Abstract
The nuclear factor of activated T cells (NFAT) family proteins are transcription factors that regulate the expression of pro-inflammatory cytokines and other genes during the immune response. Although the NFAT proteins have been extensively investigated in the immune system, their role in cancer progression remains controversial. Here, we report that NFAT3 is highly expressed in various skin cancer cell lines and tumor tissues. Knockdown of endogenous NFAT3 expression by short hairpin RNA (shRNA) significantly inhibited tumor cell proliferation, colony formation and anchorage-independent cell growth. Furthermore, results of the mammalian two-hybrid assay showed that cyclin-dependent kinase 3 (CDK3) directly interacted with NFAT3 and phosphorylated NFAT3 at serine 259 (Ser259), which enhanced the transactivation and transcriptional activity of NFAT3. The phosphorylation site of NFAT3 was critical for epidermal growth factor (EGF)-stimulated cell transformation of the HaCaT immortalized skin cell line and mutation of NFAT3 at Ser259 led to a reduction of colony formation in soft agar. We also found that overexpressing wildtype NFAT3, but not mutant NFAT3-S259A, promoted A431 xenograft tumor growth. Importantly, we showed that CDK3, NFAT3 and phosphorylated NFAT3-Ser259 were highly expressed in skin cancer compared with normal skin tissues. These results provided evidence supporting the oncogenic potential of NFAT3 and suggested that CDK3-mediated phosphorylation of NFAT3 has an important role in skin tumorigenesis.
Introduction
The nuclear factor of activated T cell (NFAT) proteins are a group of transcription factors comprising five members, NFAT1 (also called NFATp or NFATc2), NFAT2 (also called NFATc or NFATcl), NFAT3 (also called NFATc4), NFAT4 (also called NFATx or NFATc3) and NFAT5 (also called TonEBP).1 NFATs function in the development of cardiac muscle,2 skeletal muscle3 and the nervous system,4 and are also involved in cell transformation, progression, metastasis and angiogenesis during tumor development.1 Among the NFAT family members, NFAT3 was reported as a negative regulator of Ras-JNK1/2-AP-1-induced NIH3T3 cell transformation.5 Knockdown of NFAT3 enhanced 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced anchorage-independent cell transformation of JB6 Cl41 cells.6 However, NFAT3 was overexpressed in a subset of breast cancer patients and knockdown of endogenous NFAT3 reduced the growth of human breast cancer cells.7 NFAT3 was also specifically required for tumor necrosis factor-alpha (TNF-α)-induced COX-2 expression and transformation of Cl41 cells.8 Moreover, accumulating experimental evidence revealed the critical role of NFAT3 in carcinogen-induced cell transformation and tumorigenesis.9, 10, 11 These findings indicated that the function of NFAT3 in cell transformation and cancer progression is still controversial and the underlying mechanism needs further investigation.
NFAT3 can be phosphorylated at Ser168 and Ser170 by p38 MAPK12 and at Ser213 and Ser217 by JNK1 and JNK2.5 Replacement of Ser168 and Ser170 with alanine promotes nuclear localization of NFAT3 and increases NFAT3-mediated transcription activity.12 However, mutation of the two sites phosphorylated by JNK1 and JNK2 suppresses NFAT3 transactivation.5 Furthermore, phosphorylation of NFAT3 by RSK2 leads to nuclear localization of activated NFAT3 and thus induces the differentiation of muscle cells.13 These findings suggested that phosphorylation is critical for the biological functions of NFAT3, including transcription activity, but whether other kinases are also involved in the phosphorylation of NFAT3 and associated cellular functions, such as cell transformation or tumorigenesis, have not been well elucidated.
Cyclin-dependent kinases (CDKs) have a critical role in the regulation of cell cycle progression. In many human cancers, including breast, liver, melanoma and lymphoma, a series of upstream regulators and downstream substrates of CDKs are involved in abnormal CDK-related signaling.14, 15, 16 Activation of CDK3 is first observed in G1 phase,17 and was reported to be critical for G1 exit and S entry.18 The dysfunction of CDK3 leads to G1 arrest, which cannot be rescued by the G1/S-restricted CDK2, indicating that CDK3 might have distinct functions in cell cycle regulation.19 Furthermore, CDK3 was reported to enhance Myc-induced proliferation and anchorage-independent growth of Ratl cells.20 We have previously shown that CDK3 enhances transformation of JB6 cells through the phosphorylation of ATF1.21 Knockdown of CDK3 suppressed ATF1 transactivation and inhibited cell proliferation and transformation.21 Phosphorylation of c-Jun by CDK3 induces AP-1 transactivation and thus enhances Ras-induced transformation of NIH3T3 cells.22 These findings suggested that in addition to cell cycle regulation, CDK3 might be also involved in the regulation of cell transformation, which is a critical event during tumor development.
In this study, we demonstrated that NFAT3 is highly expressed in skin cancer cell lines and a novel substrate of CDK3. NFAT3 can be phosphorylated by CDK3 at Ser259, which is critical for its transactivation activity and cell transformation. We also found that CDK3, NFAT3 and phosphorylated NFAT3-Ser259 were highly expressed in human skin cancer tissues compared with adjacent normal tissues. Our findings suggested that the CDK3–NFAT3 signaling axis might have a critical role in cell transformation during cancer progression.
Results
NFAT3 is a potential oncogene in skin cancer
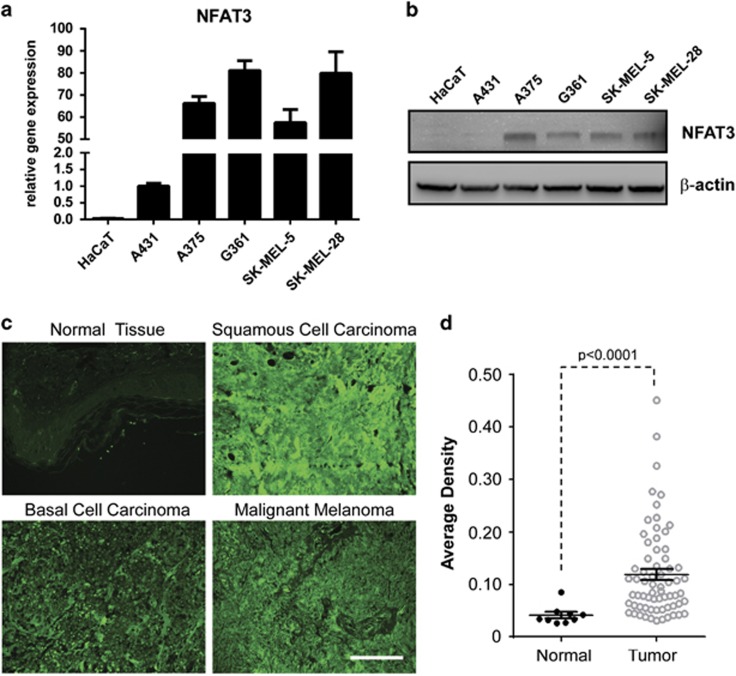
To investigate the potential role of NFAT3 in skin cancer, we first examined the expression of NFAT3 in normal and tumor skin cell lines. Results of quantitative PCR showed that the expression of NFAT3 was high in the A431, A375, G361, SK-MEL-5 and SK-MEL-28 skin cancer cell lines, but was markedly lower in the HaCaT immortalized skin cell line (Figure 1a). Consistent with the mRNA level, the NFAT3 protein level was also highly expressed in skin cancer cell lines, especially in malignant melanoma cells (Figure 1b). Next, we analyzed the expression of NFAT3 in normal and tumor skin tissues by immunofluorescence staining. As expected, compared with normal skin tissue, NFAT3 was highly expressed in tumor tissues regardless of cancer type (Figures 1c and d). These findings suggested that NFAT3 might be involved in skin cancer progression.
Figure 1.
NFAT3 is overexpressed in skin cancer cell lines. (a) NFAT3 expression was detected by quantitative PCR (qPCR) in the HaCaT immortalized skin cell line and the A431, A375, G361, SK-MEL-5 and SK-MEL-28 skin cancer cell lines. The relative gene expression was normalized by NFAT3 level in A431 cells. (b) The protein level of NFAT3 in these cell lines was analyzed by western blot. β-Actin was used as a loading control. (c) Normal and tumor skin tissues were analyzed for NFAT3 expression by immunofluorescence staining. A secondary antibody conjugated with Alexa Fluor 488 (green for NFAT3) was used for final detection. Scale bar, 100 μm. (d) NFAT3 staining was evaluated by average density and used for statistical analysis (P<0.0001).
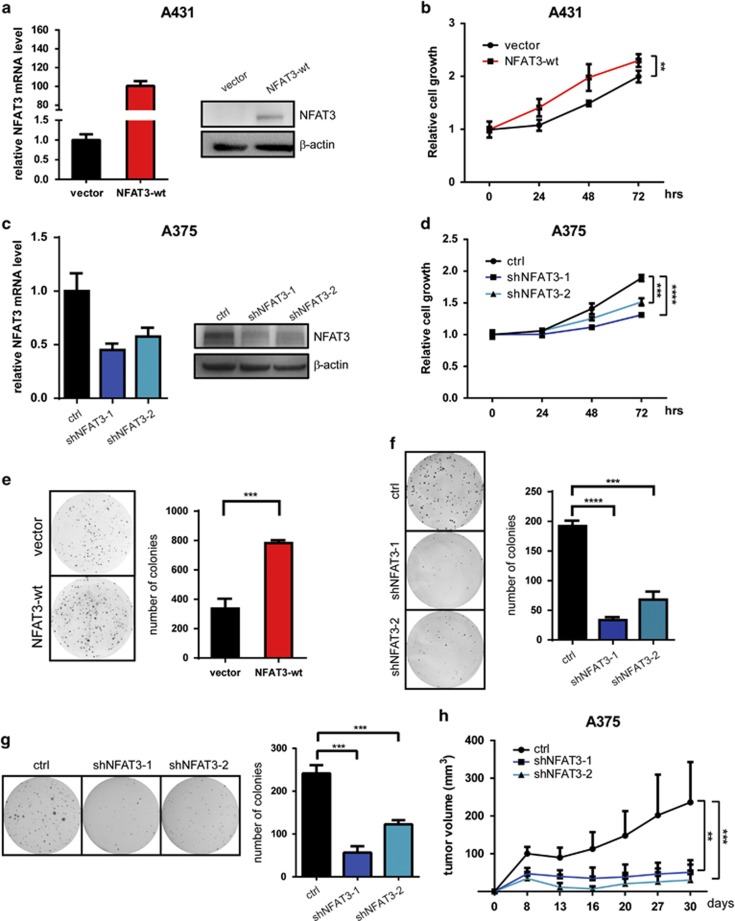
To further determine the function of NFAT3 in skin cancer progression, we then examined whether NFAT3 could regulate cell proliferation. Overexpression of wildtype NFAT3 by lentiviral infection in A431 cells increased the expression of NFAT3 at both the mRNA (Figure 2a, left panel) and protein levels (Figure 2a, right panel). Cell proliferation assessed by MTS assay showed that NFAT3 overexpression (OE) could significantly promote A431 cell growth (Figure 2b). On the other hand, knockdown (KD) of NFAT3 using two different short hairpin RNAs (shRNAs) in A375 cells with high endogenous NFAT3 levels reduced NFAT3 expression at both the mRNA (Figure 2c, left panel) and protein level (Figure 2c, right panel), and subsequently inhibited cell proliferation (Figure 2d), suggesting that NFAT3 functions in skin cancer cell proliferation and/or survival. In addition, we performed a 2-D colony formation assay in NFAT3-OE A431 cells or NFAT3-KD A375 cells. Stable expression of NFAT3 increased colony formation of A431 cells (Figure 2e), whereas NFAT3-KD by either of two different shRNAs abolished A375 cells colony formation (Figure 2f). Moreover, results of the anchorage-independent cell growth in a soft agar assay indicated that knockdown of NFAT3 expression was associated with strong inhibition of A375 cells sphere formation (Figure 2g), which was similar to the observations regarding colony formation in 2-D culture dishes. This indicated that NFAT3 is also important for the clonogeneic ability of skin cancer cell lines.
Figure 2.
NFAT3 has an oncogenic role in skin cancer cells. (a) OE of NFAT3 in A431 cells was detected by qPCR (left) or western blot (right). (b) Proliferation of stable A431 cells was measured by MTS assay. Results are expressed as mean values±s.d. from six replicate wells. (c) KD efficiency of NFAT3 in A375 cells was detected by qPCR (left) or western blot (right). (d) Proliferation of stable A375 cells was measured by MTS assay. (e and f) Representative photos and statistical analysis of 2-D colony formation in A431 stable cells with or without (e) NFAT3-OE and A375 stable cells with or without (f) NFAT3-KD. (g) Soft agar assay analysis of anchorage-independent growth of A375 stable cells with or without NFAT3-KD. Left panel shows representative photos. Right panel shows statistical analysis of colony number. (h) Xenograft tumor growth of A375 stable cells with or without NFAT3-KD in nude mice. Points indicate mean values (n=6); bars indicate s.d. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001.
To further examine NFAT3 function in tumorigenesis, we used a xenograft tumor model in which stable A375 cells were subcutaneously injected into athymic nude mice. In contrast with control mice, tumor growth of nude mice injected with NFAT3-KD A375 cells was clearly suppressed (Figure 2h) and even tumor regression was observed in several mice 2 weeks after transplantation. Altogether, these findings indicate that NFAT3 has a potential oncogenic role during skin cancer progression.
NFAT3 is a novel phosphorylation substrate of CDK3
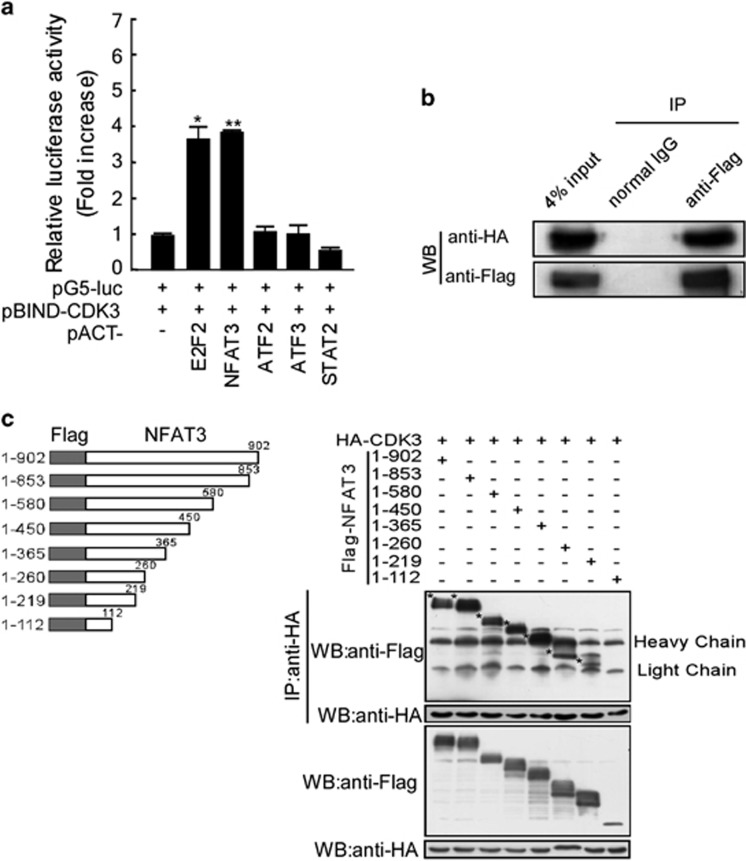
We previously reported that CDK3 interacted with and phosphorylated ATF1 at serine 63, which enhanced transformation of JB6 skin cells.21 Phosphorylation of c-Jun by CDK3 induces AP-1 transactivation and thus enhances Ras-induced transformation of NIH3T3 cells.22 These data indicate that CDK3 functions in the regulation of cell transformation and tumor development by phosphorylating some important transcription factors (TFs). To further explore the relationship of NFAT3 and CDK3, we first conducted a mammalian two-hybrid assay to determine whether NFAT3 can interact with CDK3. Briefly, each pACT-TF and the pG5-luc-reporter vector were co-transfected with pBIND-CDK3 into HEK293 cells. The interaction was evaluated by using the basal level of the pACT empty vector as a reference and E2F2 as a positive control.19 NFAT3 displayed a significant induction of luciferase activity, which was as high as that of the positive control E2F2, whereas the luciferase activities of other candidates were relatively low (Figure 3a). To verify the interaction between NFAT3 and CDK3, immunoprecipitation was performed using T98G cells transfected with HA-tagged CDK3 and Flag-tagged NFAT3 and we observed that NFAT3 was co-immunoprecipitated with CDK3 (Figure 3b). Moreover, the interaction between CDK3 and NFAT3 at their endogenous levels was further observed in melanoma cell line SK-MEL-28 (Supplementary Figure S1A). After confirming the interaction between NFAT3 and CDK3, a series of truncated NFAT3 proteins was generated and examined by in vitro immunoprecipitation to identify the specific domains that were involved in the interaction (Figure 3c, left panel). The shortest NFAT3 fragment (1–112aa) was not precipitated with HA-tagged CDK3 and the 1–219 fragment showed a relatively weak signal. This suggested that the first 112 amino acids of NFAT3 are not involved in CDK3 binding, whereas the major interacting domain of NFAT3 might be located in the fragment containing amino acids 219–902 (Figure 3c, right panel).
Figure 3.
CDK3 interacts with NFAT3. (a) The mammalian two-hybrid assay was performed to investigate the protein–protein interaction between CDK3 (as pBIND-CDK3) and transcription factors (as pACT-TFs). The binding activity was accessed as relative luciferase units normalized to the luciferase activity of pBIND-CDK3 with the empty pACT vector. The results were obtained from triplicate experiments and are shown as mean values±s.d. *P<0.05, **P<0.01. (b) HEK293 cells were transfected with Flag-tagged NFAT3 and HA-tagged CDK3 to perform the co-immunoprecipitation assay. HA-tagged CDK3 was precipitated with anti-Flag and detected with anti-HA by western blot. (c) T98G cells were transfected with HA-CDK3 and truncated Flag-NFAT3s as indicated. Anti-HA was used for immunoprecipitation and anti-Flag was introduced for the detection of precipitated truncated NFAT3 (labeled with *, right panel). The expression of truncated NFAT3 was also detected using anti-Flag.
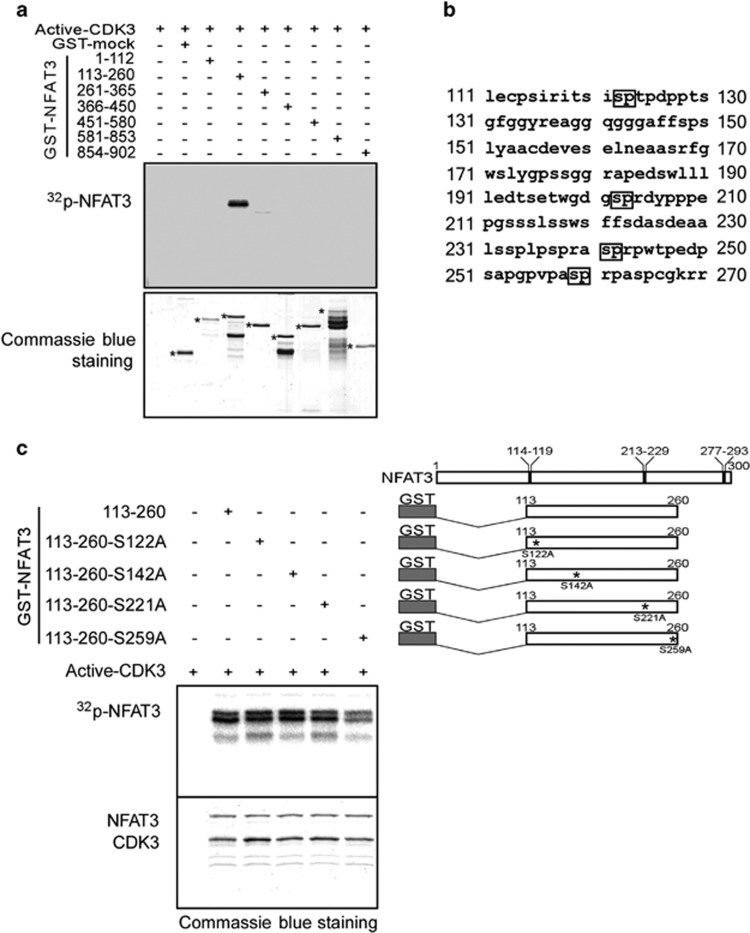
To further identify the site of NFAT3 that is phosphorylated by CDK3, a kinase assay was performed with a series of segmented NFAT3 proteins. The result revealed that amino acids 113–260 of NFAT3 are critical for CDK3 phosphorylation and NFAT3 is phosphorylated by CDK3 within this region (Figure 4a). The potential sites phosphorylated by CDK family members were analyzed for SP motifs and four putative sites in amino acids 113–260 were predicted (Figure 4b). Four mutant NFAT3 fragments were then generated and used for an in vitro kinase assay (Figure 4c, upper panel). Results indicated that the phosphorylation by CDK3 was much weaker when Ser259 was mutated (Figure 4c, lower panel). These results demonstrated that NFAT3 is a novel phosphorylation substrate of CDK3 and the phosphorylation site is Ser259. Considering CDK2 has the high-sequence identity with CDK3, and both of them are active in G1–S phase and essential for G1/S transition, we also examined whether CDK2 can phosphorylate NFAT3 on S259. However, CDK2 could neither co-immunoprecipitated with NFAT3 nor phosphorylated NFAT3 on S259 (Supplementary Figures S1B and C).
Figure 4.
CDK3 phosphorylates NFAT3 at Ser259. (a) GST-tagged truncated NFAT3 was used for an in vitro kinase assay with active CDK3. The phosphorylation was visualized by autoradiography (32P-NFAT3). Coomassie blue staining was used to verify equal loading of GST-tagged truncated NFAT3 proteins (labeled with *). (b) Sequence of NFAT3 from amino acids 111–260. The predicted SP motifs are indicated with black boxes. (c) GST-tagged wildtype NFAT3 fragments or GST-tagged mutant NFAT3 fragments were used for an in vitro kinase assay with active CDK3 as indicated (upper panel). The phosphorylation was visualized by autoradiography (32P-NFAT3) and Coomassie blue staining indicated the CDK3 protein and respective NFAT3 fragments (indicated as NFAT3 and CDK3).
CDK3 enhances the transactivation and transcription activity of NFAT3
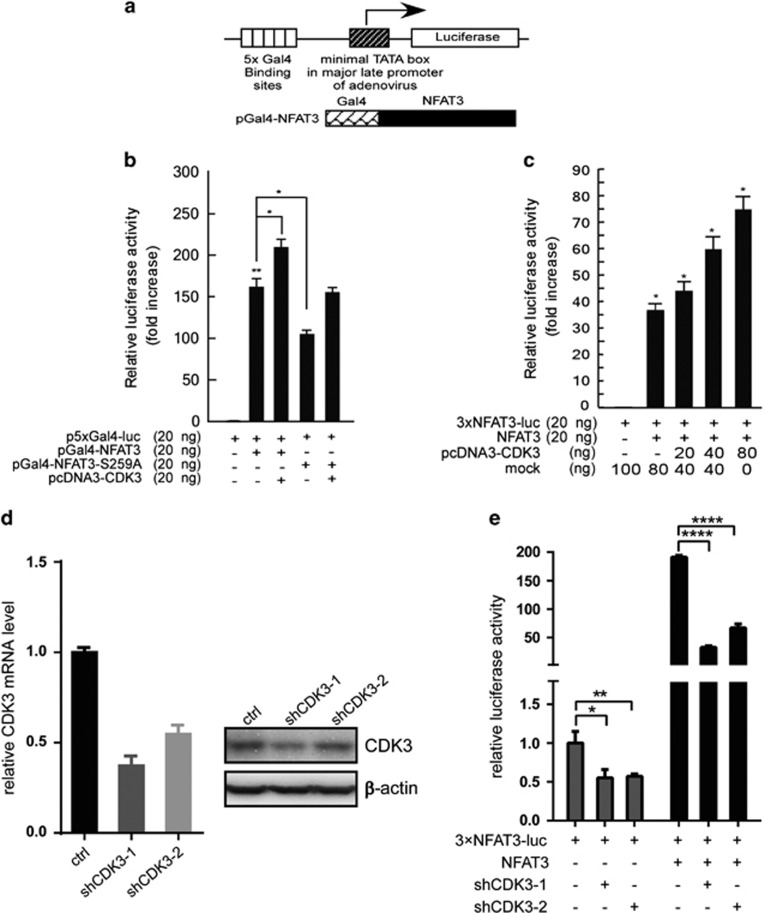
To investigate whether the phosphorylation of NFAT3 at Ser259 by CDK3 affects transactivation of NFAT3, a Gal4 DNA-binding domain–NFAT3 fusion protein was constructed and co-transfected with a luciferase reporter vector containing 5xGal4 binding sites (Figure 5a). This result revealed that co-transfection of CDK3 significantly increased the transactivation of NFAT3, whereas the transactivation was reduced when NFAT3 was mutated at Ser259 (Figure 5b). In general, the co-transfection of CDK3 increased the transactivation of mutated NFAT3 to a lesser degree compared with wildtype NFAT3.
Figure 5.
CDK3 enhances the transactivation and transcription activity of NFAT3. (a) The schematic indicates the construct of the luciferase reporter vector with 5xGal binding sites and the pGal4-NFAT3 expression vector. (b) For the luciferase assay, HEK293 cells were transfected with the p5xGal4-luc reporter vector, pGal4-NFAT3, pGal4-NFAT3-S259A and/or pcDNA3-CDK3 expression vector(s) to access the effect of phosphorylation at Ser259 on the transactivation of NFAT3. (c) The CDK3 expression vector was transfected in a dose-gradient manner with the NFAT3 expression vector and 3xNFAT3-luc reporter vector containing 3xNFAT-AP-1 binding sites. For these experiments, transactivation was accessed as relative luciferase units normalized to the luciferase activity of the mock control. (d) Knockdown efficiency of CDK3 in SK-MEL-28 cells was detected by qPCR (left) or western blot (right). (e) Stable SK-MEL-28 cells expressing CDK3 shRNAs were transiently transfected with the 3xNFAT3-luc reporter with or without NFAT3 to access NFAT3 luciferase activity. The results were obtained from triplicate experiments and are shown as mean values±s.d. *P<0.05, **P<0.01. ****P<0.0001.
To further confirm the role of CDK3 in regulating NFAT3 transcription activity, increasing amounts of CDK3 were co-transfected with NFAT3 and a 3xNFAT-luc reporter vector, which contains three NFAT-AP-1 binding sites from the promoter region of IL-2.1 A luciferase assay was performed and this result indicated that CDK3 enhanced NFAT3 transcription activity in a dose-dependent manner (Figure 5c). Moreover, the luciferase activity of NFAT3 was examined under the knockdown of endogenous CDK3 in two melanoma cell lines, SK-MEL-28 and A375. Although knockdown of CDK3 using two different shRNAs in SK-MEL-28 cells reduced CDK3 expression at both the mRNA level (Figure 5d, left panel) and protein level (Figure 5d, right panel), the luciferase activity of NFAT3 was obviously decreased, regardless of NFAT3 overexpression or not (Figure 5e). Similar results were observed in the other cell line A375 (Supplementary Figures S2A and B), supporting the important role of CDK3 on NFAT3 transcription activity. Meanwhile, the endogenous phosphorylation level of NFAT3 was increased with CDK3 overexpression in A375 cells, and its phosphorylation was decreased with CDK3 knockdown (Supplementary Figure S2C), further indicating that CDK3 is critical for NFAT3 phosphorylation.
The CDK3–NFAT3 signaling axis has an important role in cell transformation and tumor growth
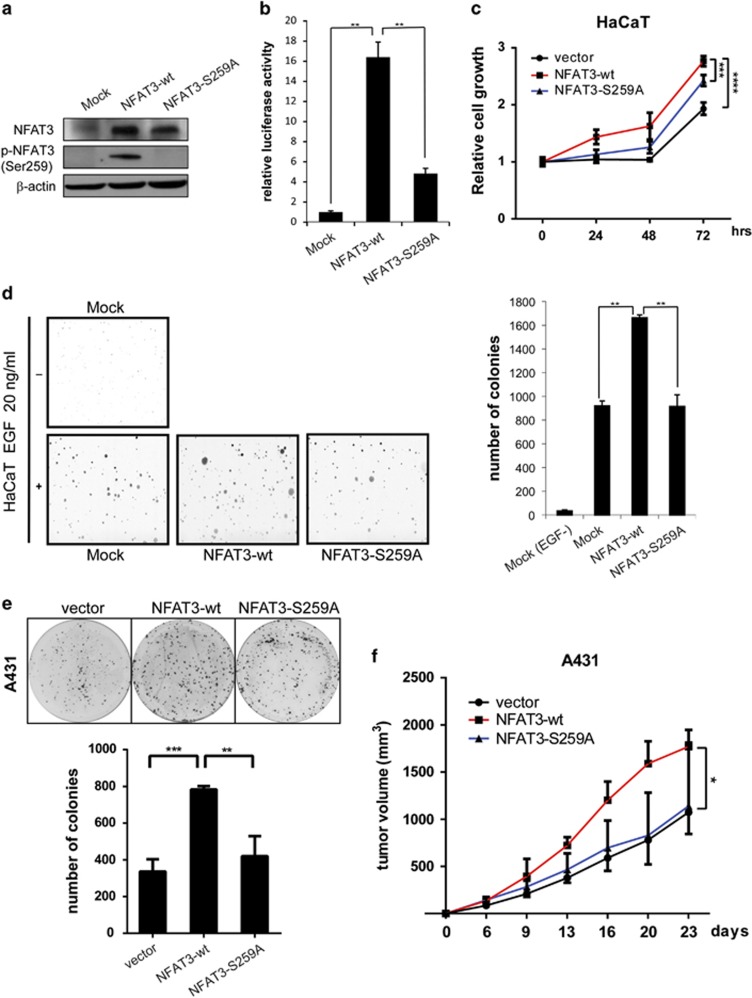
Because NFAT3 is specifically required for TNF-α-induced cyclooxygenase-2 (COX-2) expression and transformation of JB6 Cl41 cells,8 we determined whether CDK3-mediated NFAT3 phosphorylation is involved in proliferation and cell transformation. We previously reported that HaCaT cells expressed endogenous CDK322 and were sensitive to stimulation with epidermal growth factor (EGF).23 Herein, we infected HaCaT cells with lentivirus carrying wildtype NFAT3, mutant NFAT3-S259A or NFAT3 shRNAs. The expression of wildtype or mutant NFAT3 was evaluated by western blot (Figure 6a). As expected, the mutation of NFAT3 at Ser259 significantly perturbed NFAT3 transcription activity in HaCaT cells (Figure 6b). Further, we observed that the overexpression of wildtype NFAT3 significantly increased the proliferation of HaCaT cells, whereas the mutation at Ser259 reduced proliferation (Figure 6c). To further confirm the role of NFAT3 in cell proliferation, NFAT3-knockdown HaCaT cells were constructed. Results indicated that knockdown of NFAT3 expression significantly reduced cell growth compared with mock control (Supplementary Figure S3A).
Figure 6.
NFAT3 phosphorylation is responsible for cell transformation, proliferation and tumor growth. (a) HaCaT cell lysates were examined using anti-NFAT3 or anti-phosphorylated NFAT3 (Ser259). (b) Stable HaCaT cells expressing wildtype or mutant NFAT3 were transiently transfected with the 3xNFAT3-luc reporter to access NFAT3 luciferase activity. Results are shown as relative luciferase units normalized to the luciferase activity of the mock control. The results were obtained from triplicate experiments and are shown as mean values±s.d. (c) The MTS assay was performed to assess the effect of mutant NFAT3 on proliferation of HaCaT cells. (d) An EGF-stimulated (20 ng/ml) soft agar assay was performed using HaCaT cells stably expressing wildtype NFAT3 or mutant NFAT3. Left panel shows representative photos. Right panel shows statistical analysis of colony number. (e) Representative photos and statistical analysis of 2-D colony formation of A431 stable cells expressing wildtype NFAT3 or mutant NFAT3. (f) Xenograft tumor growth of A431 stable cells expressing wildtype NFAT3 or mutant NFAT3 in nude mice. Points indicate mean values (n=6); bars indicate s.d. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001.
To investigate the role of the CDK3–NFAT3 signaling axis in cell transformation, we also performed an anchorage-independent cell transformation assay. With EGF-stimulation, ~1700 colonies were observed in each well of NFAT3-transfected HaCaT cells, whereas the number of colonies was reduced to around 900 in mutant NFAT3-S259A cells (Figure 6d). Moreover, in NFAT3-knockdown HaCaT cells, the introduction of NFAT3 shRNA significantly reduced the EGF-stimulated colony formation by ~80% (Supplementary Figure S3B). Moreover, the enhanced colony formation ability of A431 cells induced by wildtype NFAT3 was almost completely abolished by mutant NFAT3-S259A (Figure 6e). To further determine whether NFAT3 phosphorylation functions in tumorigenesis, A431 cells stably expressing empty vector, NAFT3-wt or mutant NFAT3-S259A were subcutaneously injected into athymic nude mice. Notably, NFAT3-wt promoted A431 xenograft tumor growth, whereas mutant NFAT3-S259A was comparable to vector, supporting the idea that Ser259 phosphorylation is necessary for the oncogenic function of NFAT3 (Figure 6f). These results demonstrated that CDK3-mediated phosphorylation of NFAT3 at Ser259 has a critical role in cell transformation, proliferation and tumor growth.
High expression levels of CDK3 and phosphorylated NFAT3 are associated with skin cancer
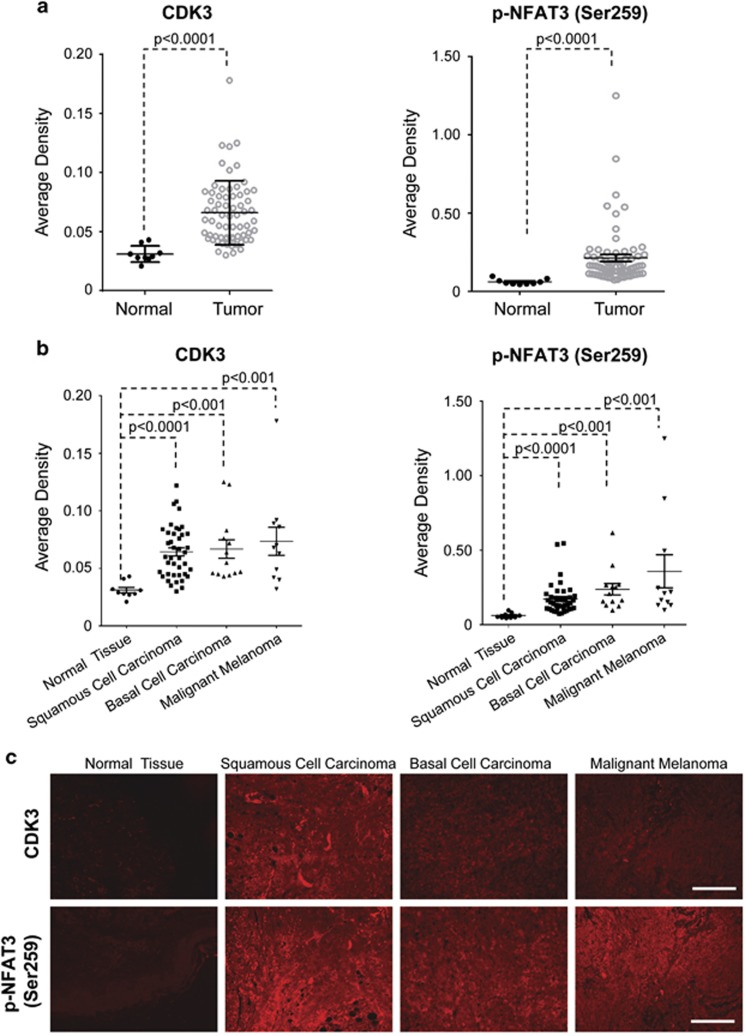
As CDK3–NFAT3 signaling is important for cell transformation, we further sought to determine whether CDK3 and NFAT3 are associated with malignancy in clinical samples. Human skin tissue arrays were examined by immunofluorescence staining for CDK3 and phosphorylated NFAT3 at Ser259 (pNFAT3-S259). We found that CDK3 was highly expressed in skin cancer tissues and high levels of NFAT3 phosphorylation at Ser259 were also detected (Figure 7a). Further analysis revealed that, compared with normal tissue, the expression level of CDK3 was enhanced in squamous cell carcinoma, basal cell carcinoma and melanoma. Interestingly, the high phosphorylation level of NFAT3 at Ser259 was observed in all three types of skin cancers, which corresponded with the expression level of CDK3 (Figures 7b and c). We then analyzed the correlation between CDK3 and phosphorylated NFAT3-Ser259, also between CDK3 and NFAT3 in human skin cancer tissues and adjacent normal tissues. The results showed that CDK3 expression was positively correlated with both NFAT3 and phosphorylated NFAT3-Ser259 (Supplementary Figure S4), further supporting the critical role of CDK3–NFAT3 signaling in cancer progression.
Figure 7.
CDK3 and phosphorylated NFAT3 are overexpressed in human skin cancer. (a) Immunofluorescence staining was used to investigate the expression of CDK3 and phosphorylated NFAT3 (Ser259) in human skin cancer (n=65) and normal tissues (n=9). The staining was evaluated by average density and used for statistical analysis (P<0.0001). (b) Tumor tissues were further divided by subtype: squamous cell carcinoma (n=41), basal cell carcinoma (n=13) and malignant melanoma (n=11). The average density was calculated by dividing the integrated density by the positive area. The data was presented as mean values of intensity score and P-value (Mann–Whitney U test). (c) Representative photos of immunofluorescence staining performed on the SK801b skin cancer tissue arrays as described in ‘Materials and methods' section. Tissues were incubated with antibodies to detect CDK3 or phosphorylated NFAT3 (Ser259). A secondary antibody conjugated with Alexa Fluor 647 (red for CDK3 or phosphorylated NFAT3) was used for final detection. Scale bar, 100 μm.
Discussion
Deregulation of cell cycle driven by abnormal cyclin/CDK activation is associated with carcinogenesis.14, 15 We previously reported that CDK3 is highly expressed in human cancer cells and CDK3-related signaling is a critical trigger for cell transformation.21 Many CDK3 substrates have been identified, including ATF1, c-Jun, pRb and IK3-1/Cables.21, 22, 24, 25 In this study, we found that NFAT3 and CDK3 physically interact with each other and NFAT3 is a novel phosphorylation substrate of CDK3, which phosphorylates NFAT3 at Ser259.
The activation of NFAT3 depends on the dephosphorylation of serine residues in a series of serine-rich motifs, SPs and SRR-1, which are highly conserved among NFAT family members.26 The SRR-1 region is a critical region responsible for nuclear import of NFATs, whereas the SP motifs regulate DNA-binding affinity and nuclear export.27, 28 NFAT3 contains three SP motifs in which SP1 is located between amino acids 213–229 and SP2 is located at amino acids 277 to 293. Phosphorylation at Ser213 and Ser217 by JNK1/2 reportedly leads to NFAT3 transactivation.13 In this study, we observed that phosphorylation at Ser259, which is located between the SP1 and SP2 motifs, is important for the transactivation of NFAT3. These findings suggested that not only are SRR-1 and SP motifs important for the transactivation of NFAT3, the phosphorylation between the SP motifs is also critical for the activation of NFAT3.
Because NFAT3 is a transcription factor and phosphorylation of NFAT3 could enhance the transactivation,21 we sought to investigate whether the phosphorylation of NFAT3 at Ser259 by CDK3 affected the transcriptional function of NFAT3. We demonstrated that the transcription induction of NFAT3 was significantly enhanced in the presence of CDK3, whereas the Ser259 NFAT3 mutant lost the enhanced transactivation. However, overexpression of CDK3 moderately increased the induction of mutant NFAT3 in which the transactivation was not fully recovered to that of wildtype NFAT3, suggesting other phosphorylation sites might also be involved in the CDK3-mediated NFAT3 transactivation. NFAT3 is known to interact with estrogen receptors α and β to up-regulate downstream pS2 and cathepsin D proteins, and knockdown of NFAT3 reduces proliferation of human breast cancer ZR75-1 cells.7 However, NFAT3 was also reported to cooperate with estrogen receptor α to inhibit cell mobility by down-regulation of lipocalin 2 in human breast cancer T-47D cells.29 These findings indicated that the role of NFAT3 in cancer might strongly depend on its downstream signaling. We previously established that CDK3 participates as a downstream kinase of the EGFR-Ras pathway21 and CDK3 is also involved in carcinogenesis by interacting with a series of downstream targets, including Myc, c-Jun, ATF-1 and AP-1.20, 21, 22 However, very little research has focused on the role of CDK3 phosphorylation of NFAT3 in cancer development.
Knockdown of NFAT3 enhanced TPA-induced transformation of JB6 Cl41 cells.8 However, NFAT3 also enhanced TNFα-induced anchorage-independent growth of JB6 Cl41 cells.8 In our anchorage-independent cell transformation assay to investigate the effect of CDK3–NFAT3 signaling in the presence of EGF, NFAT3-transfected HaCaT cells displayed a significant enhancement in colony formation compared with mutant NFAT3-S259A-transfected cells, suggesting that CDK3 phosphorylation at Ser259 is critical for the crucial role of NFAT3 in EGF-stimulated cell transformation. Further removal of endogenous NFAT3 by RNAi confirmed a critical role of CDK3/NFAT3 in cell transformation. Furthermore, we demonstrated that the overexpression of NFAT3 significantly increases proliferation, whereas knockdown of NFAT3 reduces the growth of HaCaT cells. For the first time, we demonstrated that NFAT3, as a novel CDK3 downstream target, is involved in proliferation and transformation of HaCaT cells, and blockage of CDK3–NFAT3 by the NFAT3-S259A mutant suppressed cell proliferation and transformation.
In summary, we found that NFAT3 is a novel substrate of CDK3 and that the CDK3–NFAT3 signaling axis has an important role in cell transformation and tumor growth. We also demonstrated that both CDK3 and NFAT3 are highly expressed in various types of human skin cancers and that the phosphorylation of NFAT3 (Ser259) is significantly higher in tumor tissues compared with normal skin tissue. Our findings suggest that abnormal CDK3–NFAT3 signaling may contribute to skin cancer development and a cross-talk among CDK3, EGFR, Ras, ATF-1 and NFAT3 indicate a complex signaling pathway of EGF-induced cell transformation in many human cancers, which may provide evidence for developing a series of potential drug targets for skin cancer therapy.
Materials and methods
Reagents, enzymes and antibodies
Chemical reagents and reaction buffers were obtained from Sigma-Aldrich (St. Louis, MO, USA). Restriction enzymes were purchased from Roche Diagnostics (Indianapolis, IN, USA). DNA polymerase and DNA ligase were from Qiagen (Valencia, CA, USA) and TAKARA Bio. (Kusatsu, Shiga, Japan), respectively. The Checkmate Mammalian Two-Hybrid System (Promega, Madison, WI, USA) was used in this study. Cell culture media and other reagents were obtained from Life Technologies Inc. (Grand Island, NY, USA).
Lysis buffer for the mammalian two-hybrid assay was prepared with 0.1 m potassium phosphate buffer (pH 7.8), 2 mm EDTA, 1 mm DTT and 1% Triton X-100. The NP-40 lysis buffer for western blotting was prepared with 50 mm Tris–HCl (pH 8), 150 mm NaCl and 0.5% NP-40 as previously described.21 The immunoprecipitation buffer (IP lysis buffer) was prepared with 25 mm Tris–HCl (pH 7.5), 5 mm β-glycerophosphate, 0.1 mm Na3VO4, 10 mm MgCl2, 1 mm aprotinin and 1 mm PMSF as previously reported.22
CDK3 (sc-826), pNFAT3-Ser259 (sc-32986) and HA-tag (sc-7392) antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). NFAT3 antibody (ab99431) was purchased from Abcam (Cambridge, MA, USA). Antibodies to detect the Flag-tag (TA180144Z), Myc-tag (TA150121Z) were obtained from Origene Technologies (Rockville, MD, USA). CDK2 antibody (10122-1-AP) was purchased from Proteintech (Rosemont, IL, USA).
Cell culture and transfection
HEK293 and T98G were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The normal HaCaT human keratinocyte cell line and the A431, A375, G361, SK-MEL-5, SK-MEL-28 skin cancer cell lines were kindly provided by Prof. Cong Peng (Changsha, Hunan, China). The above cells were cultured in Dulbecco's minimum essential medium (DMEM) with 10% fetal bovine serum (FBS) at 37 °C in a 5% CO2 incubator. T98G cells were cultured in minimum essential medium (MEM) supplemented with 2 mm l-glutamine, 0.1 mm non-essential amino acids, 1.0 mm sodium pyruvate and 10% FBS at 37 °C in a 5% CO2 incubator. These cells are regularly tested to ensure that they are mycoplasma free. Transfection of cells with the reporter and expression vectors was performed using the X-tremeGENE HP DNA transfection reagent (Roche Diagnostics) according to the manufacturer's instructions.
Construction of reporter and expression vectors
The expression vectors for CDK3 (pBIND-CDK3, pcDNA3-CDK3 and pRcCMV-HA-CDK3), transcription factors (pACT-E2F1, pACT-E2F2, pACT-ATF2, pACT-ATF3 and pACT-STAT2) and the luciferase reporter p5xGal4-luc were constructed and stored as previously described.21
Full-length human NFAT3 cDNA and truncated NFAT3 were amplified from a human mRNA pool generated by RT-PCR using SuperScript II RNase H Reverse Transcriptase (Life Technologies). The NFAT3 cDNA was subsequently cloned into the pACT vector as previously reported.21 The pGal4 expression vector containing the DNA-binding domain of Gal4 was used to generate the Gal4-NFAT3 fusion protein (pGal4-NFAT3). The full-length NFAT3 and a series of the truncated NFAT3 were introduced into the p3xFLAG plasmid. The sequences were confirmed by direct sequencing.
The expression vector containing the glutathione S-transferase tag (GST-tag, pGEX-5X-C) was used for the construction of the GST-tagged full-length NFAT3 and truncated NFAT3. Mutagenesis was performed to generate mutated GST-tagged truncated NFAT3 (GST-NFAT3-113-260 S122A, S142A, S221A and S259A) and pGal4-NFAT3-S259A using the Quick Change Lightning Site-directed Mutagenesis Kit (Agilent Life Sciences, Santa Clara, CA, USA) according to the manufacturer's protocol.
Mammalian two-hybrid assay
The mammalian two-hybrid assay was performed as previously reported.21 Briefly, the pG5-luc reporter vector and pBIND-CDK3 and pACT-TF expression vectors were transfected into HEK293 cells at a molecular ratio of 1:1:1 and the cells were cultured for 36 h (h). The cells were then disrupted and the cell lysate was then analyzed for luciferase activity using a luciferase assay kit (Promega). The relative luciferase activity was calculated and normalized to the luciferase activity with only pG5-luc and pBIND-CDK3 transfection.
Immunoprecipitation and western blotting
T98G cells transfected with Flag-tagged NFAT3 and HA-tagged CDK3 were cultured in a 100-mm dish and cells were collected at 80% confluence in IP lysis buffer. After clarification, the supernatant fractions were used for immunoprecipitation with anti-HA. Proteins precipitated from transfected cells in the IP assay or extracted by NP-40 lysis buffer were analyzed by western blotting. Protein concentration was determined using the Bio-Rad DC Protein Assay (Bio-Rad Laboratories, Hercules, CA, USA). The protein samples were separated by electrophoresis, transferred to polyvinylidene fluoride membranes, hybridized with corresponding antibodies and detected using the Pierce ECL Western blotting substrate (Thermo Scientific, Rockford, IL, USA).
In vitro kinase assay
GST-tagged truncated NFAT3 fusion proteins were expressed in BL21 cells induced by 0.5 mm IPTG at 25 °C for 3 h and purified using GST beads. Purified fusion proteins (200 ng) expressing each truncated NFAT3 were respectively used for an in vitro kinase assay with 100 ng of active CDK3 (Upstate Biotechnology, Lake Placid, NY, USA). The reaction was performed at 30 °C for 30 min in 1 × kinase assay buffer containing 50 μm unlabeled ATP (Upstate Biotechnology) with or without 10 μCi of [γ-32P] ATP. The proteins were subsequently analyzed by electrophoresis with 12% SDS–PAGE and visualized by autoradiography. After autoradiography, the gel was also stained with Coomassive Brilliant Blue R250 (Bio-Rad Laboratories) according to the manufacturer's instructions.
NFAT3 transactivation assay
The p5xGal4-luc reporter vector and pcDNA3-CDK3 were co-transfected with pGal4-NFAT3 or the pGal4-NFAT3-S259A mutant into T98G cells. The cells were cultured under normal conditions for 36 hrs and then disrupted using lysis buffer. The lysate was then analyzed for luciferase activity using the Dual-Luciferase assay kit (Promega) and firefly luciferase activity was normalized against Renilla luciferase activity.
Knockdown of NFAT3 or CDK3
NFAT3 or CDK3 knockdown cells were generated using lentivirus infection with expression of NFAT3 or CDK3 shRNA. The shRNA oligos were synthesized by Life Technology and cloned into the pLKO.1 expression construct (using pLKO.1-scramble shRNA as control). The pLKO.1-shRNA plasmid was co-transfected into 293 T cells together with the psPAX2 packaging plasmid and envelope plasmid pMD2.G for production of the shRNA lentivirus. After 48 h, the supernatant fractions from the cultures were collected and filtered through a 0.45 μm filter. Cells were then infected with the viral supernatant fractions supplemented with polybrene. The culture medium was replaced with fresh growth medium with puromycin for selection at 16 h post-infection. The cells were cultured in selected medium until control cells completely died and the knockdown efficiency was then evaluated by qPCR and western blot.
Anchorage-independent cell transformation assay
HaCaT cells were stably transfected with NFAT3 or mutant NFAT3-S259A for an EGF-induced cell transformation assay. Cells (8 × 103) were cultured in DMEM with 0.3% agar (Sigma-Aldrich), 20 ng/ml EGF (BD Biosciences, San Jose, CA, USA) and 10% FBS. The cells were maintained at 37 °C in a 5% CO2 incubator for 10 days and then colonies were counted and scored using Image-Pro Plus software as described by Colburn et al.30
MTS assay
A CellTiter 96 MTS assay (Promega) was performed according to the manufacturer's instructions. Briefly, HaCaT or tumor cells were seeded in 96-well plates and viability was evaluated at 0, 24, 48 or 72 h of incubation.
Immunofluorescence staining
A human skin tissue array SK801b (US Biomax, Rockville, MD, USA) was deparaffinized and rehydrated before retrieving the antigen with 10 mm sodium citrate buffer. After blocking with 5% normal goat serum at room temperature for 1 h, tissues were incubated overnight with antibodies to detect CDK3, NFAT3 or pNFAT3 (Ser259) at 4 °C. On the second day, the tissues were incubated for 2 h at room temperature with the Alexa Fluor 647 (red for CDK3) or Alexa Fluor 488 (green for NFAT3) conjugated secondary antibody and protected from light. Images were captured by laser scanning confocal microscopy (NIKON C1si Confocal Spectral Imaging System, NIKON Instruments Inc., Tokyo, Japan) and analyzed with ImageJ software as previously described.21 The images were converted to 8-bit format and the mean value of gray levels was used as the threshold for positive area and the average density was calculated by dividing the integrated density by the positive area.31
Xenografts in athymic nude mice
Forty 6-week-old male BALB/c nu/nu mice were randomized into five groups. A431 cells (5 × 106) with NFAT3-wt or NFAT3-S259A overexpression or A375 cells with NFAT3-knockdown were subcutaneously injected at right back of mice. Tumor volume was monitored twice a week and mice were killed when tumors were >1.5 cm in diameter at the widest dimension of the tumor. All mice were sacrificed 4 weeks later for tumor weight analysis. The investigator who injected the mice was blinded to these groupings. All mice were housed in a specific pathogen free environment at Shenzhen University Health Science Center and treated in strict accordance with protocols approved by the Institutional Animal Use Committee of the Health Science Center, Shenzhen University.
Statistical analysis
All in vitro experiments were performed at least in triplicate. The results of each experiment are shown as the mean of experimental replicates. Data are presented as the mean±s.d. Two-tailed Student's t-test was used to compare the difference between two groups of data sets with similar variance. For two groups of standard deviations about 1 and average differences larger than 3, it would be sufficient (power 0.7) to detect the effect at a sample size of 3. For all tests, P<0.05 was considered statistically significant.
Linear regression curves were plotted by GraphPad Prism Version 6.01 (GraphPad Software Inc., La Jolla, CA, USA). using the X–Y scatter graph function. A linear trendline was established and the correlation coefficient was determined by Pearson test for the correlation coefficient (r). The P-value was determined using a significance calculator examining the r-value and the number of trials.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81071655, 81372149, 81401894, 81402289), the National Science Foundation Projects of Guangdong Province (2014A030313547), the High level Talents Project of Guangdong Province (2013), the Shenzhen Municipal Government of China (KQCX20140519104925300, JCYJ20140418193546118, JCYJ20140418091413510, ZDSY20130329101130496) and Natural Science Foundation of SZU (201406). We thank Dr Barrett J. Rollins for the pRcCMV-cdk3 (pCMV-cdk3); Dr Yong-Yeon Cho for the pACT-TFs and Dr Ke Yao for pGal4-NFAT3 and p3xFlag-NFAT3.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Mancini M, Toker A. NFAT proteins: emerging roles in cancer progression. Nat Rev Cancer 2009; 9: 810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef IA, Chen F, Chen L, Kuo A, Crabtree GR. Signals transduced by Ca(2+)/calcineurin and NFATc3/c4 pattern the developing vasculature. Cell 2001; 105: 863–875. [DOI] [PubMed] [Google Scholar]
- Friday BB, Pavlath GK. A calcineurin- and NFAT-dependent pathway regulates Myf5 gene expression in skeletal muscle reserve cells. J Cell Sci 2001; 114: 303–310. [DOI] [PubMed] [Google Scholar]
- Kao SC, Wu H, Xie J, Chang CP, Ranish JA, Graef IA et al. Calcineurin/NFAT signaling is required for neuregulin-regulated Schwann cell differentiation. Science 2009; 323: 651–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao K, Cho YY, Bergen HR 3rd, Madden BJ, Choi BY, Ma WY et al. Nuclear factor of activated T3 is a negative regulator of Ras-JNK1/2-AP-1 induced cell transformation. Cancer Res 2007; 67: 8725–8735. [DOI] [PubMed] [Google Scholar]
- Li J, Song L, Zhang D, Wei L, Huang C. Knockdown of NFAT3 blocked TPA-induced COX-2 and iNOS expression, and enhanced cell transformation in Cl41 cells. J Cell Biochem 2006; 99: 1010–1020. [DOI] [PubMed] [Google Scholar]
- Zhang H, Xie X, Zhu X, Zhu J, Hao C, Lu Q et al. Stimulatory cross-talk between NFAT3 and estrogen receptor in breast cancer cells. J Biol Chem 2005; 280: 43188–43197. [DOI] [PubMed] [Google Scholar]
- Yan Y, Li J, Ouyang W, Ma Q, Hu Y, Zhang D et al. NFAT3 is specifically required for TNF-alpha-induced cyclooxygenase-2 (COX-2) expression and transformation of Cl41 cells. J Cell Sci 2006; 119: 2985–2994. [DOI] [PubMed] [Google Scholar]
- Ding J, Li J, Xue C, Wu K, Ouyang W, Zhang D et al. Cyclooxygenase-2 induction by arsenite is through a nuclear factor of activated T-cell-dependent pathway and plays an antiapoptotic role in Beas-2B cells. J Biol Chem 2006; 281: 24405–24413. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Hu Y, Li J, Ding M, Lu Y, Zhang D et al. Direct evidence for the critical role of NFAT3 in benzo[a]pyrene diol-epoxide-induced cell transformation through mediation of inflammatory cytokine TNF induction in mouse epidermal Cl41 cells. Carcinogenesis 2007; 28: 2218–2226. [DOI] [PubMed] [Google Scholar]
- Tang H, Sun Y, Xiu Q, Lu H, Han H. Cyclooxygenase-2 induction requires activation of nuclear factor of activated T-cells in Beas-2B cells after vanadium exposure and plays an anti-apoptotic role. Arch Biochem Biophys 2007; 468: 92–99. [DOI] [PubMed] [Google Scholar]
- Yang TT, Xiong Q, Enslen H, Davis RJ, Chow CW. Phosphorylation of NFATc4 by p38 mitogen-activated protein kinases. Mol Cell Biol 2002; 22: 3892–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YY, Yao K, Bode AM, Bergen HR 3rd, Madden BJ, Oh SM et al. RSK2 mediates muscle cell differentiation through regulation of NFAT3. J Biol Chem 2007; 282: 8380–8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carcer G, Perez de Castro I, Malumbres M. Targeting cell cycle kinases for cancer therapy. Curr Med Chem 2007; 14: 969–985. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer 2001; 1: 222–231. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci 2005; 30: 630–641. [DOI] [PubMed] [Google Scholar]
- Keezer SM, Gilbert DM. Evidence for a pre-restriction point Cdk3 activity. J Cell Biochem 2002; 85: 545–552. [DOI] [PubMed] [Google Scholar]
- van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science 1993; 262: 2050–2054. [DOI] [PubMed] [Google Scholar]
- Hofmann F, Livingston DM. Differential effects of cdk2 and cdk3 on the control of pRb and E2F function during G1 exit. Genes Dev 1996; 10: 851–861. [DOI] [PubMed] [Google Scholar]
- Braun K, Holzl G, Soucek T, Geisen C, Moroy T, Hengstschlager M. Investigation of the cell cycle regulation of cdk3-associated kinase activity and the role of cdk3 in proliferation and transformation. Oncogene 1998; 17: 2259–2269. [DOI] [PubMed] [Google Scholar]
- Zheng D, Cho YY, Lau AT, Zhang J, Ma WY, Bode AM et al. Cyclin-dependent kinase 3-mediated activating transcription factor 1 phosphorylation enhances cell transformation. Cancer Res 2008; 68: 7650–7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YY, Tang F, Yao K, Lu C, Zhu F, Zheng D et al. Cyclin-dependent kinase-3-mediated c-Jun phosphorylation at Ser63 and Ser73 enhances cell transformation. Cancer Res 2009; 69: 272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Cho YY, Ma WY, Bode AM, Dong Z. Effects of MAP kinase inhibitors on epidermal growth factor-induced neoplastic transformation of human keratinocytes. Mol Carcinog 2006; 45: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S, Rollins BJ. Cyclin C/cdk3 promotes Rb-dependent G0 exit. Cell 2004; 117: 239–251. [DOI] [PubMed] [Google Scholar]
- Yamochi T, Semba K, Tsuji K, Mizumoto K, Sato H, Matsuura Y et al. ik3-1/Cables is a substrate for cyclin-dependent kinase 3 (cdk 3). Eur J Biochem 2001; 268: 6076–6082. [DOI] [PubMed] [Google Scholar]
- Salazar C, Hofer T. Activation of the transcription factor NFAT1: concerted or modular regulation? FEBS Lett 2005; 579: 621–626. [DOI] [PubMed] [Google Scholar]
- Okamura H, Aramburu J, Garcia-Rodriguez C, Viola JP, Raghavan A, Tahiliani M et al. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell 2000; 6: 539–550. [DOI] [PubMed] [Google Scholar]
- Beals CR, Clipstone NA, Ho SN, Crabtree GR. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes Dev 1997; 11: 824–834. [DOI] [PubMed] [Google Scholar]
- Fougere M, Gaudineau B, Barbier J, Guaddachi F, Feugeas JP, Auboeuf D et al. NFAT3 transcription factor inhibits breast cancer cell motility by targeting the Lipocalin 2 gene. Oncogene 2010; 29: 2292–2301. [DOI] [PubMed] [Google Scholar]
- Colburn NH, Wendel EJ, Abruzzo G. Dissociation of mitogenesis and late-stage promotion of tumor cell phenotype by phorbol esters: mitogen-resistant variants are sensitive to promotion. Proc Natl Acad Sci USA 1981; 78: 6912–6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasbey C. An analysis of histogram-based thresholding algorithms. CVGIP. Graph Models Image Process 1993; 55: 532–537. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.