Abstract
Crosstalk between transforming growth factor beta (TGF-β) signaling and p53 has a critical role in cancer progression. TGF-β signals via Smad and non-Smad pathways. Under normal conditions, wild-type p53 forms a complex with Smad2/3 and co-activates transcription of a variety of tumor suppressor genes, resulting in tumor suppressive effects. Thus, p53 stability is essential in progression of tumor suppressive responses mediated by TGF-β signaling. However, it remains unknown whether p53 stability is regulated by TGF-β. In the current study, we identify that USP15 binds to and stabilizes p53 through deubiquitination in U2OS and HEK293 cells. TGF-β promotes the translation of USP15 through activation of mammalian target of rapamycin by the phosphoinositide 3-kinase/AKT pathway. Upregulation of USP15 translation links the crosstalk between TGF-β signaling and p53 stability, allowing this cytokine to have a critical role in cancer progression.
Introduction
Transforming growth factor beta (TGF-β) is a pleiotropic cytokine with a dual role in cancer progression. In the early stages of progression, TGF-β acts as a tumor suppressor that regulates the transcription of tumor suppressor genes, inhibiting cell proliferation and promoting cell cytostasis, apoptosis and autophagy. Paradoxically, TGF-β can also function as a tumor promoter in the advanced stages of cancers, supporting tumor cell motility, survival, invasion, metastasis and immune evasion.1, 2, 3, 4, 5 Thus, to understand the molecular mechanisms for TGF-β and the regulation of cancer progression will provide vital information and help to develop targeted therapeutics. TGF-β signaling is initiated upon binding of TGF-β to its receptor (TGF-βRII), leading to the formation of a heterotetrameric complex with TGF-βR1 and to subsequent phosphorylation of TGF-βR1 by TGF-βRII. The cytosolic transcription factors, Smad2 and Smad3, are recruited and phosphorylated by the active TGF-β receptors.6, 7, 8 After dissociation from the TGF-β receptors, phosphorylated Smad2/3 form a complex with the co-Smad (Smad4) and are translocated into the nucleus where the transcription complex regulates the expression of numerous target genes.9, 10, 11 In addition, TGF-β signaling also includes several non-Smad pathways. The active TGF-β receptors can activate downstream ligands. These non-Smad pathways include the phosphoinositide 3-kinase (PI3K)/AKT, INK/p38, Ras-ERK and RhoA pathways.4, 12 The cooperation between Smad and non-Smad pathways determines the ultimate consequence of cellular reactions to TGF-β.
The linkage between p53 and TGF-β signaling was first pointed out by Wyllie et al.13 Inactivation of p53 disrupted the cellular response to TGF-β treatment. More recent studies have demonstrated crosstalk between p53 and TGF-β signaling, indicating that p53 can act as a component of Smad complexes, participate in the stabilization of Smad-DNA complexes and regulate a myriad of tumor suppressor genes.14, 15, 16, 17, 18 Therefore, stabilization of p53 is essential in maintaining cooperation with TGF-β signaling.
However, it remains unknown whether TGF-β can regulate the stability of p53. In this study, we demonstrate that TGF-β can upregulate the translation of USP15 through the PI3K/AKT pathway, in turn resulting in stabilizing p53 through deubiquitination.
Results
USP15 binds to and stabilizes p53
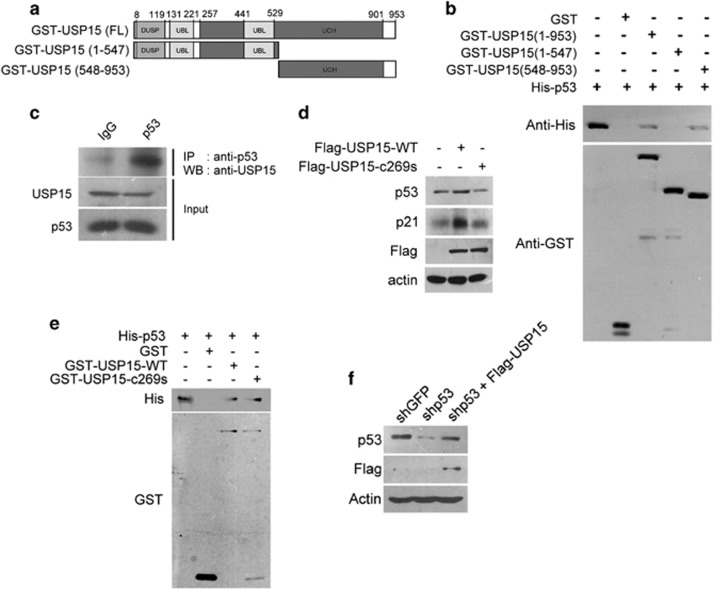
In our yeast two-hybrid screen and glutathione S-transferase (GST) pull-down assay, we have identified peptidylarginine deiminase 4 (PADI4) in association with USP15 (Supplementary Figure S1). PADI4 is a p53-binding protein which regulates the expression of p53 target genes.19 We wished to determine whether USP15 also binds to p53. The interaction between USP15 and p53 was verified by GST pull-down assays. The GST-USP15 fusion protein was prepared (Figure 1a) and incubated with His6-p53. The GST-USP15:His6-p53 complex co-precipitated by glutathione-Sepharose, suggesting that USP15 directly associates with p53 (Figure 1b). The p53-binding site was located at the C-terminal region of USP15, as evidenced by the GST pull-down assay (Figure 1b). USP15 binding to p53 in cells was also confirmed by co-immunoprecipitation (Figure 1c), suggesting that USP15 forms a complex with p53. To determine whether USP15 can stabilize p53 in cells, Flag-USP15 was transiently expressed in HEK293 cells. Western blot analysis indicated that overexpression of Flag-USP15 increased the stability of p53, in turn leading to an increased expression of p21, a p53 regulated target gene (Figure 1d). The residue Cys-269 is the active site of USP15.20 Replacement of Cys-269 with Ser results in loss of the USP15 enzymatic activity. Overexpression of an inactive USP15 C269S did not increase the stability of p53 (Figure 1d). This replacement did not disrupt the complex formation of USP15 with p53 (Figure 1e), suggesting that USP15 stabilization of p53 is dependent on its enzyme activity. The levels of endogenous p53 in HEK293 cells were reduced by treatment with shRNA (Figure 1f). However, co-overexpression of USP15 in the p53-knockdowned cells can increase the levels of p53 (Figure 1f).
Figure 1.
USP15 binds to and stabilizes p53. (a) Schematic representation of USP15 and its deletions. (b) The C-terminal domain of USP15 binds to p53 in GST pull-down assays. An aliquot (0.015 μg) of GST, GST-USP15(1–953), GST-USP15(1–547) or GST-USP15(548–953) was incubated with His6-p53 (0.3 μg) in 0.7 ml of buffer, followed by precipitation using Glutathione-Sepharose beads. All precipitated components were analyzed by western blotting and probed for GST and His6. (c) The endogenous USP15 co-immunoprecipitated with endogenous p53 from crude lysates of HEK293 cells. Crude proteins (2000 μg) extracted from HEK293 cells were incubated with the anti-p53 antibody (2 μl) that pre-immobilized on protein A Sepharose (30 μl). After centrifugation, all components in the pellet were analyzed by western blotting using anti-USP15 antibody. (d) USP15 promoting p53 stability is dependent on its enzymatic activity. Flag-USP15 or Flag-USP15 C269S was overexpressed in HEK293 cells for 24 h. Cells lysates were prepared and probed for p53, p21, actin and Flag. Actin was used for the loading control. (e) The wild-type USP15 or mutant USP15 binds to p53 in GST pull-down assays. An aliquot (0.015 μg) of GST, GST-USP15(WT) or GST-USP15(c269s) was incubated with His6-p53 (0.3 μg) in 0.7 ml of buffer, followed by precipitation using Glutathione-Sepharose beads. All precipitated components were analyzed by western blotting and probed for GST and His6. (f) Overexpression of USP15 increases the levels of p53 in p53-knockdowned cells. HEK293 cells with p53 knockdown by shRNA were transfected with the plasmid harboring USP15 cDNA for 24 h. Crude protein extracts were prepared and probed for p53, Flag and actin.
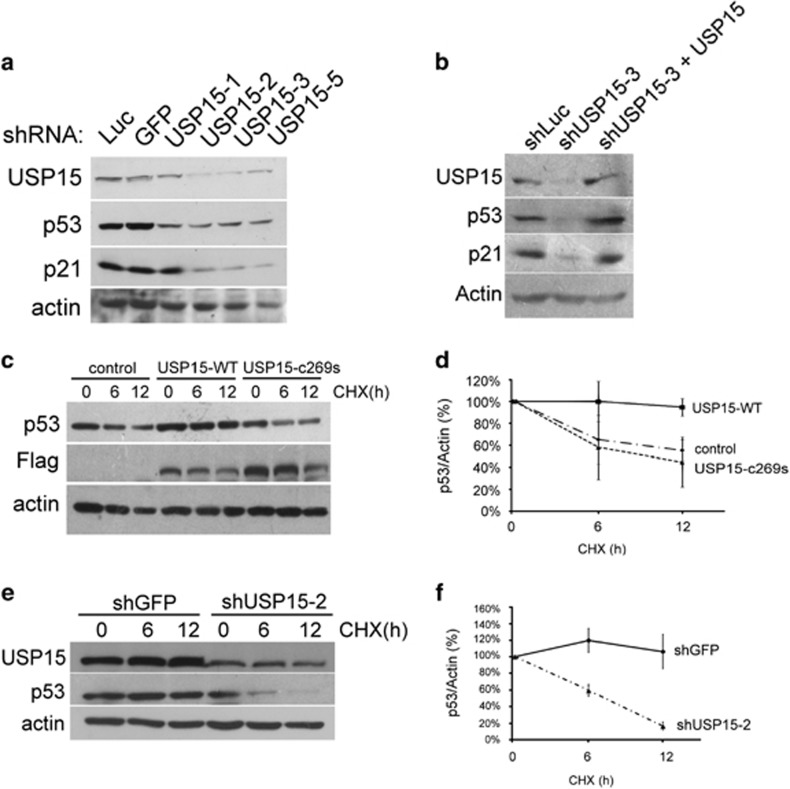
Cells were pre-treated with shRNAs to knockdown USP15. The levels of endogenous USP15 in HEK293 cells were reduced after treatment with shRNAs, concomitant with the reduction of cellular p53 (Figure 2a). This scenario was also observed in U2OS cells (Figure 2b). In addition, co-overexpression of shRNA-insensitive USP15 cDNA in USP15-knockdowned cells can increase the levels of the recombinant USP15 and, in turn, upregulate the levels of the cellular p53 (Figure 2b). Overexpression of USP15 increased the half-life of endogenous p53, but overexpression of the inactive USP15 C269S failed to increase the half-life of p53 (Figures 2c and d). Moreover, pre-treatment with shRNA, to knockdown the endogenous USP15, significantly reduced p53 stability (Figures 2e and f). Taken together, the results suggest that USP15 can bind to p53 and regulate its stability.
Figure 2.
USP15 knockdown by shRNA disrupts the p53 stability. (a) USP15 knockdown by shRNA destabilizes p53. USP15 knockdown of HEK293 cells were carried out by the different shRNAs. Cell lysates were prepared and probed for USP15, p53, p21 and actin. (b) Overexpression of the shRNA-insensitive USP15 cDNA increases the p53 stability in USP15-knockdowned U2OS cells. (c) USP15 overexpression stabilizes p53. Twenty-four hours after transfection with the indicated constructs, HEK293 cells were incubated with cycloheximide (100 μg/ml) for the indicated times. Total cell lysates were prepared and probed for p53, Flag and actin. (d) Quantification of c. The results are shown as a percentage of the p53/actin ratios in the absence of cycloheximide and are averaged from three independent experiments (means±s.d.). (e) USP15 knockdown accelerates the p53 degradation. HEK293 cells with USP15 knockdown by shRNA were cultured in the presence of cycloheximide (100 μg/ml) for the indicated time. Crude protein extracts were prepared and probed for USP15, p53 and actin. (f) Quantification of e. The results are averaged from three independent experiments (means±s.d.).
USP15 deubiquitinates p53
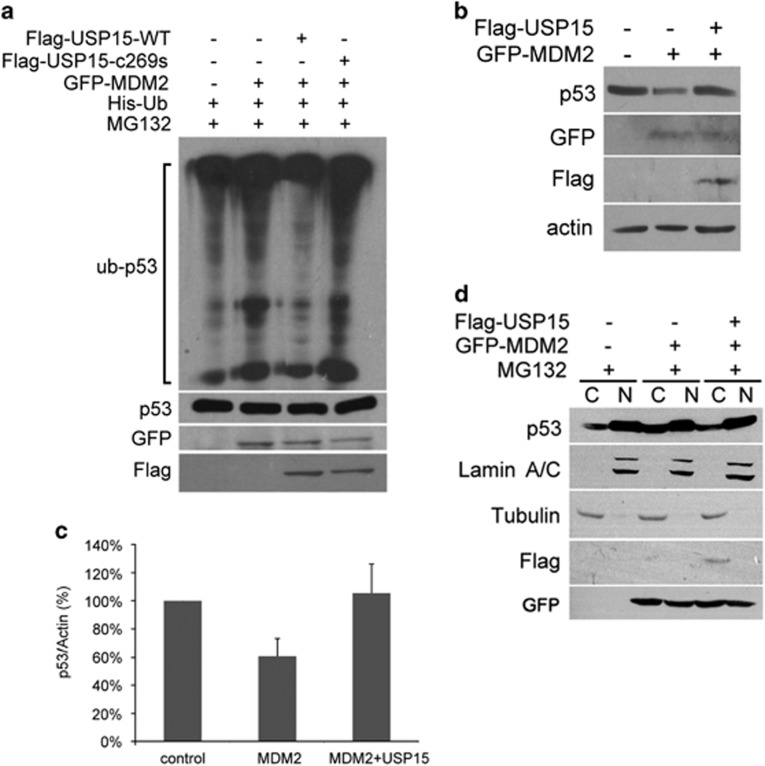
To examine whether USP15 can deubiquitinate p53, the effect of USP15 on p53 that had been ubiquitinated by overexpressed MDM2 was determined. Ubiquitination of p53 increased when ubiquitin and MDM2 were co-transfected into HEK293 cells (Figure 3a). A reduction of MDM2-dependent ubiquitination of p53 was observed following overexpression of USP15 in cells. However, co-transfection of an inactive USP15 C269S failed to show this function (Figure 3a). MDM2 overexpression reduced p53 stability whereas the stability was recovered following co-transfection with USP15 (Figures 3b and c). In addition, p53 localization in cells was also regulated by USP15 through deubiquitination of p53. The polyubiquitinated forms of p53 in cytosol were increased when HEK293 cells were transfected with MDM2. However, these forms of p53 were reduced in cytosol but increased in nucleus following co-transfection of USP15 (Figure 3d).
Figure 3.
USP15 deubiquitinates p53. (a) USP15 overexpression deubiquitinates p53. HEK293 cells were co-transfected with the indicated constructs. Twenty-four hours after transfection, cells were treated with MG132 (20 μg/ml) for 5 h. Cell lysates were prepared and precipitated by Ni2+-Sepharose beads and probed for p53, GFP and Flag. (b) USP15 antagonizes the destabilization of p53 by MDM2. HEK293 cells were co-transfected with the indicated constructs. Twenty-four hours after transfection, cell lysates were prepared and antibodies were used to probe for p53, GFP, Flag and actin. (c) Quantification of b. The results are shown as a percentage of the p53/actin ratios with the control, and are averaged from three independent experiments (means±s.d.). (d) Regulation of subcellular localization of p53 by USP15. HEK293 cells transfected with the indicated constructs were treated with MG132 (10 μM). After 16 h, cells were harvested and fractionated as described in 'Materials and methods' section. Cellular fractions were analyzed by western blotting using the indicated antibodies. A cytoplasmic marker protein, tubulin and a nuclear markers protein, lamin A/C, were used as controls to confirm the quality of fractions. Abbreviations: C, cytoplasmic fractions; N, nuclear fractions.
Competitive recognition of both MDM2 and p53 by USP15
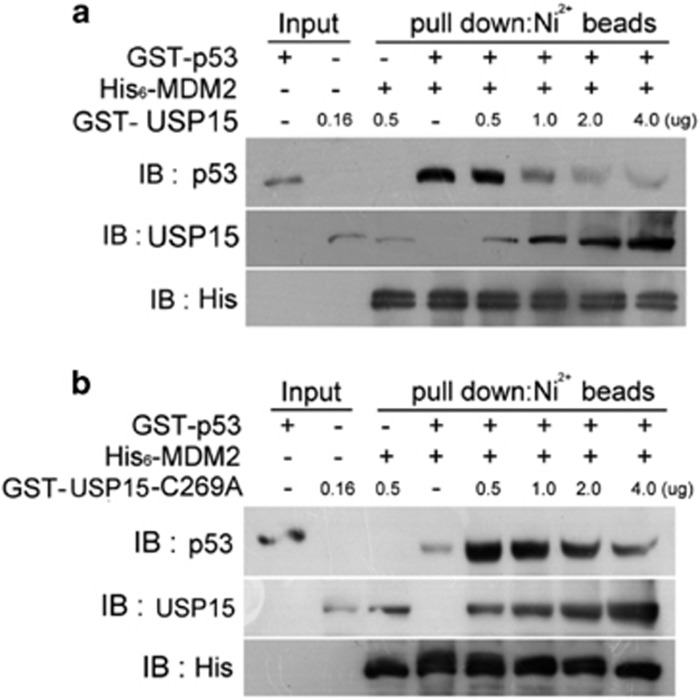
The mechanism for USP15 to block p53-ubiquitination mediated by MDM2 was analyzed. MDM2 can form a complex with p53 and catalyze p53-ubiquitination. When the MDM2/p53 complex was incubated with USP15, the original complex was disassembled and the new complex, MDM2/USP15, was formed (Figure 4a). Inactivation of USP15 by replacement of the active site, Cys-269, with Ala, did not affect the competitive recognition of both MDM2 and p53 by USP15 (Figure 4b).
Figure 4.
Competitive recognition of both MDM2 and p53 by USP15. GST-p53 (0.1 μg) was incubated with His6-MDM2 (0.6 μg) to form the complex, followed by addition of GST-USP15 or GST-USP15-C269A as the indicated amount. All components were subjected to precipitation by the Ni2+-Sepharose. The precipitated products were analyzed by western blotting and probed for p53, USP15 and His6 tag. (a) USP15 disrupts the MDM2/p53 complex. (b) Replacement of the active site, Cys269, by Ala does not affect the competitive recognition of both MDM2 and p53 by USP15. Input: GST-p53 (0.033 μg); GST-USP15 (0.16 μg); GST-USP15-C269A (0.16 μg).
TGF-β promotes USP15 production
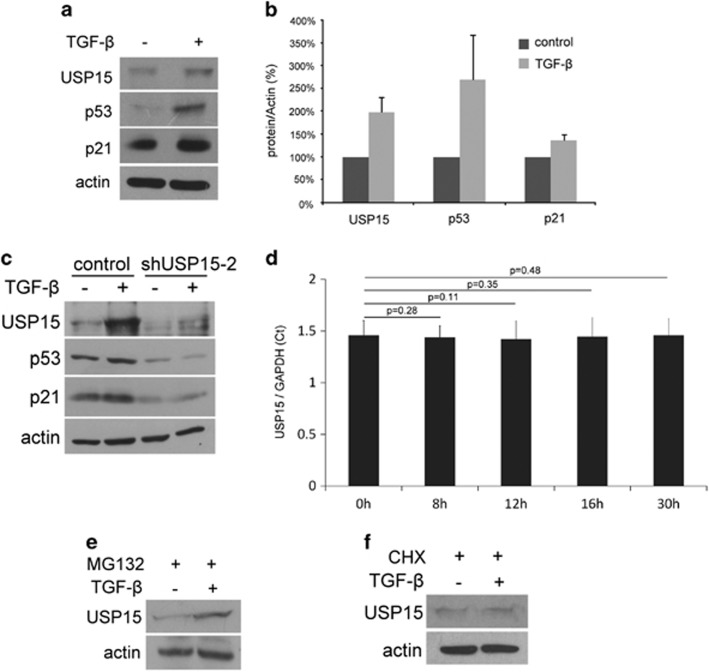
It is important to elucidate the signaling mechanism that modulates production of USP15. This could explain how enhanced p53 stability mediates the cell cycle and cell apoptosis. DNA damage induced by doxorubicin can promote the accumulation of p53.21 However, when HEK293 cells were treated with doxorubicin, the levels of USP15 were decreased in the time-course assays (Supplementary Figure S2), suggesting that USP15 is not involved in p53 accumulation induced by DNA damage. The cytokine(s) which upregulates the production of USP15 remains unknown. There are several reports implying that TGF-β may upregulate USP15.22, 23 Indeed, when U2OS cells were treated with TGF-β, the levels of USP15 were increased (Figures 5a and b), concomitant with an increase in p53 stability and synthesis of p21. USP15 knockdown disrupted p53 stability and p21 synthesis mediated by TGF-β, indicating that TGF-β modulated p53 stability through upregulation of USP15 synthesis (Figure 5c). Upregulation of USP15 by TGF-β is not due to an increase in USP15 transcription. No effect on the mRNA levels of USP15 was observed after TGF-β treatment in the time-course assays (Figure 5d and Supplementary Figure S3). Upregulation of USP15 by TGF-β was also observed in HEK293 cells (Figure 5e). In the presence MG132 that inhibits proteasome activity, treatment with TGF-β increases USP15 production (Figure 5e). In addition, in the presence of cycloheximide that blocks translation, the levels of USP15 increased by TGF-β treatments were blocked (Figure 5f). Taken together, TGF-β enhanced USP15 production was due to upregulation of its translation and not by suppression of USP15 degradation.
Figure 5.
TGF-β upregulates USP15 translation. (a) TGF-β promotes the USP15 synthesis in U2OS cells. U2OS cells were cultured in the absence or presence of TGF-β for 16 h. Cell lysates were prepared and probed for USP15, p53, p21 and actin. (b) Quantification of a. The results are shown as a percentage of the protein/actin ratios without TGF-β treatment and are averaged from three independent experiments (means±s.d.). (c) USP15 knockdown abolishes p53 stability and p53 transcriptional activation mediated by TGF-β. U2OS cells with USP15 knockdown were cultured in the absence or presence of TGF-β for 24 h. Cell lysates were prepared and probed for USP15, p53, p21 and actin. (d) TGF-β does not affect USP15 transcription. USP15 mRNA of HEK293 cells was quantitated by real-time PCR as described in 'Materials and methods' section and normalized to GAPDH mRNA. (e) TGF-β enhances USP15 translation. HEK293 cells pre-treated with MG132 (20 μM) for 5 h were cultured for 16 h in the absence or presence of TGF-β (2 ng/ml). (f) TGF-β does not affect USP15 degradation. HEK293 cells pre-treated with cycloheximide (100 μg/ml) for 5 h were treated with or without TGF-β (2 ng/ml) for 16 h. Cell lysates were analyzed by western blotting and probed for USP15 and actin.
TGF-β upregulates the translation of USP15 through a non-smad pathway
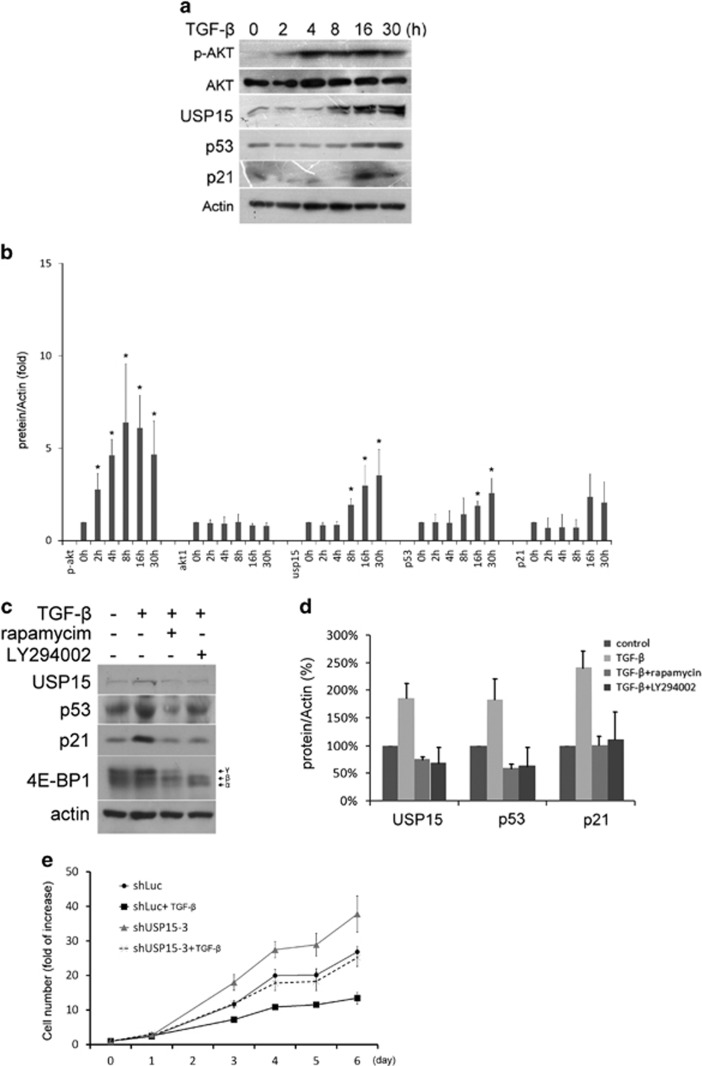
TGF-β signaling can regulate protein synthesis through the PI3K/AKT pathway.4, 24, 25, 26, 27, 28, 29, 30 TGF-β treatment can activate the AKT activity, as judged by an increased level of phospho-AKT (ser-473) in the time-course assays (Figures 6a and b). The key regulator of protein synthesis is mammalian target of rapamycin (mTOR), a downstream effector in the PI3K/AKT pathway mediated by TGF-β signaling, which activates S6 kinase and initiation factor 4E-binding protein 1 (4E-BP1) via phosphorylation, thereby promoting translational capacity.29 Indeed, TGF-β can increase the phosphorylation of 4E-BP1 and upregulate the synthesis of USP15 (Figures 6c and d). Pre-treatment with LY294002, an inhibitor of PI3K, or with MK2206, an inhibitor of AKT (Supplementary Figure S4), or with rapamycin or RAD001 (Supplementary Figure S4), the inhibitor of mTOR, blocked the phosphorylation of 4E-BP1 mediated by the PI3K/AKT pathway, and disrupted upregulation of USP15 synthesis mediated by TGF-β (Figures 6c and d). In addition, the biological effects of TGF-β signaling on cell proliferation were also analyzed. TGF-β treatment suppressed the cell growth (Figure 6e). Knockdown of USP15 without TGF-β treatment promoted the cell proliferation. The cell proliferation of USP15-knockdowned cells was suppressed after treatment with TGF-β (Figure 6e). Taken together, our results demonstrate that TGF-β signaling can regulate p53 stability through upregulation of USP15 translation mediated by the PI3K/AKT pathway and partly elucidate why TGF-β can suppress cancer progression in early stages when cells express wild-type p53.
Figure 6.
TGF-β promotes USP15 synthesis through the PI3K/AKT pathway. (a) AKT is activated by treatment with TGF-β in time-course assays. HEK293 cells were treated with TGF-β (2 ng/ml) and harvested at the indicated time. Cell lysates were analyzed by western blotting and probed for phospho-AKT (Ser-473), AKT, USP15, p53, p21 and actin. (b) Quantification of a. The results are shown as a fold of the protein/actin ratios with the initial time of TGF-β treatment and are averaged from three independent experiments (means±s.d.). *P<0.05. (c) Upregulation of USP15 synthesis by TGF-β is inhibited by treatment with LY294002 or with rapamycin. HEK293 cells pre-treated with rapamycin (500 nM) or LY294002 (20 μM) for 5 h were cultured for 16 h in the absence or presence of TGF-β (2 ng/ml). Cell lysates were analyzed by western blotting and probed for USP15, p53, p21, 4E-BP1 and actin. Phosphorylated forms of 4E-BP1 are marked with β and γ non-phosphorylated forms are marked with α. (d) Quantification of c. The results are shown as a percentage of the protein/actin ratios without any treatment and are averaged from three independent experiments (means±s.d.). (e) TGF-β treatment suppresses the cell proliferation. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay was used to measure the cell proliferation of U2OS cells. The results were averaged from three independent experiments.
Discussion
In this study, we have demonstrated that USP15 binds to p53 (Figures 1b and c). USP15 can stabilize p53 through deubiquitination (Figures 3a–d) and upregulate the p53 transcriptional activity, in turn promoting the expression of p21 (Figure 1d). Our results confirmed a previous report that overexpression of USP15 can upregulate the transcriptional activity of p53.31 We have also demonstrated that TGF-β signaling can upregulate USP15 synthesis (Figure 5a). Upregulation of USP15 synthesis results from activation of the PI3K/AKT pathway. Thus, the time for the augmented USP15 expression in TGF-β signaling is later than that of the activation for the PI3K/AKT pathway, but it is earlier than that of the increased levels of p53, followed by an increased expression of p21 in the time-course assay (Figures 6a and b). In the presence of the inhibitor of PI3K, LY294002, or an inhibitor of AKT, MK2206, or an inhibitor of mTOR, rapamycin or RAD001, to block the PI3K/AKT pathway, TGF-β upregulated phosphorylation of 4E-BP1 was reduced and levels of USP15 synthesis were suppressed (Figure 6c and Supplementary Figure S4). Phosphorylation of 4E-BP1 is one of the key steps for mTOR to upregulate protein translation.29 USP15 knockdown abolished p53 stabilization by TGF-β signaling (Figure 5c) and suppressed the transcriptional activity of p53 (Figure 5c).
Wild-type p53 has a key role in the anti-tumor effects of TGF-β signaling, which sends a signal to promote p53 stability, allowing this anti-tumor protein to be accumulated and consequently recruited to its DNA-binding sites on chromatin. This specific TGF-β signaling for stabilization of p53 was identified in our study. U2OS and HEK293 cells displaying the wild-type p53 are considered as the cell lines more close to the early stages of cancer. TGF-β signaling in U2OS and HEK293 cells promotes p53 stability through upregulation of USP15 translation by the PI3K/AKT pathway. The wild-type p53 can be sustained at a higher concentration by TGF-β signaling to promote TGF-β-induced tumor suppression. That USP15 has a key factor in TGF-β signaling to suppress the cell growth was also observed in the immortalized HaCaT keratinocytes.23 Cell growth of HaCaT cells was arrested by treatment with TGF-β and knockdown of USP15 impairs this TGF-β-dependent growth arrest.23
However, in advanced cancer, TGF-β signaling can also mediate USP15 to promote oncogenesis.22 In glioblastoma, USP15 associates with the SMAD7-SMAD specific E3 ubiquitin protein ligase 2 to form a ternary complex. USP15 in this trimeric complex deubiquitinates and stabilizes type 1 TGF-β receptor, upregulating the TGF-β signaling and providing a critically pathogenic factor for glioblastoma. In MDA-MB-231 breast cancer cells, USP15 is required for metastasis triggered by TGF-β signaling.23 The role of USP15 in TGF-β signaling is complicated and has a dual function in regulation of cancer progression. Switch of USP15 function in TGF-β signaling from a tumor suppressor to a promoter is highly dependent on the stages of cancer cells. Apparently, USP15 appears to be an important therapeutic target for cancer.
Domain-mapping studies revealed that the first ubiquitin-like (Ubl) domain ranging from residue-114 to −278 of USP15 associates with MDM2,32 whereas p53 binds to the C-terminal region (residues-548–953) of USP15 (Figure 1b). Although MDM2 and p53 interact with the different regions of USP15, USP15/p53/MDM2 cannot form a ternary complex. On the contrary, USP15 competitively recognizes both p53 and MDM2 (Figure 4a). The deubiquitination activity of USP15 is not essential in this competitive recognition (Figure 4b), but is required in stabilization of p53 (Figure 1d). Taken together, we suggested that USP15 disrupts the MDM2/p53 complex and forms two new complexes, USP15/MDM2 and USP15/p53. The herpesvirus-associated ubiquitin-specific protease (also named as USP7) can be activated by ATM to stabilize p53 response to DNA damage.33, 34 However, DNA damage cannot increase the USP15 production to stabilize p53 (Supplementary Figure S3). USP7 can also competitively recognize both p53 and MDM2.35, 36, 37 However, USP7 and USP15 may carry out the different mechanism in this action. The molecular mechanism for competitive recognition of p53 and MDM2 by USP7 has been well elucidated by the co-crystal structure.36, 37 USP7 contains one unique tumor necrosis factor-receptor associated factor (TRAF)-like domain that is not found in other members of USP family in human.38 Both MDM2 and p53 bind to the TRAF-like domain of USP7 in a mutually exclusive manner.36, 37 Especially, both MDM2 and p53 binds to the same key sequence, 164DWGF167, of TRAF-like domain in USP7. Other different residues of TRAF-like domain also interact with MDM2, but not with p53, and vice versa, accounting for USP7 binding to MDM2 and p53 with the different affinities. Indeed, USP7 has a higher affinity with MDM2 than with p53. USP15 lacks the TRAF-like domain.38 The first Ubl domain of USP15 does not contain the DWGF sequence.37 It remained unknown whether USP15 shows a different affinity with MDM2 and p53. Molecular recognition of MDM2 and p53 by USP15 needs to be characterized.
Materials and methods
Antibodies
Anti-p53 (Cat. No. sc-98; 1: 3000 dilution in use), anti-p21 (Cat. No. 397; 1: 1000 dilution in use) and anti-AKT1 (Cat. No. sc-5298; 1: 3000 dilution in use) antibodies were obtained from Santa Cruz Biotechnology (Dallas, TX, USA). Anti-USP15 antibody (Cat. No. NBP1–46805; 1: 3000 dilution in use) was obtained from Novus Biologicals (Littleton, CO, USA). Anti-4E-BP1 (Cat. No. 9644; 1: 1000 dilution in use) and anti-phospho-AKT (ser-473) (Cat. No. 9271-S; 1: 3000 dilution in use) antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA), anti-PARP (Cat. No. 1051-S; 1: 20000 dilution in use) and anti-caspase 3 (Cat. No. 1476-1; 1: 2000 dilution in use) antibodies were obtained from Epitomics Inc. (Burlingame, CA, USA). Anti-tubulin (Cat. No. NB100-690SS; 1: 5000 dilution in use) and anti-lamin (Cat. No. 612262; 1: 3000 dilution in use) antibodies were obtained from BD Biosciences (Franklin Lakes, NJ, USA). Anti-Flag (Cat. No. F1804; 1: 3000 dilution in use) and anti-GFP (Cat. No. G6539; 1:3000 dilution in use) antibodies were purchased from Sigma-Aldrich (St Louis, MO, USA). Anti-actin antibody (Cat. No. MAB1501; 1: 3000 dilution in use) was ordered from Merck Millipore (Darmstadt, Germany).
Plasmids
The constructed plasmids (pET32a-p53, pFLAG-CMV-USP15, pGEX-4T-1-USP15, pcDNA 3.1-His6-Ubiquitin, pEGFP-C1-MDM2 and pET32a-MDM2), the used primers and cloning sites are summarized in Supplementary Figure S5. The p53 cDNA of pET32a-p53 was digested by BamHI/SalI and subcloned into the pGEX-4T-1 plasmid to generate the plasmid, pGEX-4T-1-p53.
Protein purification
Escherichia coli BL21(DE3) was transformed with recombinant pET32a encoded with His6-p53 or His6-MDM2 fusion protein. The transformed bacteria were grown in 1 l of Luria-Bertani broth with ampicillin (0.1 g/l) and induced by isopropyl-beta-D-1-thiogalactopyranoside (final concentration of 1 mM) for 12 h at 16 °C. Cells were harvested by centrifugation and resuspended in 30 ml 20 mM Tris-HCl buffer, pH 7.9, containing 5 mM imidazole, 0.5 M NaCl, 0.2 mM phenylmethylsulfonyl fluoride, 0.02% NaN3 and 4 mM benzamidine. Bacteria were lysed by a French press and the p53 or MDM2 fusion protein was purified from the lysate by nickel-chelating Sepharose. Full-length (or truncated) USP15 or p53 was cloned into the pGEX4T-1 vector and the resulting plasmid transformed into E.coli BL21(DE3). Protein expression was induced by 1 mM isopropyl-beta-D-1-thiogalactopyranoside for 12 h at 16 °C. Bacteria were isolated by centrifugation and resuspended in 0.2 M Tris-HCl, pH 7.5, containing 0.2 M ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol (DTT), 0.02% NaN3, 0.3 M NaCl and 4 mM benzamidine. The bacteria were ruptured by a French press and the recombinant protein was purified from the lysate by GST-Sepharose.
Site-directed mutagenesis
The active site, C269, of USP15 was replaced by serine or alanine. pFlag-CMV plasmids encoding USP15 were carried out for mutagenesis by using polymerase chain reaction (PCR)with the specific primer. The primers used for replacement of C269 by Ser and Ala were 5′-CCT AAG TAA CTT GGG AAA TAC GAG TTT CAT GAA CTC AGC TAT TCA G-3′ and 5′-CCT AAG TAA CTT GGG AAA TAC GGC TTT CAT GAA CTC AGC TAT TC-3′, respectively. The PCR product was digested with DpnI and transformed into E.coli DH5α for amplification.
Knockdown of USP15 and p53
pLKO.1 plasmids expressing USP15 shRNA were purchased from the National RNAi Core Facility platform located at the Institute of Molecular Biology/Genomic Research Center, Academia Sinica (Taipei, Taiwan). Preparation of the recombinant lentivirus and virus infection followed the methods as described by the manufacturer. pLKO.1 plasmid expressing GFP shRNA served as a negative control. DNA sequences of the shRNA used for USP15 or p53 knockdowns are shown in Supplementary Figure S6.
Preparation of shUSP15-3 insensitive cDNA of USP15
The shUSP15-3 insensitive cDNA of USP15 was generated by three-step PCR. Fragments containing the mutant shUSP15-3 targeting sequence at the 3′-end and 5′-end were cloned by PCR using primer 1 (GAG CGG CCG CGA TGG CGG AAG GCG GA) /primer 2 (ATT GTC GAT CGG TCC AGG ATA CAC ATT TTG) and primer 3 (CCGATCGACAAT TCT GGA CTT CTC AAA GAT G)/ primer 4 (GCC AAT TGT ATC TAT TGT GTC AGC TTT GC), respectively. The mutated sequence of shUSP15-3 is underlined and the mutant sites are marked by bold. The fragment 1 and fragment 2 were annealed and the annealed full-length was cloned by PCR using primer 1 and primer 4. The resulting product was cleaved by NotI/MfeI and subcloned to pFlag-CMV-USP15.
GST pull-down assay
GST-USP15 or GST-USP15 truncated proteins were incubated with His6-p53 in buffer containing 0.1 M Tris-HCl, pH 7.5, 5 mM EDTA, 2 mM DTT, 0.1 M L-arginine, 0.1 M Glutamic acid, at 4 °C for 3 h, followed by addition of 40 μl Glutathione-Sepharose 4B (GE Healthcore, San Francisco, CA, USA). All components were allowed to incubate at 4 °C for 1 h, and centrifuged at 10 000g for 1 min. The pelleted beads were washed three times with 1 ml buffer containing 10 mM Tris-HCl, pH 7.5, 0.2 mM EDTA, 4 mM benzamidine, 0.02% NaN3, 150 mM NaCl and 0.1% NP-40. After centrifugation, the supernatant was discarded and the pelleted beads were resuspended in the sodium dodecyl sulfate (SDS) sample buffer and analyzed by SDS–polyacrylamide gel electrophoresis (PAGE) and western blotting.
Effect of USP15 on the MDM2/p53 complex
GST-p53 (0.1 μg) was incubated with His6-MDM2 (0.6 μg) for 5 h at 4 °C in the buffer (20 mM Tris-HCl buffer, pH 7.9, containing 5 mM imidazole, 0.5 M NaCl, 0.05% NP-40), followed by addition of GST-USP15 with the indicated amount from 0.5 to 4 μg. All components were allowed to incubate at 4 °C for 16 h and then were added with Ni2+ beads (50 μl). After incubation for 1 h, all components were subjected to centrifugation (13 000g) for 1 min at 4 °C. The pellet was washed by the same buffer for four times, analyzed by western blot and probed for p53, His6 and USP15.
Immunoprecipitation
HEK293 cells (6 × 105 cells) were seeded into 10-cm petri dishes, grown to confluence, and harvested by centrifugation. The pelleted cells were ruptured in buffer containing 5 mM Tris-HCl, pH 7.4, 15 mM NaCl, 1 mM EDTA and 0.1% NP-40. The lysate was added to anti-p53 antibody and incubated at 4 °C for 1 h, followed by addition of rabbit anti-mouse IgG (1 μl) immobilized on protein G-Sepharose (30 μl). After incubation at 4 °C for 3 h, beads were isolated by centrifugation and washed three times with the same buffer. Proteins bound to the beads were extracted by SDS sample buffer and analyzed by SDS–PAGE and western blotting.
Measurement of protein stability
HEK293 cells (2 × 105 cells) were seeded into 6-cm petri dishes and maintained in Dulbecco's modified eagle medium (DMEM) with 10% fetal bovine serum and 1% penicillin/streptomycin. After 16 h, cells were transfected with the indicated constructs using lipofectamine 2000 (Invitrogen, Carlsbad, CA) for 6 h and cultured for an additional 24 h. For the protein stability assay, 100 μg/ml cycloheximide (Sigma-Aldrich, St Louis, MO) were added. Cells were harvested at indicated time by centrifugation at 1500g for 5 min. Pelleted cells were resuspended in 0.1 ml of 1% SDS and lysed by ultrasonication. Aliquots of cell extracts were analyzed by SDS–PAGE and western blotting.
In vivo deubiquitination assay
HEK293 cells (6 × 105 cells) were seeded into 10-cm petri dishes and maintained in DMEM. After 16 h, cells were transfected with the indicated constructs using lipofectamine 2000 (Invitrogen) for 6 h and cultured for an additional 24 h. The transfected cells were treated with proteasome inhibitor MG132 (20 μM) for 5 h. Cells were harvested by centrifugation and ruptured with RIPA buffer. The lysate (2500 μg) was incubated with nickel-chelating Sepharose (90 μl) at 4 °C for 16 h. Beads were collected by centrifugation at 10 000g for 1 min and washed three times with 1 ml RIPA buffer. Ubiquitinated p53 was analyzed by SDS–PAGE and western blotting.
Subcellular fractionation
HEK293 cells (2 × 105 cells) were seeded into 6-cm petri dishes and maintained in DMEM for 16 h. Cells were transfected with the indicated constructs using jetPRIME (polyplus) and maintained in DMEM with MG132 (20 μM) for 8 h. Cells were harvested by centrifugation at 1200g for 5 min, resuspended in hypotonic buffer containing 10 mM Tris-HCl, pH 7.9, 10 mM KCl, 0.05% NP-40 and 1.5 mM MgCl2 and incubated on ice for 10 min. Proteins in the cytoplasmic and nuclear fractions were separated by centrifugation at 760 × g at 4 °C for 10 min. The supernatant containing the cytoplasmic proteins was transferred into a new microcentrifugation tube. The pellet containing the nuclear proteins was resuspended in 1% SDS. Both protein fractions were subjected to SDS–PAGE and western blot analysis. The cytoplasmic and nuclear fractions were identified with anti-tubulin and anti-lamin antibodies, respectively.
Real-time PCR
HEK293 cells were cultured in DMEM medium with or without TGF-β (2 ng/ml) for 0, 8, 12, 16 or 30 h. Cells were harvested by centrifugation at 1200g for 5 min and resuspended in 1 ml Trizol buffer plus 0.2 ml chloroform. Total RNA extraction, RT–PCR and real-time PCR were performed using kits (Promega, Madison, WI, USA) following the manufacturer's instructions. The following primers were used for the real-time PCR: for USP15, 5′-CTG CTC AAA ACC TCG CTC C-3′ and 5′-CAA TGG GTC CAG GAT ACA CA-3′ and for GAPDH, 5′-CTC TGC TCC TCC TGT TCG AC-3′ and 5′-TTA AAA GCA GCC CTG GTG AC-3′.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay
U2OS cells were seeded into six-well plates. After 16 h culture at 37 °C, the medium was replaced with fresh McCoy's 5A medium. Cells were cultured at 37 °C for 24, 72, 96, 120 or 144 h. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide was added to the McCoy's 5A medium (0.5 mg/ml) at the indicated time and incubated for 4 h. The medium was removed and 1 ml dimethyl sulfoxide was added to each well. After shaking for 1 min, the optical density was measured at 570 nm.
TGF-β treatment
HEK293 or U2OS cells were cultured for 16 h. Cells pre-treated with LY294002 (20 μM), rapamycin (500 nM), MG132 (20 μM) or cycloheximide (100 μg/ml) for 5 h were cultured for 16 h in the absence or presence of TGF-β (2 ng/ml). Cells were harvested by centrifugation at 1500g for 5 min. The cell pellets were ruptured by 1% SDS with ultrasonication, and cell lysates were analyzed by western blotting.
Statistical analysis
Data are displayed as means±s.d. Statistical significance was analyzed by unpaired Mann–Whitney U-tests from comparing numerical data between different groups. P-values less than 0.05 were considered statistically significant.
Acknowledgments
This work was supported by grants from the DaLin Tzu-Chi Buddhist Hospital (DTCRD 97-14, DTCRD 102-E-06 and DTCRD103(2)-I-10) and from the National Science Council, ROC (NSC 102-2320-B194-002).
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Massague J. TGFbeta in Cancer. Cell 2008; 134: 215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston R, Inman GJ. Crosstalk between p53 and TGF-beta Signalling. J Signal Transduct 2012; 2012: 294097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman GJ. Switching TGFbeta from a tumor suppressor to a tumor promoter. Curr Opin Genet Dev 2011; 21: 93–99. [DOI] [PubMed] [Google Scholar]
- Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res 2009; 19: 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester E, Ten DP. The dynamic roles of TGF-beta in cancer. J Pathol 2011; 223: 205–218. [DOI] [PubMed] [Google Scholar]
- Huang T, David L, Mendoza V, Yang Y, Villarreal M, De K et al. TGF-beta signalling is mediated by two autonomously functioning TbetaRI, TbetaRII pairs. EMBO J 2011; 30: 1263–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. A very private TGF-beta receptor embrace. Mol Cell 2008; 29: 149–150. [DOI] [PubMed] [Google Scholar]
- Groppe J, Hinck CS, Samavarchi-Tehrani P, Zubieta C, Schuermann JP, Taylor AB et al. Cooperative assembly of TGF-beta superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol Cell 2008; 29: 157–168. [DOI] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev 2005; 19: 2783–2810. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Moustakas A. Role of Smads in TGFbeta signaling. Cell Tissue Res 2012; 347: 21–36. [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol 2005; 21: 659–693. [DOI] [PubMed] [Google Scholar]
- Mu Y, Gudey SK, Landstrom M. Non-Smad signaling pathways. Cell Tissue Res 2012; 347: 11–20. [DOI] [PubMed] [Google Scholar]
- Wyllie FS, Dawson T, Bond JA, Goretzki P, Game S, Prime S et al. Correlated abnormalities of transforming growth factor-beta 1 response and p53 expression in thyroid epithelial cell transformation. Mol Cell Endocrinol 1991; 76: 13–21. [DOI] [PubMed] [Google Scholar]
- Cordenonsi M, Dupont S, Maretto S, Insinga A, Imbriano C, Piccolo S. Links between tumor suppressors, p53 is required for TGF-beta gene responses by cooperating with Smads. Cell 2003; 113: 301–314. [DOI] [PubMed] [Google Scholar]
- Takebayashi-Suzuki K, Funami J, Tokumori D, Saito A, Watabe T, Miyazono K et al. Interplay between the tumor suppressor p53 and TGF beta signaling shapes embryonic body axes in Xenopus. Development 2003; 130: 3929–3939. [DOI] [PubMed] [Google Scholar]
- Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature 1997; 389: 85–89. [DOI] [PubMed] [Google Scholar]
- Cordenonsi M, Montagner M, Adorno M, Zacchigna L, Martello G, Mamidi A. Integration of TGF-beta and Ras/MAPK signaling through p53 phosphorylation. Science 2007; 315: 840–843. [DOI] [PubMed] [Google Scholar]
- Dupont S, Zacchigna L, Adorno M, Soligo S, Volpin D, Piccolo S et al. Convergence of p53 and TGF-beta signaling networks. Cancer Lett 2004; 213: 129–138. [DOI] [PubMed] [Google Scholar]
- Li P, Yao H, Zhang Z, Li M, Luo Y. Regulation of p53 target gene expression by peptidylarginine deiminase 4. Mol Cell Biol 2008; 28: 4745–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos RM, Altreuter J, White EA, Howley PM. The ubiquitin-specific peptidase USP15 regulates human papillomavirus type 16 E6 protein stability. J Virol 2009; 83: 8885–8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz EU, Douglas P, Lees-Miller SP. Doxorubicin activates ATM-dependent phosphorylation of multiple downstream targets in part through the generation of reactive oxygen species. J Biol Chem 2004; 279: 53272–53281. [DOI] [PubMed] [Google Scholar]
- Eichhorn PJ, Rodón L, Gonzàlez-Juncà A, Dirac A, Gili M, Martínez-Sáez E et al. USP15 stabilizes TGF-beta receptor I and promotes oncogenesis through the activation of TGF-beta signaling in glioblastoma. Nat Med 2012; 18: 429–435. [DOI] [PubMed] [Google Scholar]
- Inui M, Manfrin A, Mamidi A, Martello G, Morsut L, Soligo S et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol 2011; 13: 1368–1375. [DOI] [PubMed] [Google Scholar]
- Lee H, Kim J, Lee B, Chang JW, Ahn J, Park JO et al. Oncolytic potential of E1B 55 kDa-deleted YKL-1 recombinant adenovirus, correlation with p53 functional status. Int J Cancer 2000; 88: 454–463. [DOI] [PubMed] [Google Scholar]
- Bakin AV, Tomlinson AK, Bhowmick NA, Moses HL, Arteaga CL. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J Biol Chem 2000; 275: 36803–36810. [DOI] [PubMed] [Google Scholar]
- Shin I, Bakin AV, Rodeck U, Brunet A, Arteaga CL. Transforming growth factor beta enhances epithelial cell survival via Akt-dependent regulation of FKHRL1. Mol Biol Cell 2001; 12: 3328–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinals F, Pouyssegur J. Transforming growth factor beta1 (TGF-beta1) promotes endothelial cell survival during in vitro angiogenesis via an autocrine mechanism implicating TGF-alpha signaling. Mol Cell Biol 2001; 21: 7218–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes MC, Mitchell H, Penheiter SG, Doré JJ, Suzuki K, Edens M et al. Transforming growth factor-beta activation of phosphatidylinositol 3-kinase is independent of Smad2 and Smad3 and regulates fibroblast responses via p21-activated kinase-2. Cancer Res 2005; 65: 10431–10440. [DOI] [PubMed] [Google Scholar]
- Lamouille S, Derynck R. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol 2007; 178: 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JY, Shin I, Arteaga CL. Type I transforming growth factor beta receptor binds to and activates phosphatidylinositol 3-kinase. J Biol Chem 2005; 280: 10870–10876. [DOI] [PubMed] [Google Scholar]
- Zhang X, Berger FG, Yang J, Lu X. USP4 inhibits p53 through deubiquitinating and stabilizing ARF-BP1. EMBO J 2011; 30: 2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q, Jin J, Hu H, Li HS, Romano S, Xiao Y et al. USP15 stabilizes MDM2 to mediate cancellcell survival and inhibit antitumor T cell responses. Nat Immunol 2014; 15: 562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoronenkova SV, Dianov GL. Regulation of USP7/HAUSP in response to DNA damage: yet another role for ATM. Cell Cycle 2012; 11: 2409–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoronenkova SV, Dianov II, Ternette N, Kessler BM, Parsons JL, Dianov GL. ATM-dependent downregulation of USP/HAUSP by PPM1G activates p53 response to DNA damage. Mol Cell 2012; 45: 801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell 2004; 13: 879–886. [DOI] [PubMed] [Google Scholar]
- Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH et al. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol 2006; 13: 285–291. [DOI] [PubMed] [Google Scholar]
- Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol 2006; 4: e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Clagu MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 2009; 10: 550–563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.