Our study characterized the richness of cyanophage g20 genes in a much wider range of environments than was previously examined. Although distinct sequence clusters occurred in distinct environments, some sequences with >99% sequence identity were also amplified from vastly different environments. This suggests that similar hosts and their viruses are found in freshwater and marine environments or that high sequence homology in this gene may not be indicative of viruses that infect closely related hosts. In addition, g20 sequences were amplified from samples from a depth of 3,246 m in the Chuckchi Sea, consistent with the possibility of CPS primers not being specific for cyanophages.
MATERIALS AND METHODS
Samples originated from the Gulf of Mexico, the Arctic Ocean, the Southern Ocean, the Southeast Pacific Ocean, several inlets in the Northeast Pacific Ocean, two lakes in British Columbia (Chilliwack Lake and Cultus Lake), one lake in Germany (Lake Constance), a commercial catfish production pond (Limco, Inc.) in Crowley, La., and a shallow meltwater pond located on the Ward Hunt Island ice shelf in the Canadian high Arctic. A small plug was cut from a cyanobacterial mat growing in the Arctic meltwater pond. The details of the sites sampled and the labels assigned to each sample are listed in Table 1.
TABLE 1.
Details of samples used for this study
| Sample no. or type | Origin | Latitude and longitude | Date collected | Depth (m)a | Temp (°C)a | Salinity (ppt)a |
|---|---|---|---|---|---|---|
| Marine | ||||||
| GOM01 | Gulf of Mexico | 27°30.04′ N, 88°24.11′ W | 18 Jul 2001 | 110 | 22.5 | 36.6 |
| BES02A | Beaufort Sea, Arctic Ocean | 72°30.0′ N, 151°20.0′ W | 14 Sep 2002 | 35 | −1.0 | 34.0 |
| CHS02 | Chuckchi Sea, Arctic Ocean | 73°30.0′ N 157°00.0′ W | 9 Sep 2002 | 3,246 | −0.31 | 35.0 |
| BES02B | Beaufort Sea, Arctic Ocean | 70°12.0′ N, 137°0.0′ W | 26 Sep 2002 | 25 | −0.82 | 30.0 |
| ANT98 | Antarctic peninsula, Southern Ocean | 61°11.68′ S, 54°36.25′ W | 3 Nov 1998 | 0.5 | −0.24 | 34.1 |
| COL00 | Coast of Colombia, SE Pacific | 3°00.60′ N, 82°37.90′ W | 4 Aug 2000 | 1-3 | 26.8 | 33.5 |
| CHI00 | Coast of Chile, SE Pacific | 29°00′ S, 77°00′ W | 27 Jul 2000 | 1-3 | 16.6 | 34.4 |
| SAI99 | Salmon Inlet, NE Pacific | 49°36.25′ N, 123°48.18′ W | 17 Aug 1999 | 0.5 | 16.7 | 17.9 |
| PES99 | Pendrell Sound, NE Pacific | 50°16.24′ N, 124°42.80′ W | 20 Aug 1999 | 6.2 | 18.3 | 22.3 |
| MAI00 | Malaspina Inlet, NE Pacific | 50°04.77′ N, 124°42.85′ W | 2 Aug 2000 | 1 | 17.6 | 25.6 |
| TEA00 | Teakerne Arm, NE Pacific | 50°11.35′ N, 124°51.14′ W | 1 Aug 2000 | 7-10 | 18 | 24 |
| Freshwater | ||||||
| CUL02M | Cultus Lake, BC, Canada | 49°03.45′ N, 121°59.05′ W | 25 Sep 2002 | 10 | 15 | NA |
| CUL02H | Cultus Lake, BC, Canada | 49°03.45′ N, 121°59.05′ W | 25 Sep 2002 | 18 | 7.3 | NA |
| CHL02E | Chilliwack Lake, BC, Canada | 49°04.22′ N, 121°25.54′ W | 29 Jun 2002 | 3 | 12.7 | NA |
| CAT02 | Catfish pond, Crowley, La | 30°13.00′ N, 92°23.00′ W | 12 Sept 2002 | NA | NA | NA |
| SPM02 | Shore Pond cyanobacterial mat, Arctic Ocean | 83°00.00′ N, 77°00.00′ W | 7 Aug 2001 | NA | NA | NA |
| LAC95A | Lake Constance, Germany | 47°35.00′ N, 9°25.00′ W | 10 May 1995 | 3 | NA | NA |
| LAC95B | Lake Constance, Germany | 47°35.00′ N, 9°25.00′ W | 16 May 1995 | 3 | NA | NA |
NA, not applicable.
Catfish pond water (2 liters) was cleared of particles by centrifugation at 20,250 × g for 15 min followed by filtration through 47-mm-diameter, 0.45-μm-pore-size cellulose acetate filters (Nalgene, Rochester, N.Y.). Viruses in the filtrate were concentrated to ca. 10 to 15 ml by use of a 10-kDa-cutoff Amicon cartridge (Millipore, Bedford, Mass.). For the cyanobacterial mat, DNAs were extracted (20) from a 1-cm-diameter, 1-mm-thick plug that had been stored wet at 4°C in the dark for ca. 19 months. The extracted DNAs were stored at −20°C in 10 mM Tris-HCl (pH 8.5).
For other samples, organisms and particles that were larger than viruses were removed by filtration through a 142-mm-diameter, 1.2-μm-pore-size (GC50; Advantec MFS, Dublin, Calif.) or 0.6-μm-pore-size (GFF; Whatman, Clifton, N.J.) glass-fiber filter followed by a 0.45-μm-pore-size (GVWP; Millipore) or 0.2-μm-pore-size (Gelman, East Hills, N.Y.) filter. Virus-sized particles in the filtrate were concentrated ca. 50- to 500-fold by ultrafiltration (30) with a 10- or 30-kDa-cutoff Amicon (S1Y10/S1Y30/S10Y30) cartridge. Subsamples (100 ml) from the Arctic Ocean were further concentrated to ca. 500 μl by the use of Centricon Plus-20 centrifugal filters (Millipore) and then stored at −20°C. Other concentrates were stored at 4°C in the dark until used for analysis. DGGE g20 profiles of virus communities stored in this way for >18 months were invariant (7).
The upstream primer CPS4 was designed previously (40). To amplify a larger fragment for phylogenetic analysis, we aligned the available g20 sequences in GenBank (for the cyanomyoviruses S-PM2, S-BnM1, and S-WHM1; the coliphage T4; and the marine vibriophage KVP40) to design a new downstream primer (G20-2) that targeted cyanophages but not other phages. The primers were tested on four cyanomyoviruses (S-PWM1, S-PWM2, S-PWM3, and S-PWM4), a cyanopodovirus (S-BBP1), a cyanosiphovirus (S-BBS1) (28), and 20 uncharacterized cyanophages isolated from the Gulf of Mexico. Also, the primer specificity was tested against two myoviruses that infect Vibrio parahaemolyticus (VpV 360 and VpV HS1) and against the coliphages T4, RB33, RB69, and LZ4.
Subsamples (35 ml) of viral concentrates from the Gulf of Mexico, the Southern Ocean, and British Columbian lakes were centrifuged at 85,000 × g for 3.5 h; subsamples (38 ml) from the Southeast Pacific were centrifuged at 104,000 × g for 3 h. Either 100 or 500 μl of virus-free water was added to the virus pellets. An established hot-cold technique (4) was used to extract nucleic acids from the diluted virus pellets and from 100-μl subsamples of the viral concentrates from the Northeast Pacific inlets, Lake Constance, the Arctic Ocean, and filtered culture lysates. Three microliters of each template was added to a 47-μl PCR mixture containing Taq DNA polymerase assay buffer (50 mM KCl, 20 mM Tris-HCl [pH 8.4]), 1.5 mM MgCl2, a 0.20 mM concentration of each deoxyribonucleoside triphosphate, 40 pmol each of the primers CPS4 (40) and G20-2 [5′-(G/C)(A/T)(A/G)AAATA(C/T)TT(inosine)CC(A/G)AC(A/G)(A/T)A(G/T)GGATC-3′], and 1.0 U of Platinum Taq polymerase (Invitrogen Life Technologies, Carlsbad, Calif.). Negative controls contained all reagents and sterile water instead of a template. PCRs were performed with the following cycle parameters: denaturation at 94°C for 90 s, 35 cycles of denaturation at 94°C for 45 s, annealing at 50°C for 1 min, and extension at 72°C for 45 s, and a final extension at 72°C for 5 min. PCR products were electrophoresed in 2% agarose in 0.5× TBE buffer (45 mM Tris-borate, 1 mM EDTA [pH 8.0]) at 100 V for 75 min. Gels were stained with ethidium bromide, visualized on a UV transilluminator, and photographed with a Nikon Coolpix 950 digital camera. Digital images were manipulated with Adobe Photoshop 5.0 LE. After the PCR products were purified from agarose gels, they were used as templates for a second round of PCRs. With a clean glass pipette, a plug of agarose containing amplified DNA was removed from each lane. One hundred microliters of 0.5× TBE was added to each plug in a sterile microcentrifuge tube and heated to 65°C to elute the DNA. Two microliters of the eluted DNA was added to a 98-μl PCR mixture, and the PCR was conducted as described above, except that the number of cycles of amplification was reduced to 25. Amplification in second-stage PCRs was confirmed by agarose gel electrophoresis as described above.
Second-stage PCR products (20 to 35 μl) were separated by DGGE. Gels with 20 to 40% linear denaturing gradients (100% denaturant is defined as 7 M urea and 40% deionized formamide) and 7 to 8% polyacrylamide were run for 15 h in 1× TAE buffer (40 mM Tris base, 20 mM sodium acetate, 1 mM EDTA [pH 8.5]) at 80 V and 60°C in a D-code electrophoresis system (Bio-Rad Laboratories, Hercules, Calif.). The gels were stained in a 0.1× SYBR Gold (Molecular Probes, Eugene, Oreg.) solution for 3 h, visualized, and photographed as described above.
Ninety-nine bands were excised from the gels, added to 100 μl of 1× TAE buffer in sterile microcentrifuge tubes, and heated to 95°C for 10 min. PCRs were conducted with 2 μl of the eluted DNA as a template under the same conditions as the first-stage PCRs, but with 28 cycles of amplification. PCR products were cloned by use of a pGEM-T Vector System I (Promega, Madison, Wis.) TA cloning kit, and the resulting reactions were used to transform competent Escherichia coli JM109 (Promega). Colonies were screened for correct inserts by PCRs with the primers CPS4 and G20-2. Plasmid DNAs were harvested from overnight cultures by the use of QIAprep spin miniprep kits (Qiagen, Valencia, Calif.). The clones were sequenced by use of an AmpliTaq FS BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.), and excess dye terminators were removed by the use of Centri-Sep spin columns (Princeton Separations, Adelphia, N.J.). Reactions were run in an ABI model 373 stretch or ABI Prism 377 automated sequencer.
Bioedit (version 5.0.7) was used for sequence editing, translation, and the generation of pairwise DNA identity matrices (10). For sequences from a single location with >99% identical nucleotides, only one was used for phylogenetic analysis. Inferred amino acids of the unknown sequences were aligned with other g20 sequences from GenBank by the use of ClustalX (version 1.81) and the protein weight matrix Gonnet (32). The alignment was used to construct neighbor-joining trees with the Treecon program (33). Evolutionary distances were calculated by use of the correction of Tajima and Nei (31a), and the node reproducibility was estimated by a bootstrap analysis which included 1,000 replicates. Phylogenetic trees were drawn and visualized with Treecon (33).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been submitted to GenBank and assigned the accession numbers AY705091 to AY705146.
RESULTS
The CPS4 and G20-2 primer pair amplified g20 fragments from 3 of 4 cyanomyovirus isolates and 17 of 20 uncharacterized cyanophage isolates from the Gulf of Mexico, but not from 2 vibrophages, 4 coliphages, or 2 cyanophages belonging to the Podoviridae or Siphoviridae (data not shown). Also, the primer pair amplified gene fragments from almost every marine and freshwater sample screened. Most samples were collected from the euphotic zone, although one sample was collected from ca. 3,246 m deep in the Chuckchi Sea (Arctic Ocean). Temperatures in the environments ranged from <0°C in the Arctic and Southern Oceans to 26.8°C near the equator. DGGE community patterns resulted in the separation of 3 to 18 bands (Fig. 1).
FIG. 1.
Negative images of denaturing gradient gels of PCR-amplified g20 gene fragments. Labeled bands were excised from the gels, cloned, and then sequenced. Bands in the same lane with the same letter beside them (a, b, or c) had >99% identical nucleotides. From within each lane, only one sequence with >99% identical nucleotides was included in further analyses. Bands with labels that have an asterisk and are shown in bold were included in the phylogenetic analysis.
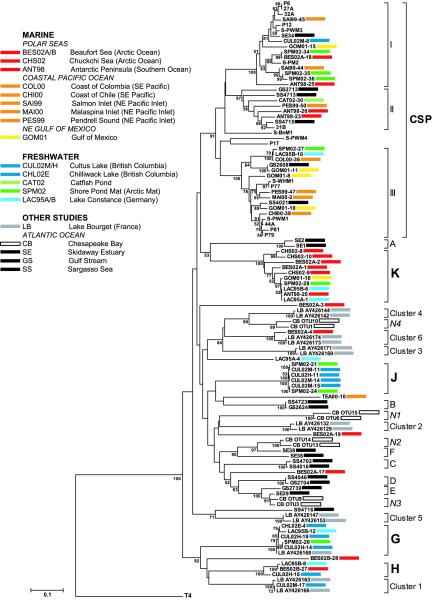
All g20 sequences amplified from cyanomyovirus isolates (including S-PWM3 and S-PWM4) clustered within the cultured Synechococcus phage (CSP) group (Fig. 2). Also, 22 of our 54 environmental sequences with <99% identical nucleotides fell into existing clades within the CSP group, while the remaining 32 sequences were either associated with four new clades or were not closely related to g20 sequences obtained in other studies. Only 2 of 15 sequences from the three lakes and 1 of 13 sequences from the Arctic Ocean clustered within the CSP group. Higher fractions of sequences from the Gulf of Mexico (four of five), the Southern Ocean (three of four), the Northeast Pacific inlets (five of six), the cyanobacterial mat (four of eight), the Southeast Pacific (two of two), and the catfish pond (one of one) were closely related to sequences in the CSP group.
FIG. 2.
Neighbor-joining tree of g20 fragments. Bootstrap values are shown as percentages to the lower left of the appropriate nodes. Sequences of cyanophage isolates do not have a bar next to them, and sequences with a black, white, or gray bar to the right are environmental g20 sequences that were obtained from GenBank. Environmental sequences obtained during the present study are indicated by colored bars, and the sequences of the cyanophage isolates obtained during the present study are named S-PWM3 and S-PWM4. The three letters in the sequence name refer to the location and the two numbers refer to the year of collection. Clusters A to F and I to III were assigned by Zhong et al. (41), clusters N1 to N4 were assigned by Wang et al. (35), and clusters 1 to 6 were assigned by Dorigo et al. (5). We assigned clusters G, H, J, K, and CSP. All clusters were assigned on the basis of phylogenetic relatedness. The scale bar represents the number of amino acid substitutions per residue.
Two of the four new clades were composed entirely of sequences from freshwater. One freshwater group (group G) included two sequences from Cultus Lake, British Columbia, one sequence each from the Arctic cyanobacterial mat, Lake Constance, Germany, and Chilliwack Lake, British Columia, and a sequence amplified from Lake Bourget, France (5). The other freshwater group (group J) contained four sequences from Cultus Lake and two from the Arctic cyanobacterial mat. In addition, a g20 sequence from Cultus Lake (CUL02M-17) and two sequences from Lake Bourget fell within cluster 1. The two other groups consisted of sequences from both freshwater and marine environments. Group H contained three sequences, from Cultus Lake, the Beaufort Sea, and Lake Constance. Group K contained three sequences from the Chuckchi Sea, two from the Beaufort Sea, and one sequence each from the northeast Gulf of Mexico, the Arctic cyanobacterial mat, Lake Constance, and the Southern Ocean.
One of the most surprising results was that sequences with >99% identical nucleotides were recovered from environments that differed substantially in temperature, salinity, and location. For example, sequences GOM01-16, SPM02-28, LAC95A-1, and ANT98-20 shared 99% identity and were recovered from the Gulf of Mexico, the Arctic cyanobacterial mat, Lake Constance, and the Southern Ocean, respectively. Similarly, sequences COL00-36, LAC95B-10, and SPM02-27 had >99% identity and occurred in samples from the Southeast Pacific, Lake Constance, and the Arctic cyanobacterial mat, respectively, as did sequences BES02-27 and LAC95B-8, which were recovered from the Beaufort Sea and Lake Constance.
DISCUSSION
Viruses are extremely abundant in lakes and oceans, yet little is known about the composition of these viral communities. In particular, the distribution of genotypes is virtually unexplored for bacteriophages. This study examined the distribution and genetic variation of a gene (g20) that occurs in viruses that infect cyanobacteria, one of the most important aquatic primary producers. This gene occurred in environments ranging from polar oceans and lakes to the Gulf of Mexico. Some groups of sequences with >99% identities occurred in environments that varied greatly in location, temperature, and salinity, while others were restricted to a single type of environment (e.g., freshwater). We also provided evidence that the g20 primers used for this study, and likely those used by other investigators, amplify DNAs from viruses other than cyanophages. These findings and their implications are discussed below.
We designed a degenerate downstream primer (G20-2) that, when used with a previously designed (40) nondegenerate upstream primer (CPS4), amplifies a g20 fragment from myoviruses infecting Synechococcus spp. but not from T4 or other myoviruses infecting heterotrophic bacteria. Limited testing showed that these primers were specific to cyanomyoviruses, consistent with the results of other investigators who tested similar primers (CPS1 and CPS8) (41). G20-2 and CPS4 target the same location on g20 as primers (CPS1 and CPS8) used by other investigators to amplify g20 sequences from the Atlantic Ocean (41), Chesapeake Bay (35), and Lake Bourget (5). These two primer pairs amplified some nearly identical sequences, but there were also g20 sequence groups that were amplified by only one of the primer pairs. The differences may have been because of differences in primer degeneracy. CPS4 is nondegenerate, while CPS1 has fourfold degeneracy, CPS8 has 12-fold degeneracy, and G20-2 has 256-fold degeneracy and includes one inosine residue, which increases the degeneracy of this primer to 1,024-fold without increasing the number of different primers.
As found previously (41), a group (CSP) of three clades contained all of the g20 sequences from cyanophage isolates. In addition, 22 of 54 environmental sequences from our study fell into this group, including most sequences from the Northeast Pacific inlets and the Gulf of Mexico. This was consistent with the high levels of abundance of Synechococcus and infectious cyanophages at these locations (7, 27, 28). Because of the presence of cyanobacteria, it was also not surprising that many sequences from the Arctic cyanobacterial mat and the sequence from a catfish production pond fell within the CSP group.
The majority of g20 sequences (32 of 54) were not closely related to sequences in the CSP group or to sequences from other studies. Many sequences that clustered outside the CSP group were from the Arctic Ocean and lakes in British Columbia and Germany. They formed four new phylogenetic groups (G, H, J, and K), two of which were entirely composed of sequences from freshwater. It seems likely that sequences outside the CSP group were not from cyanophages. For example, three Arctic Ocean sequences were from a depth of >3,000 m. The picocyanobacterial abundance in the Arctic Ocean is <102 cells ml−1 due to low growth rates and high losses from grazing, advection, and mixing (34). This is well below the abundance that would be expected to support significant lytic virus production (27). In addition, the half-life of virus particles in the sea is typically a few days due to adsorption to particles, solar radiation, and flagellate grazing (31). Given the low levels of abundance of cyanobacteria in Arctic surface waters, the inability of cyanobacteria to produce viruses below the photic zone, and the high turnover of viruses in the sea, it is most likely that these and other g20 sequences outside the CSP group are from bacteriophages that do not infect cyanobacteria. It is possible that infected cyanobacteria sink out of the photic zone or that particle-associated cyanophages were transported to these depths, but these should have been removed by prefiltration (0.2-μm-pore-size filter).
Three groups of indistinguishable sequences were recovered from environments that differed greatly in location, salinity, and temperature. The group with representatives from the widest range of environments included sequences from the Gulf of Mexico, the Southern Ocean, an Arctic ice-shelf meltwater pond, and Lake Constance. Nearly identical g20 gene sequences have also been obtained from cyanophage isolates from the Altamaha River estuary in Georgia (P77) (16) and Woods Hole, Mass. (S-WHM1) (38), and S-RIM1 from coastal Rhode Island (17) clusters close to S-PWM1 (28) from the Gulf of Mexico (17). The presence of almost identical sequences at the ends of long branches argues against genetic drift, which would be expected to produce a continuum of sequences with similar differences to each other. Similarly, >99% identical podophage DNA polymerase sequences occur in marine, freshwater, and terrestrial environments (2), and genetically related vibriophages are widely distributed in waters off Florida and Hawaii (14).
If closely related g20 genes are indicative of viruses that infect closely related hosts, as is the case for DNA polymerase genes of algal viruses (3, 23), then the results imply that closely related host cells and the viruses which infect them are distributed across a very broad range of environments. Alternatively, if these sequences are from phages that infect distantly related or unrelated hosts, it implies a broad horizontal exchange of g20 among phage communities (12). Evidence for the latter is provided by the coliphage T4 and the cyanophage S-PM2, which both contain a 10-kb genetic module that includes g20 (11). Alternatively, it was hypothesized that nearly identical T7-like DNA polymerase sequences in marine, freshwater, and terrestrial environments suggested that phages moved among these environments (2). Although the origin of identical gene sequences in different environments is open to speculation, evidence of broad mosaicism among tailed bacteriophages provides a compelling argument for horizontal exchange (12).
The present study provides the first data on the widespread distribution and sequence diversity of T4-like g20 genes in nature. The extent of this diversity is remarkable, as is the observation that nearly identical gene sequences can be found in very different environments. Our data strongly suggest that g20 sequences outside the CSP group are not from cyanophages, but confirmation of this awaits the isolation of representative phages from these groups. Further work will need to explore the mechanisms responsible for controlling the distribution and maintenance of specific g20 genotypes in nature.
Acknowledgments
We thank the crew and scientists aboard the research vessels Walton Smith, Mirai, Lawrence M. Gould, Nathaniel B. Palmer, and Vector for their help with sample collection. We are grateful to Amy Chan, Jessica Clasen, Barry Hurlburt, Derek Mueller, Steven Short, Warwick Vincent, and Steven Wilhelm for providing samples, Elizabeth Kutter for providing bacteriophages (T4, RB33, RB69, and LZ4), Nina Nemcek for help with sample preparation, Steven Short for help with phylogenetic analysis, Emma Hambly for comments on the manuscript, and Eddy Carmack for organizing the JWACS 2002 cruise.
This study was supported by an NSERC grant to C.A.S., by the Japan/Canada Western Arctic Climate Study (JWACS), and by the Department of Fisheries and Oceans Strategic Fund.
REFERENCES
- 1.Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 2.Breitbart, M., J. H. Miyake, and F. Rohwer. 2004. Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol. Lett. 236:249-256. [DOI] [PubMed] [Google Scholar]
- 3.Chen, F., and C. A. Suttle. 1996. Evolutionary relationships among large double-stranded DNA viruses that infect microalgae and other organisms as inferred from DNA polymerase genes. Virology 219:170-178. [DOI] [PubMed] [Google Scholar]
- 4.Chen, F., C. A. Suttle, and S. M. Short. 1996. Genetic diversity in marine algal virus communities as revealed by sequence analysis of DNA polymerase genes. Appl. Environ. Microbiol. 62:2869-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorigo, U., S. Jacquet, and J. F. Humbert. 2004. Cyanophage diversity, inferred from g20 gene analyses, in the largest natural lake in France, Lake Bourget. Appl. Environ. Microbiol. 70:1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fogg, G. E. 1995. Some comments on picoplankton and its importance in the pelagic ecosystem. Aquat. Microb. Ecol. 9:33-39. [Google Scholar]
- 7.Frederickson, C. M., S. M. Short, and C. A. Suttle. 2003. The physical environment affects cyanophage communities in British Columbia inlets. Microb. Ecol. 46:348-357. [DOI] [PubMed] [Google Scholar]
- 8.Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 9.Fuller, N. J., W. H. Wilson, I. R. Joint, and N. H. Mann. 1998. Occurrence of a sequence in marine cyanophages similar to that of T4 g20 and its application to PCR-based detection and quantification techniques. Appl. Environ. Microbiol. 64:2051-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall, T. A. 1999. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 11.Hambly, E., F. Tetart, C. Desplats, W. H. Wilson, H. M. Krisch, and N. H. Mann. 2001. A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc. Natl. Acad. Sci. USA 98:11411-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendrix, R. W., M. C. M. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hennes, K. P., C. A. Suttle, and A. M. Chan. 1995. Fluorescently labeled virus probes show that natural virus populations can control the structure of marine microbial communities. Appl. Environ. Microbiol. 61:3623-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellogg, C. A., J. B. Rose, S. C. Jiang, J. M. Thurmond, and J. H. Paul. 1995. Genetic diversity of related vibriophages isolated from marine environments around Florida and Hawaii, USA. Mar. Ecol. Progr. Ser. 120:89-98. [Google Scholar]
- 15.Li, W. K. W. 1994. Primary production of prochlorophytes, cyanobacteria, and eukaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol. Oceanogr. 39:169-175. [Google Scholar]
- 16.Lu, J., F. Chen, and R. E. Hodson. 2001. Distribution, isolation, host specificity, and diversity of cyanophages infecting marine Synechococcus spp. in river estuaries. Appl. Environ. Microbiol. 67:3285-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marston, M. F., and J. L. Sallee. 2003. Genetic diversity and temporal variation in the cyanophage community infecting marine Synechococcus species in Rhode Island's coastal waters. Appl. Environ. Microbiol. 69:4639-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Middelboe, M., A. Hagstrom, N. Blackburn, B. Sinn, U. Fischer, N. H. Borch, J. Pinhassi, K. Simu, and M. G. Lorenz. 2001. Effects of bacteriophages on the population dynamics of four strains of pelagic marine bacteria. Microb. Ecol. 42:395-406. [DOI] [PubMed] [Google Scholar]
- 19.Proctor, L. M., and J. A. Fuhrman. 1990. Viral mortality of marine bacteria and cyanobacteria. Nature 343:60-62. [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Scanlan, D. J. 2003. Physiological diversity and niche adaptation in marine Synechococcus. Adv. Microb. Physiol. 47:1-64. [DOI] [PubMed] [Google Scholar]
- 22.Short, S. M., and C. A. Suttle. 2000. Denaturing gradient gel electrophoresis resolves virus sequences amplified with degenerate primers. BioTechniques 28:20-22. [DOI] [PubMed] [Google Scholar]
- 23.Short, S. M., and C. A. Suttle. 2002. Sequence analysis of marine virus communities reveals that groups of related algal viruses are widely distributed in nature. Appl. Environ. Microbiol. 68:1290-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Short, S. M., and C. A. Suttle. 2003. Temporal dynamics of natural communities of marine algal viruses and eukaryotes. Aquat. Microb. Ecol. 32:107-119. [Google Scholar]
- 25.Suttle, C. A. 2000. The ecological, evolutionary and geochemical consequences of viral infection of cyanobacteria and eukaryotic algae, p. 248-286. In C. J. Hurst (ed.), Viral ecology. Academic Press, London, United Kingdom.
- 26.Suttle, C. A. 1994. The significance of viruses to mortality in aquatic microbial communities. Microb. Ecol. 28:237-243. [DOI] [PubMed] [Google Scholar]
- 27.Suttle, C. A., and A. M. Chan. 1994. Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl. Environ. Microbiol. 60:3167-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suttle, C. A., and A. M. Chan. 1993. Marine cyanophages infecting oceanic and coastal strains of Synechococcus—abundance, morphology, cross-infectivity and growth-characteristics. Mar. Ecol. Progr. Ser. 92:99-109. [Google Scholar]
- 29.Suttle, C. A., A. M. Chan, and M. T. Cottrell. 1990. Infection of phytoplankton by viruses and reduction of primary productivity. Nature 347:467-469. [Google Scholar]
- 30.Suttle, C. A., A. M. Chan, and M. T. Cottrell. 1991. Use of ultrafiltration to isolate viruses from seawater which are pathogens of marine phytoplankton. Appl. Environ. Microbiol. 57:721-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suttle, C. A., and C. Feng. 1992. Mechanisms and rates of decay of marine viruses in seawater. Appl. Environ. Microbiol. 58:3721-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Tajima, F., and M. Nei. 1984. Estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol. 1:269-285. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 34.Vincent, W. F. 2000. Cyanobacterial dominance in the polar regions, p. 321-340. In B. A. Whitton and M. Potts (ed.), The ecology of cyanobacteria: their diversity in time and space. Kluwer Academic Publishers, Boston, Mass.
- 35.Wang, K., and F. Chen. 2004. Genetic diversity and population dynamics of cyanophage communities in the Chesapeake Bay. Aquat. Microb. Ecol. 34:105-116. [Google Scholar]
- 36.Waterbury, J. B., and F. W. Valois. 1993. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl. Environ. Microbiol. 59:3393-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilhelm, S. W., and C. A. Suttle. 1999. Viruses and nutrient cycles in the sea—viruses play critical roles in the structure and function of aquatic food webs. Bioscience 49:781-788. [Google Scholar]
- 38.Wilson, W. H., I. R. Joint, N. G. Carr, and N. H. Mann. 1993. Isolation and molecular characterization of five marine cyanophages propagated on Synechococcus WH7803. Appl. Environ. Microbiol. 59:3736-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson, W. H., J. F. Nicholas, I. R. Joint, and N. H. Mann. 1999. Analysis of cyanophage diversity in a North-South transect of the Atlantic Ocean. Bull. Inst. Oceanogr. 19:209-216. [Google Scholar]
- 40.Wilson, W. H., J. F. Nicholas, I. R. Joint, and N. H. Mann. 2000. Analysis of cyanophage diversity in the marine environment using denaturing gradient gel electrophoresis, p. 565-570. In C. R. Bell, M. Brylinsky, and P. Johnson-Green (ed.), Microbial biosystems: new frontier. Proceedings of the 8th International Symposium on Microbial Ecology, Halifax, Nova Scotia, Canada. Atlantic Canada Society for Microbial Ecology, Halifax, Nova Scotia.
- 41.Zhong, Y., F. Chen, S. W. Wilhelm, L. Poorvin, and R. E. Hodson. 2002. Phylogenetic diversity of marine cyanophage isolates and natural virus communities as revealed by sequences of viral capsid assembly protein gene g20. Appl. Environ. Microbiol. 68:1576-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]