ABSTRACT
Widespread antibiotic resistance among bacterial pathogens is providing the impetus to explore novel sources of antimicrobial agents. Recently, the potent antibacterial activity of certain clay minerals has stimulated scientific interest in these materials. One such example is Kisameet glacial clay (KC), an antibacterial clay from a deposit on the central coast of British Columbia, Canada. However, our understanding of the active principles of these complex natural substances is incomplete. Like soils, clays may possess complex mixtures of bacterial taxa, including the Actinobacteria, a clade known to be rich in antibiotic-producing organisms. Here, we present the first characterization of both the microbial and geochemical characteristics of a glacial clay deposit. KC harbors surprising bacterial species richness, with at least three distinct community types. We show that the deposit has clines of inorganic elements that can be leached by pH, which may be drivers of community structure. We also note the prevalence of Gallionellaceae in samples recovered near the surface, as well as taxa that include medically or economically important bacteria such as Actinomycetes and Paenibacillus. These results provide insight into the microbial taxa that may be the source of KC antibacterial activity and suggest that natural clays may be rich sources of microbial and molecular diversity.
KEYWORDS: Actinobacteria, clay mineral, geochemical characteristics, Kisameet, microbiome, antimicrobial activity, microbial communities
IMPORTANCE
Identifying and characterizing the resident microbial populations (bacteria, viruses, protozoa, and fungi) is key to understanding the ecology, chemistry, and homeostasis of virtually all sites on Earth. The Kisameet Bay deposit in British Columbia, Canada, holds a novel glacial clay with a history of medicinal use by local indigenous people. We previously showed that it has potent activity against a variety of antibiotic-resistant bacteria, suggesting it could complement our dwindling arsenal of antibiotics. Here, we have characterized the microbiome of this deposit to gain insight into what might make the clay antibacterial. Our analyses suggest that the deposit contains a surprising diversity of bacteria, which live in at least three distinct environments. In addition, the clay harbors bacteria that may have interesting potential as biocontrol/bioremediation agents or producers of novel bioactive compounds.
INTRODUCTION
Microbes are essential components of most of the environments on Earth. They survive, and even thrive, in a variety of niches and factor centrally in metabolic pathways that shape geologic, ecological, and human health. A holistic understanding of how microbes cycle different elements, such as carbon in the soil and oceans, is essential to understanding global homeostatic processes (1), and the microbiome is likewise being accepted as an essential component of the human system. An understanding of microbial diversity is therefore central to environmental, evolutionary, and biomedical sciences. Cultivation-independent molecular surveys have revealed the extent of this diversity and have identified some factors that may influence microbial community structures. This knowledge is revolutionizing our understanding of the contribution of the microbiome to human health and disease (2, 3). However, these studies are only now beginning to reveal the phylogeny, gene content, and phenotypic potential of the so-called microbial dark matter, and comprehensive surveys of microbes in novel environments are necessary (1, 4).
There is also a pressing, if not desperate, need for new strategies to replace or complement existing antimicrobial agents in the face of ever-increasing (multi)antibiotic resistance (5). Much research is focused on identifying “wild” microbes that might produce novel antimicrobials (6–8). However, processing new candidates through the research and regulatory pipelines is slow. As such, “historical” agents are increasingly being investigated in modern empirical studies for treatment of infectious disease. The medicinal applications of natural substances such as clays, muds, and other natural terrestrial products have been known for thousands of years. More recently, the use of poultices of French green clay for treatment of Mycobacterium ulcerans skin infections in Africa by the humanitarian Line Brunet de Courssou has renewed interest in the antibacterial potential of clays (9, 10).
Clays are a diverse group of economically important natural materials, made up of clay minerals—silicates with a repeating layer structure and a small particle size (<2 μm) (11). Their small particle size and large surface area provide novel adsorptive and ion-exchange characteristics, which may underlie their therapeutic and cosmetic properties (12). Pioneering work has demonstrated the broad spectrum of activity of several natural mineral clays in vitro (13–16; reviewed in reference 17). The mechanism(s) of action of different clays appears diverse and/or multifactorial and may include pH and redox buffering of metal ion toxicity (Fe2+ and Al3+), adsorption and release of compounds in clay interlayers, absorption of micronutrients required for bacterial growth, or proliferation of bacterial species that produce antibacterial compounds (14, 18, 19).
Kisameet glacial clay (KC), located in a deposit on the northwestern coast of British Columbia, Canada, has been used historically by the local Heiltsuk First Nation for therapeutic purposes. KC has potent antibacterial activity against multidrug-resistant ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species) pathogens (20), which are common nosocomial species that frequently escape current antibiotic treatments (21). KC is also strongly active against extensively resistant and multidrug-resistant opportunistic pathogens, such as members of the Burkholderia cepacia complex, isolated from cystic fibrosis patients (S. Behroozian, J. E. A. Zlosnik, and J. Davies, submitted for publication). The clay is also active against fungal pathogens such as Candida albicans and Cryptococcus neoformans (S. Behroozian, S. L. Svensson, W. Xu, L. Li, and J. Davies, unpublished data). KC is relatively rich in iron (22), and therefore, pH-dependent metal ion toxicity, as described for other clays (13, 14), may underlie some of its activity (Behroozian et al., unpublished). Active fractions can also be extracted with organic solvents (J. Tan and J. Davies, unpublished data), suggesting that the mechanism of action might be multifactorial. Much remains to be identified about the mechanism of antimicrobial activity of KC to facilitate the production of consistent, safe, and potent preparations for future medical applications.
The overall expressed properties of many natural products may be the result of biochemical interactions with their resident microbiota. A comprehensive microbiome study of a clay deposit in situ has not yet, to our knowledge, been reported. Previous microbiological studies of clays have been limited to small sample sizes of processed samples or clay-rich soils. Here, we describe investigations of the bacterial communities of the KC deposit by 16S rRNA metagenome characterization from five vertical cores. The deposit has three general types of bacterial community, which correlate with depth and pH. We found surprisingly high bacterial species diversity, with a dominance of Betaproteobacteria. Identification of bacterial taxa that make up the dominant core of each community revealed differences in the distribution of putative iron-oxidizing Gallionella spp. Bacterial community structure did not correlate with antibacterial activity. However, we found evidence that the KC microbiome includes potentially valuable species, such as Actinobacteria. This initial microbiological survey of a novel environment provides a framework for understanding the basis for the unique properties of KC.
RESULTS
Description of the Kisameet Bay deposit and experimental design.
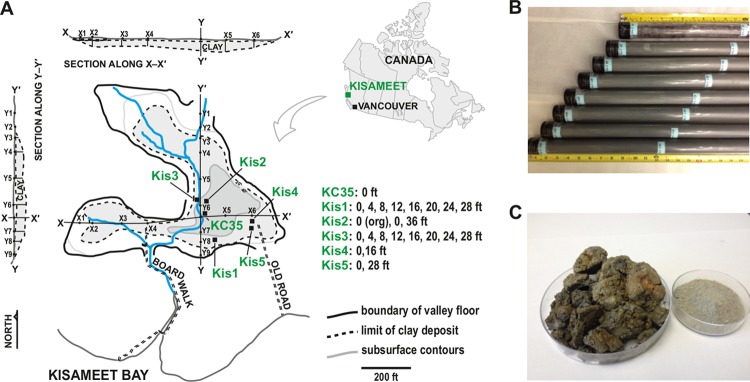
Kisameet glacial clay is found in a dry-land (i.e., nonmarine) deposit on the Central Coast of British Columbia in Canada, 450 km northwest of Vancouver (51°58′20″ N/127°52′50″ W) (23, 24) (Fig. 1A, map in the top right corner). The deposit covers ~2 ha and is estimated to contain 181,000 tons of clay in a depression sitting on sand, gravel, and bedrock to a maximum depth of 40 ft (Fig. 1A). The clay itself is Quaternary in age, resulting from glacial expansion and retreat during past ice ages but sits upon gray and black schist and gneiss of the Jurassic to Tertiary Coast Plutonic Complex (23). A layer of organic overburden of up to 2 meters thick covers the deposit, and the clay itself is dark greenish-gray when moist and light gray when dry (Fig. 1B). Areas of reddish-brown material can also be present, especially upon exposure to air (Fig. 1C). The KC deposit remains mostly untouched, but it was drilled in 1946, and samples were submitted to the British Columbia Mines Branch. About 25 tons of clay were sold as a natural pharmaceutical and cosmetic. Initial mineralogical studies indicated that KC is an iron-rich (4.5% by weight) bentonite clay, which is fine-grained (54.6% with a grain diameter of 1.7 to 3 μm and 30.7% with a diameter of <1.7 μm) (22, 23).
FIG 1 .
The Kisameet Bay glacial clay deposit, spatial description of core sampling, and physical appearance of KC. (A) The approximate location of Kisameet Bay on King Island on the central coast of British Columbia, ∼450 km northwest of Vancouver, Canada, is shown in the top right inset. The topographical layout of the deposit showing the approximate locations of the five vertical core samples (Kis1 to Kis5) and KC35 makes up the main part of panel A. The KC deposit map is simplified/modified from reference 23. (B) Examples of KC vertical core samples (Kis5), showing the green-gray color of the clay in situ, as well as organic overburden present in some shallow (i.e., 0 to 4 ft) samples from the top of the deposit. (C) KC following exposure to the air, showing reddish-brown areas, as well as a dried and powdered sample.
Five vertical cores were extracted from the deposit at five locations (Kis1 to Kis5 [Fig. 1A]). Because the depth of the deposit is variable, Kis1 extended from 0 ft to ~32 ft, for example, whereas the bottom of the Kis4 core was only ~20 ft below the surface. Each core was then divided into 4-ft-long sections and labeled according to the depth at the top of the section. For example, the Kis3 core was separated into samples Kis3-0 (0 ft ≤ depth < 4 ft), Kis3-4 (4 ft ≤ depth < 8 ft), Kis3-8 (8 ft ≤ depth < 12 ft), and so on, where the first number indicates the core and the second number indicates the depth. Twenty-three of these sections were selected for physicochemical and microbiome characterization (listed in Table S1 in the supplemental material). Two of the samples, Kis3-0 Org and Kis5-0, consisted primarily of organic material from overburden (Kis3-0 was separated into “organic” and “clay” fractions). We also analyzed KC35, a sample from the surface that has been previously shown to have strong antimicrobial activity (20).
Library and read statistics for next-generation sequencing. Download TABLE S1, DOCX file, 0.03 MB (33KB, docx) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Physicochemical characteristics of the deposit.
We first performed a general characterization of the physicochemical properties of KC. Mineralogical analysis of selected core samples showed that KC contains phyllosilicates such as biotite (~3 to 15%) and relatively small amounts of clay minerals such as illite (~5%) and clinochlore (~5 to 13%) (Table S2). We did not observe any major differences between the selected core samples and KC35. Likewise, analysis of acid-digested elements in KC core samples and elemental analysis of bulk clay did not reveal major variations (Table S3) but confirmed previous observations of significant amounts of Fe (~104 mg/kg). Because speciation could presumably affect the antibacterial activity of toxic metals, including Fe, we also determined the redox potential of each sample (Table S3). The measured potential of the core samples varied relatively widely from +19 mV to a maximum of +373 mV, with the redox of KC35 being +427 mV.
Mineralogical composition of selected KC core samples. Download TABLE S2, DOCX file, 0.03 MB (36KB, docx) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Acid-digested elemental composition of KC core sample bulk clay. Download TABLE S3, DOCX file, 0.04 MB (43.9KB, docx) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
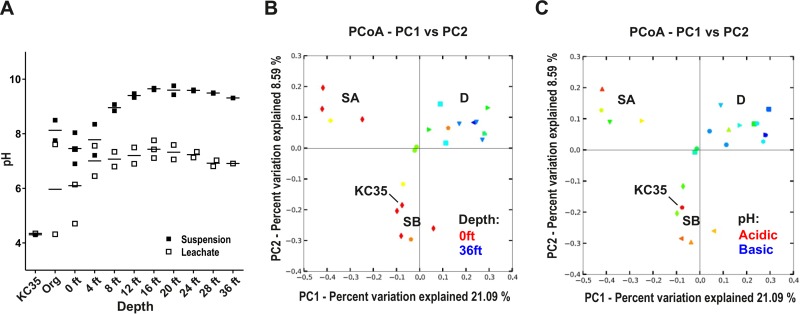
In contrast, comparison of the pH of core sample suspensions revealed more marked differences. While previous studies suggested that KC is mildly alkaline (pH ~8) (15), the low pH of KC35 we measured previously (20) suggests that some samples could be markedly more acidic. Measurement of different preparations of KC core samples (fresh aqueous suspensions, aqueous leachates, or suspensions of clay that had been stored exposed to air for approximately 1 month) suggested that the clay pH can vary depending on (i) depth of the sample in the deposit and (ii) exposure to air. First, the pH of fresh KC suspensions revealed a strong trend with depth (Fig. 2A and Table S4), with the pH of clay from greater depths significantly higher than for samples from depths of 0 to 8 ft (pH 9.5 versus pH 8.0; P < 0.0001). Also, while the pH of prepared aqueous leachates showed the same positive correlation between increasing pH and depth, we also noted that in general, aqueous leachates showed a lower pH than their suspension counterparts (Fig. 2A). This may reflect a lowering of pH during the 24 h of incubation under aeration during leachate preparation. In support of this, remeasurement of suspension pHs a few months after core cutting resulted in markedly higher levels of acidity. In addition, the pH of KC35, which was harvested at least 2 years before the core samples and stored at 4°C, had an extremely low suspension pH of 4.3. Together, this suggests that the pH of KC can vary markedly depending on depth-related factors, as well as exposure to air.
FIG 2 .
KC communities cluster into three types (SA, SB, and D) according to depth and pH. (A) The pH of KC samples varies with depth. The pH of either fresh aqueous suspensions or prepared aqueous leachates of each sample was measured. The measurement for each individual sample, as well as the mean, is plotted. (B and C) Principal-coordinate analysis of core sample communities. Communities for each core sample were clustered using principal-coordinate analysis (PCoA) of unweighted UniFrac distances obtained from 16S rRNA amplicon sequencing. In panel B, symbols representing each KC sample are colored according to a gradient representing depth. In panel C, symbols are colored according to a gradient representing their aqueous leachate pH (acidic [pH ~4] to basic [pH ~9]). PC1, principal component 1.
Physicochemical characteristics of KC core samples. Download TABLE S4, DOCX file, 0.03 MB (32.3KB, docx) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
KC microbiomes cluster into three distinct community types that correlate with depth and pH.
DNA was extracted from each core sample, and community profiling was performed by sequencing 16S rRNA amplicons. We identified 5,032 unique operational taxonomic units (OTUs) (3% genetic distance definition), with a range of 281 (Kis1-28) to 2,410 (Kis3-0 Org). Principal-coordinate analysis (PCoA) of unweighted UniFrac distances (25) was performed to compare beta diversity of KC communities. The first two components accounted for 21.09% and 8.59% of the difference between the communities (Fig. 2B). The KC communities grouped into three clusters (Table S1). The PCoA plot was first overlaid with depth information (i.e., 0 ft, 4 ft, and so on). This suggested that two of the clusters included most of the shallower samples (0 to 8 ft), as well as KC35 (also from the surface) (Fig. 2B). We therefore refer to these “shallow” clusters as SA and SB. In contrast, the third cluster, termed D (“deep/interior”), included mostly samples from 12 ft and below, although some surface samples, such as Kis1-0 and Kis3-4, were in this cluster. For surface (0 to 8 ft) samples in clusters SA and SB, there may also be an effect of the horizontal location of the sample. For example, cluster SA samples were all from Kis1, Kis3, and Kis5 (edge of the deposit [Fig. 1A]), whereas cluster SB samples were from Kis2 and Kis4 (closer to the interior). When the leachate pH was overlaid onto PCoA analyses, we saw a trend reminiscent of depth analysis, where more acidic samples were part of either cluster SA or SB (Fig. 2C). Statistical analyses also suggested that both cluster SA and cluster SB samples had a significantly lower pH than cluster D (P = 0.047 and P = 0.0073, respectively), while the pH of clusters SA and SB was not significantly different (P > 0.5). This suggests that a depth-driven cline in pH affected the microbiome of KC. The KC deposit has a significant organic overburden of macroflora (Fig. 1B), which might decrease the pH at the surface. The KC deposit therefore harbors several different bacterial communities, likely shaped by environmental parameters that change with depth.
KC harbors rich bacterial diversity that is dominated by Betaproteobacteria.
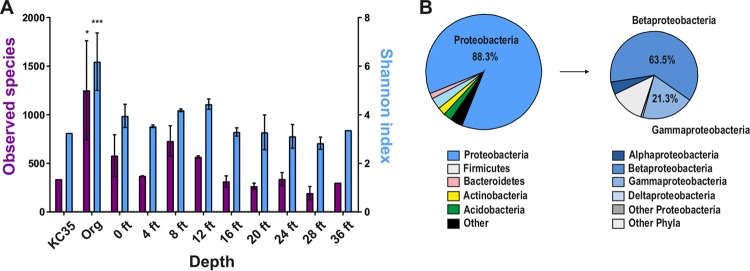
As noted above, we identified more than 5,000 unique OTUs, with a range of 281 (Kis1-28) to 2,410 (Kis3-0 Org). Alpha rarefaction analysis of the sequence data showed that our survey is likely an underestimation of the diversity of some samples (Fig. S1). Nonetheless, we compared the bacterial diversity of each KC sample (observed species and Shannon index) (26) at the same sequencing depth, not including singleton and doubleton OTUs (Fig. 3A). The KC deposit has a layer of overburden (Fig. 1B), and samples that were composed of almost entirely organic material (Kis3-0 Org and Kis5-0; greater than 90% amorphous by X-ray diffraction [XRD]) had a higher mean observed species (mean 1,251 OTUs versus 431 for clay samples; P = 0.0015) and Shannon index (6.2 versus 3.6 for clay samples; P = 0.0003) metrics. Among the clay samples, there was also a minor trend toward lower diversity measures at greater depths (Fig. 3A). We next identified the major bacterial phyla in KC samples. Most of the reads (88.3%) were assigned to Proteobacteria OTUs. Other significant phyla identified included Bacteroidetes (3.2%), Actinobacteria (2.4%), Acidobacteria (1.8%), and Firmicutes (1.3%) (Fig. 3B). Further separation of the Proteobacteria into constituent classes revealed that approximately 63.5% of reads for this phylum could be assigned to Betaproteobacteria, followed by Gammaproteobacteria (21.3%), Alphaproteobacteria (4.0%), and Deltaproteobacteria (0.4%). These results are consistent with other surveys of bacteria in soil environments, which have shown that most are dominated by Proteobacteria (27). Acidobacteria and Actinobacteria are also common in soils. Thus, KC harbors a surprising number of different bacterial species, with a large contribution from Proteobacteria.
FIG 3 .
Bacterial diversity of KC core samples. (A) Mean measures of alpha diversity (observed species [OTUs] and Shannon index) for KC35, core samples composed mostly of organic (Org) material (Kis3-0 Org and Kis5-0), and clay samples from different depths as identified by 16S rRNA amplicon sequencing. The mean ± standard error of the mean (SEM) (error bar) for each indicated sample group for OTUs detected at the same sequencing depth (41,294 reads), not including doubleton and singleton OTUs, are plotted. The values for Org samples that were significantly different from the values for all other samples by Student’s t test are shown by asterisks: *, P ≤ 0.05; ***, P ≤ 0.001. (B) Phylum- and class-level analysis of total assigned reads for all KC samples. OTUs with only one or two reads are not included in this analysis.
Alpha rarefaction for sequencing data. Download FIG S1, PDF file, 0.1 MB (134.6KB, pdf) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The core bacterial communities of KC.
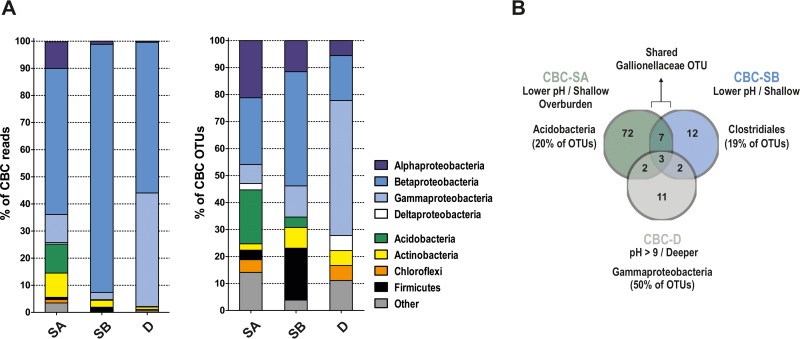
In general, Betaproteobacteria and Gammaproteobacteria species dominated the sequences obtained for all KC samples (Fig. 3B). However, three distinct communities may be present in KC (Fig. 2B and C). To identify taxa that might reveal differences between these communities and/or their environments, we defined a core bacterial community (CBC) for each of the three PCoA clusters. We defined CBCs based on common membership (i.e., an OTU was part of the cluster CBC if at least one sequence read was present in all samples of that cluster). CBC OTUs were also limited to those that made up at least 0.1% of the total reads in the cluster being compared. We chose this approach because of limitations in sample harvesting and storage (see Materials and Methods), and thus, we did not take into account the proportion of the community made up by an OTU, as has been done previously (28).
Under this definition, only a handful of the species we detected (5,032 OTUs) made up the CBC of each KC community (Table S5). For example, the CBC of cluster D, which included 14 of the KC samples, included only 18 OTUs. For comparison between communities, CBC OTUs were separated into major phyla (Proteobacteria, Acidobacteria, Actinobacteria, Chloroflexi, or Firmicutes). Proteobacterial OTUs were also separated according to class (i.e., Alpha-, Beta-, Gammaproteobacteria, etc.). We then compared the proportion of either (i) reads or (ii) OTUs from each phylum/class between the CBCs (Fig. 4A). Betaproteobacteria dominated CBC reads for all three clusters (53.8%, 91.5%, and 55.6% for CBC-SA, CBC-SB, and CBC-D, respectively) (Fig. 4A, left panel). However, while a significant proportion of CBC-SA reads were also assigned to Acidobacteria (10.7%), Gammaproteobacteria (10.4%), Alphaproteobacteria (9.8%), and Actinobacteria (9%), CBC-SB was dominated by Betaproteobacteria. Finally, reads for CBC-D showed a more even distribution between Betaproteobacteria and Gammaproteobacteria (55.6% versus 41.9%, respectively).
FIG 4 .
The core bacterial community (CBC) of KC clusters. For each cluster, OTUs were identified. These OTUs had at least one read in each sample designated part of the cluster and that also made up at least 0.1% of all reads for the cluster. (A, left) Distribution of reads among phyla and proteobacterial classes for OTUs determined to be part of the CBC of each cluster (SA, SB, or D). (Right) Distribution of CBC OTUs among different phyla and proteobacterial classes. Complete OTU listings for each CBC are shown in Table S5 in the supplemental material. (B) Shared and distinct OTUs of cluster CBCs. Details of shared OTUs are listed in Table S5.
Core bacterial community (CBC) OTUs for SA, SB, and D, as well as shared OTUs. Download TABLE S5, DOCX file, 0.05 MB (49KB, docx) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Next, the CBCs of clusters SA, SB, and D were compared based on the percentage of CBC OTUs belonging to different phyla (Fig. 4A, right panel). This revealed that CBC-SA, with the majority of reads from Betaproteobacteria (53.8%), actually had a similar proportion of OTUs from this group (21/85 CBC OTUs [24.7%]) as from Alphaproteobacteria (18/85 [21.1%]) or Acidobacteria (17/85 [20%]). Likewise, CBC-SB, while having more than 90% of reads from Betaproteobacteria, had a comparatively large proportion of Firmicutes (Clostridiales accounted for 5/26, or 19.2%, of OTUs). Finally, CBC-D had more Gammaproteobacteria species (9/18 OTUs) compared to the CBCs of SA and SB. Definition of the core bacterial species of each KC community therefore suggested that the Betaproteobacteria dominated all three communities but with significant differences between the constituent families of species.
We next identified shared or unique species between the three CBCs (Table S5). Only three OTUs were shared by all three CBCs (one Oxalobacteraceae and two pseudomonads). Both taxa include common soil bacteria with diverse metabolic capabilities (29, 30). CBC-SA and -SB (both shallow/low pH) shared more OTUs with each other (10 OTUs) than either one shared with cluster D (5 OTUs each [Fig. 4B]). Three of the OTUs shared by only the CBCs of SA and SB (but absent from CBC-D) were from Alphaproteobacteria lineages, including nitrogen-fixing Rhizobiales normally associated with plant roots, which suggested that the shallower communities may be influenced by the rhizosphere. A notable difference between CBC-SA and CBC-SB was the number of Acidobacteria OTUs (20% versus 4%, respectively). In contrast, we observed more Clostridiales OTUs in CBC-SB (20% versus 3.5% for CBC-SA). The five Clostridiales OTUs of CBC-SB included two potential Desulfosporosinus species and one Thermosinus species, both of which include sulfate-reducing bacteria (31–33). For the CBC of cluster D, there was greater representation from Gammaproteobacteria (such as xanthomonads and pseudomonads), Acinetobacter, and Serratia. Also uniquely present in CBC-D was a member of the Anaerolineae, a Chloroflexi whose cultivated members appear to be strictly anaerobic (34, 35).
Iron bacteria mark the KC microbiome near the surface.
Acid-soluble metal ions, such as iron species, are important for the activity of other antibacterial clays (13), and may be responsible for the activity of KC (S. Behroozian and J. Davies, unpublished data), which is relatively rich in iron (15, 22). To gain insight into the ferruginous environment of KC, we focused on clay bacteria that may play roles in iron cycling. Interestingly, many iron bacteria (those that perform dissimilatory iron oxidization at near neutral pH) that have been characterized are Betaproteobacteria (36, 37), which were dominant in KC. Numerous comamonads (142 OTUs), which may include iron-oxidizing Leptothrix spp., were identified in the total sequence data set, and several were defined as part of CBC-SA, CBC-SB, and CBC-D (Table S5). We also identified genera of iron bacteria or bacteria that can tolerate heavy metals such as Delftia (38) and Acidivorax (36).
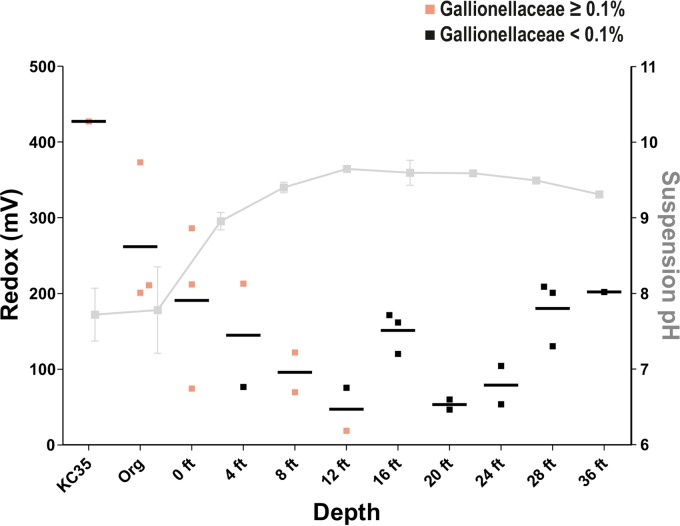
Notably, a Gallionellaceae OTU was shared only by CBC-SA and CBC-SB but was not present in the CBC we defined for cluster D. The type species of this family (Gallionella ferruginea) colonizes the transition zone between anaerobic and aerobic environments (redox potential of +200 to +320 mV and low oxygen tensions of 0.1 to 1 mg/liter) and participates in dissimilatory iron oxidation, requiring Fe(II) and oxygen for growth (39; reviewed in references 36 and 37). For KC samples, we observed a trend of decreasing redox potential from 0 to 12 ft (approximately +300 to +50 mV) and an increase in redox from 20 to 36 ft (+50 to +200 mV) (Fig. 5). Interior samples tended to have a lower potential (+50 to +150 mV). Samples with at least 0.1% of reads from Gallionellaceae OTUs (all 14 identified in our studies) were from 12 ft and above. The absence of Gallionella below 12 ft is consistent with redox measurements of these samples, which were below +200 mV and the previously reported redox niche of these bacteria (39). When the pH of each clay sample was included, all samples with Gallionella had circumneutral, or even slightly acidic pH (Fig. 5). In addition, all samples that had at least 0.1% of reads as Gallionellaceae species were part of clusters SA and SB, except for Kis1-12 (Fig. S2).
FIG 5 .
Prevalence of iron-oxidizing bacteria in KC compared to depth, pH, and redox. Redox measured for KC core samples (left y axis) versus depth (x axis) is shown. Communities with ≥0.1% reads mapping to the order Gallionellaceae are shown. The mean pH of each depth is shown in gray.
Presence of Gallionellaceae sequences in KC sample amplicons, shown according to PCoA clusters (SA, SB, and D). Download FIG S2, PDF file, 0.2 MB (173.1KB, pdf) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Antibacterial and economic potential of KC.
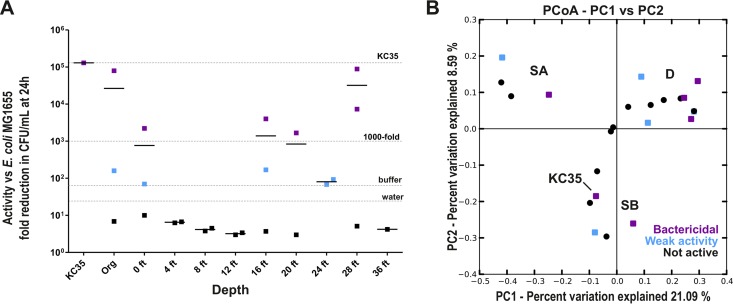
KC exhibits potent inhibitory activity against a variety of bacteria and other microbes, including multidrug-resistant ESKAPE pathogens (20) and pathogenic fungi (Behroozian et al., unpublished). To gain insight into what might contribute to the antibacterial activity of KC and how this activity might shape its resident communities, we also determined the antibacterial activity of each sample used for microbiome analysis against E. coli MG1655. The activity of KC core samples was highly variable (Fig. 6A). While KC35 caused an approximately 5-log-unit decrease in CFU during incubation, the KC core samples varied from bactericidal (purple squares show greater than 3-log-unit killing) to more weakly active (blue squares show less than 1,000-fold decrease in CFU, but higher than water alone) (Fig. 6A). Some samples actually enhanced viability of the test inoculum compared to the H2O control (black squares) (Fig. 6A). While we did not identify a correlation between community type and activity, even though depth appeared to affect microbiome composition, all samples from the interior (i.e., 4 to 12 ft) showed little or no activity. There was also no correlation between activity and core (i.e., Kis3 versus Kis1 [not shown]). Finally, we sought to determine whether activity correlated with the type of bacterial community type identified by PCoA analysis (Fig. 2B and C), as we hypothesized, for example, that communities marking a particular redox environment could affect the speciation of antibacterial Fe ions. However, there was an almost random distribution of high-activity samples (Fig. 6B). There was a very weak correlation between redox potential and antibacterial activity, with samples with higher redox potential tending to show greater reduction of CFU (Fig. S3), while the presence of Gallionella showed no correlation with activity.
FIG 6 .
Antibacterial activity of KC core samples. Suspensions (10 mg/ml) were prepared from dried clay in water from each core sample and tested for antibacterial activity against E. coli MG1655. Bacteria in log phase were added to suspensions and incubated at 37°C with shaking. After 24 h, samples were serially diluted and plated for CFU on LB agar. (A) Higher antibacterial activity is detected at the edges of the deposit. The mean fold reduction (n = 4) for each depth (short lines) and the value for each individual sample (symbols) are plotted. Organic samples and KC35 are plotted separately. Samples with weak activity (blue squares) were defined as those resulting in at least a 66-fold decrease in viable CFU after 24 h, the reduction observed for phosphate buffer (pH 4.3). “Bactericidal” samples (Purple squares) were defined as those reducing viable CFU by at least 1,000-fold. For comparison, the activity of KC35 and water alone are indicated. (B) KC community structure is not predictive of clay activity. The results of unweighted UniFrac PCoA analysis of core sample communities with respect to activity against E. coli MG1655 are indicated as follows: weak activity (>66-fold reduction in CFU); bactericidal (>1,000-fold reduction); not active (less activity than phosphate buffer).
Redox potential, antibacterial activity, and Gallionella sequences. Download FIG S3, PDF file, 0.1 MB (78.4KB, pdf) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Soils are rich sources of economically valuable bacterial species, such as those that produce novel antimicrobials or with plant growth-promoting activities. Up to 3% of reads for KC samples were assigned to the Actinomycetales (Fig. S4A). This group includes many well-known producers of bioactive and antimicrobial secondary metabolites (40). Several species of Paenibacillus, a genus that includes nitrogen fixers with plant growth-promoting ability such as Paenibacillus polymyxa (41), are also present in KC (Fig. S4A). Preliminary culturing studies isolated viable bacteria on Actinomycete-selective agar at relatively high CFU (Fig. S4B).
Potentially economically or medically interesting bacteria in Kis3 samples. Download FIG S4, PDF file, 0.1 MB (126.2KB, pdf) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Characterizing resident microbial populations is essential for a complete understanding of terrestrial environments and systems and identification of microbes with economically useful capabilities. Here, we have characterized the bacteria that colonize the Kisameet Bay glacial clay deposit, which is the source of potently antibacterial KC. The deposit harbors surprising bacterial diversity, with greater than 300 species in most samples and thousands of species in samples mixed with the organic overburden. In fact, our estimates may be an underestimate due to sequencing depth. Previous analyses of the bacterial diversity present in Boom clay borehole water found that this environment contained only 100 to 150 OTUs (42). Diversity in the Boom clay microbiome correlated weakly with total organic carbon measures (42). While we did not measure organic carbon in this study, the organic overburden is likely responsible for the increase in diversity in some samples from the deposit. Soil pH is also a strong predictor of bacterial diversity, with neutral environments harboring more diversity and species richness than environments with either low or high pH (43). The high pH of the interior of the KC deposit is consistent with its slightly lower bacterial diversity. In iron-rich freshwater wetlands, bacteria such as Gallionella tend to be more abundant in the spring compared to summer or fall (44, 45). Sampling of KC in different seasons may reveal variable temporal bacterial community dynamics. Profiling of resident Archaea, Eukarya, or bacteriophages will also further an understanding of the microbial diversity of KC.
To date, there have been few studies of the bacterial content of native clays. Bacteria enriched from a desiccated sample of commercially available Wyoming bentonite clay were found to be closely related to Desulfovibrio africanus, a sulfate-reducing deltaproteobacterium (46). We did not detect this species in our survey; however, the deeper samples of cluster D did share a deltaproteobacterium OTU. Two studies have addressed the microbiome of the deep subsurface Boom clay. A 16S rRNA clone library approach tentatively identified 11 different OTUs in the clay (47). Interestingly, Proteobacteria made up 76% of the species, with Betaproteobacteria, such as the comamonad Acidovorax, accounted for 46%. Our survey of KC bacteria likewise suggests a dominance of Betaproteobacteria. In addition, both the CBCs defined in our survey and the species identified in the Boom clay revealed Gammaproteobacteria such as Pseudomonas and Acinetobacter.
We note that KC harbors at least three major bacterial community types. Two different communities (SA and SB) were identified in more shallow/lower pH samples. Comparison of the CBCs of these clusters showed that cluster SA had more Acidobacteria (27, 48, 49) than cluster SB. In contrast, acidobacteria were absent from the CBC defined for the deeper cluster D. The Acidobacteria is a relatively new, poorly characterized, and diverse phylum, although members of this group have been identified in many different environments, where they may constitute up to 20% of bacteria (27, 49, 50). We note that the two samples composed of mostly organic material were part of cluster SA. Cluster SA may represent communities in soil like KC under higher organic material or oxygen tension. Acidobacteria were not reported in the Boom clay (42, 47) but were found to be relatively abundant in 16S rRNA libraries prepared from low-pH, clay-rich Cerrado soil (51). The presence of this clade in CBC-SA may therefore reflect a more soil-like, rather than clay-like, environment. Certainly, the correlation between the horizontal location of the core and the PCoA cluster for shallow samples (i.e., the tendency of surface samples from cores extracted closer to the edge of the deposit, such as Kis1, Kis3, and Kis5, to be part of cluster SA) provides an impetus to explore correlations of these communities with their macroflora neighbors.
In contrast to CBC-SA, CBC-SB included four potential sulfate-reducing clostridia—three Desulfosporosinus species and a Thermosinus species. Boom clay likewise included numerous clostridia such as sulfate-reducing Desulfotomaculum (42). Sulfate-reducing bacteria participate in anaerobic dissimilatory reduction of sulfate to compounds such as hydrogen sulfide, and are often associated with decaying organic matter (52). Thus, cluster SB may reveal environments that are more anaerobic, while still in proximity to the overburden layer of the deposit. Interestingly, cyclic S8 extracted from KC with organic solvents (J. Tan and J. Davies, unpublished observations), has antibacterial activity (53–55). Therefore, microbial sulfur cycling may contribute to the complexity of KC. Furthermore, Desulfosporosinus and Thermosinus strains have been identified that can reduce not only sulfate but also Fe(III) (56, 57), supporting the presence of both these Clostridia and iron bacteria in KC.
KC is iron-rich (15, 22), and potential iron bacteria were identified in our survey. In particular, Gallionella-like iron bacteria are found in shallower samples of KC. Gallionella spp. are microaerobic iron oxidizers that live at iron-rich oxic/anoxic interfaces (36). While they have been described as inhabiting neutral pH zones, acid-tolerant species of Gallionella have been identified, and Gallionella-like organisms are increasingly found in heavy metal-rich acid mine drainage (58–64). Bulk clay iron levels are relatively uniform across the deposit (data not shown), but the low pH of some regions may affect the solubility and availability of iron species. KC provides an interesting environment for studying iron bacteria, which are associated with biofouling and the deterioration of pipes and water distribution systems (36), and the 14 Gallionellaceae OTUs identified in KC may represent species with novel characteristics. Iron-oxidizing bacteria have been shown to have potential bioagents removing heavy metals (65).
What makes KC antibacterial? Previous characterization of density-separated KC fractions showed that antibacterial activity was higher in those fractions that had higher water-leachable metal content (W. Xu, S. Behroozian, W. Yenjaichon, J. Grace, J. Davies, and L. Li, unpublished data). Low pH also affects KC activity, which may contribute to the solubility of ions such as Fe(II), Fe(III), or Al(III) (Behroozian et al., unpublished). We found a weak relationship between redox condition, which might affect the speciation or solubility of antibacterial ions, such as iron, and antibacterial activity. Alternatively, specific redox regions may indicate regions of the clay where antibacterial activity resides. For example, Jordan’s red clay harbors bacteria of the genus Lysobacter, xanthomonads that are important biocontrol agents and sources of exoenzymes and antibiotics (66), which appear to be responsible for its antibacterial activity (19). However, we found no strong correlation between bacterial community type and activity of the clay. Highly active KC35 had an increased number of Gallionella reads, but we found no correlation between Gallionella and activity in core samples. Acidobacteria are known to produce antibiotics such as polyketides (67). KC awaits more-focused culturing studies for the identification of useful bacteria. While we cannot rule out the possibility that particular bacterial species contribute to its inhibitory properties, the bioactivity of KC does not strongly shape community structure.
KC is a very complex natural material, and many different factors may contribute to its unique properties, including the resident bacterial communities. If clays and other natural products are to be used as therapeutic agents, their resident bacteria must be characterized, as they may alter clay structure and affect the efficacy and/or consistency of preparations. KC bacteria should be examined in more detail for the production of novel bioactive compounds or useful metabolic capabilities. In addition, future microbiome studies of natural environments, such as the KC deposit with the unique chemical, absorptive, diffusion, and microbial properties of KC, may uncover novel microbial interactions involving small bioactive molecules, antimicrobial metal ions, nutrients, and oxygen availability. The KC deposit may serve as an environment in which to observe novel chemical and genetic interactions between microbes, to study ecological succession, and to reveal new bacterial types with valuable economic properties.
MATERIALS AND METHODS
Harvesting and storage of core samples.
Five vertical cores (Kis1 to Kis5) were harvested from the Kisameet Bay deposit (51°58′16″ N/127°52′56″ W) on 24 October 2012. Following transport to the University of British Columbia, cores were stored sealed at 4°C under normal atmospheric conditions in the dark. The cores were opened, and the top 10 cm from each 4 ft length of core was saved for analyses of physical, chemical, and biotic characteristics. To avoid cross-contamination between depths, only the interior undisturbed part of each core was used for analysis. To measure antibiotic activity, a portion of each core sample was dried in a desiccator at room temperature under vacuum for 2 to 4 days, ground to a fine powder with a mortar and pestle, autoclaved for 20 min, and stored at room temperature in the dark prior to testing. For metagenome analysis and culturing studies, unautoclaved, native clay was used. The sample activity, physicochemical characteristics, and bacterial community were compared to that of the previously characterized, highly active sample KC35 (20).
Measurement of KC mineralogical and physicochemical characteristics.
Details of the analysis of the abiotic properties of KC core samples (mineralogy by X-ray diffraction, pH, redox, and elemental analyses by inductively coupled plasma optical emission spectroscopy) are presented in Text S1 in the supplemental material.
Supplemental methods. Download TEXT S1, DOCX file, 0.04 MB (47.8KB, docx) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Extraction of DNA from KC.
Total DNA was extracted from wet, unautoclaved clay from each core sample using the FastDNA kit for soil (MP Biomedicals, Santa Ana, CA) according to the manufacturer’s instructions with minor modifications. Briefly, ~400 mg of wet, unautoclaved clay was added to a tube with ~500 mg of silica beads (1-mm diameter; Sigma). Samples were shaken on a FastPrep homogenizer (MP Biomedicals) for 40 s at a speed of 6.0. Following removal of particulate matter at 14,000 × g for 10 min, the clarified extract was applied to a spin column and purified according to the manufacturer’s recommendations. DNA was eluted with water and quantified using a Nanodrop spectrophotometer.
Next-generation sequencing of KC 16S rRNA amplicons and sequence data analysis.
Sequencing of the 16S rRNA gene variable 3 (V3) region, amplified by PCR with primers 341F (F stands for forward) and 518R (R stands for reverse) from total DNA extracted from KC core samples, was performed as previously described (68, 69) on an Illumina Miseq platform. Details of library preparation and sequencing can be found in Text S1. Details of data quality control and analysis using Qiime (26) are likewise outlined in detail in Text S1.
Isolation of KC-resident bacteria.
Viable bacteria from KC core samples and KC35 were cultured from unautoclaved clay. To determine viable counts of Actinomycete-like bacteria, equal weights of wet unautoclaved clay from each core sample that had been stored under native atmospheric conditions at 4°C (approximately 100 mg) were suspended in 10-ml portions of sterile saline, vortexed, and serially diluted in sterile saline. Dilutions were plated on Actinomycete isolation agar (AIA) and incubated at 30°C for 4 to 6 days until colonies were visible.
Antibacterial activity of core samples.
Quantification of antibacterial activity was performed essentially as previously described (20) using the wild-type Escherichia coli K-12 strain MG1655 (70), grown aerobically at 37°C in Luria-Bertani broth (LB). Each clay sample was tested at a concentration of 10 mg/ml, as described in Text S1.
ACKNOWLEDGMENTS
We especially acknowledge the generosity of the British Columbia Heiltsuk First Nation for providing access to the Kisameet deposit, as well as Kisameet Glacial Clay, Inc., and Lawry Lund. We also thank Vivian Miao and Ivan Villanueva for helpful discussions and Jarvis Li for technical assistance.
Support for these studies comes from a Mitacs Elevate Fellowship to S.L.S., a Mitacs (UBC) student fellowship (grant 22R07416) to S.B., Kisameet Glacial Clay, Inc., and the Tally fund (J.D.). Work in J.D.’s laboratory is also supported by the National Sciences and Engineering Research Council of Canada.
Footnotes
Citation Svensson SL, Behroozian S, Xu W, Surette MG, Li L, Davies J. 2017. Kisameet glacial clay: an unexpected source of bacterial diversity. mBio 8:e00590-17. https://doi.org/10.1128/mBio.00590-17.
REFERENCES
- 1.Alivisatos AP, Blaser MJ, Brodie EL, Chun M, Dangl JL, Donohue TJ, Dorrestein PC, Gilbert JA, Green JL, Jansson JK, Knight R, Maxon ME, McFall-Ngai MJ, Miller JF, Pollard KS, Ruby EG, Taha SA, Unified Microbiome Initiative Consortium . 2015. A unified initiative to harness Earth’s microbiomes. Science 350:507–508. doi: 10.1126/science.aac8480. [DOI] [PubMed] [Google Scholar]
- 2.Blaser M, Bork P, Fraser C, Knight R, Wang J. 2013. The microbiome explored: recent insights and future challenges. Nat Rev Microbiol 11:213–217. doi: 10.1038/nrmicro2973. [DOI] [PubMed] [Google Scholar]
- 3.Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. 2010. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 4.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 5.Carlet J, Pulcini C, Piddock LJ. 2014. Antibiotic resistance: a geopolitical issue. Clin Microbiol Infect 20:949–953. doi: 10.1111/1469-0691.12767. [DOI] [PubMed] [Google Scholar]
- 6.Sengupta S, Chattopadhyay MK, Grossart HP. 2013. The multifaceted roles of antibiotics and antibiotic resistance in nature. Front Microbiol 4:47. doi: 10.3389/fmicb.2013.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schaberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. 2015. A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nichols D, Cahoon N, Trakhtenberg EM, Pham L, Mehta A, Belanger A, Kanigan T, Lewis K, Epstein SS. 2010. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl Environ Microbiol 76:2445–2450. doi: 10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunet de Courssou L. 2002. Study Group Report on Buruli ulcer treatment with clay, 5th WHO Advisory Group Meeting on Buruli ulcer, World Health Organization, Geneva, Switzerland. [Google Scholar]
- 10.Williams LB, Haydel SE, Giese RF, Eberl DD. 2008. Chemical and mineralogical characteristics of French green clays used for healing. Clays Clay Miner 56:437–452. doi: 10.1346/CCMN.2008.0560405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giese RF, van Oss CJ. 2002. Clay minerals, colloid and surface properties of clays and related minerals. Marcel Dekker, New York, NY. [Google Scholar]
- 12.Carretero MI. 2002. Clay minerals and their beneficial effects upon human health. A review. Appl Clay Sci 21:155–163. doi: 10.1016/S0169-1317(01)00085-0. [DOI] [Google Scholar]
- 13.Cunningham TM, Koehl JL, Summers JS, Haydel SE. 2010. pH-dependent metal ion toxicity influences the antibacterial activity of two natural mineral mixtures. PLoS One 5:e9456. doi: 10.1371/journal.pone.0009456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrison KD, Underwood JC, Metge DW, Eberl DD, Williams LB. 2014. Mineralogical variables that control the antibacterial effectiveness of a natural clay deposit. Environ Geochem Health 36:613–631. doi: 10.1007/s10653-013-9585-0. [DOI] [PubMed] [Google Scholar]
- 15.Williams LB, Metge DW, Eberl DD, Harvey RW, Turner AG, Prapaipong P, Poret-Peterson AT. 2011. What makes a natural clay antibacterial? Environ Sci Technol 45:3768–3773. doi: 10.1021/es1040688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams LB, Haydel SE, Ferrell RE. 2009. Bentonite, bandaids, and borborygmi. Elements (Que) 5:99–104. doi: 10.2113/gselements.5.2.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams LB, Haydel SE. 2010. Evaluation of the medicinal use of clay minerals as antibacterial agents. Int Geol Rev 52:745–770. doi: 10.1080/00206811003679737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Londono SC, Williams LB. 2016. Unraveling the antibacterial mode of action of a clay from the Colombian Amazon. Environ Geochem Health 38:363–379. doi: 10.1007/s10653-015-9723-y. [DOI] [PubMed] [Google Scholar]
- 19.Falkinham JO III, Wall TE, Tanner JR, Tawaha K, Alali FQ, Li C, Oberlies NH. 2009. Proliferation of antibiotic-producing bacteria and concomitant antibiotic production as the basis for the antibiotic activity of Jordan’s red soils. Appl Environ Microbiol 75:2735–2741. doi: 10.1128/AEM.00104-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behroozian S, Svensson SL, Davies J. 2016. Kisameet clay exhibits potent antibacterial activity against the ESKAPE pathogens. mBio 7:e01842-15. doi: 10.1128/mBio.01842-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rice LB. 2008. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis 197:1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 22.Hauser EA, Colombo U. 1953. Colloid science of montmorillonites and bentonites. Clays Clay Miner 2:439–461. doi: 10.1346/CCMN.1953.0020136. [DOI] [Google Scholar]
- 23.Minister of Mines, Province of British Columbia 1952. Annual report for the year ended 31st December 1951. British Columbia Department of Mines, Victoria, British Columbia, Canada. [Google Scholar]
- 24.British Columbia Geological Survey 2011. MINFILE no. 092M 007. Kisameet Bay, Canadian Canamin, Ray-Vite, Hvidsten Inlet. Ministry of Energy and Mines, Victoria, British Columbia, Canada: http://minfile.gov.bc.ca/Summary.aspx?minfilno=092M++007. [Google Scholar]
- 25.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janssen PH. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shade A, Handelsman J. 2012. Beyond the Venn diagram: the hunt for a core microbiome. Environ Microbiol 14:4–12. doi: 10.1111/j.1462-2920.2011.02585.x. [DOI] [PubMed] [Google Scholar]
- 29.Baldani J, Rouws L, Cruz L, Olivares F, Schmid M, Hartmann A. 2014. The family Oxalobacteraceae, p 919–974. In Rosenberg E, DeLong E, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes. Springer, Berlin, Germany. doi: 10.1007/978-3-642-30197-1_291. [DOI] [Google Scholar]
- 30.Silby MW, Winstanley C, Godfrey SA, Levy SB, Jackson RW. 2011. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35:652–680. doi: 10.1111/j.1574-6976.2011.00269.x. [DOI] [PubMed] [Google Scholar]
- 31.Vatsurina A, Badrutdinova D, Schumann P, Spring S, Vainshtein M. 2008. Desulfosporosinus hippei sp. nov., a mesophilic sulfate-reducing bacterium isolated from permafrost. Int J Syst Evol Microbiol 58:1228–1232. doi: 10.1099/ijs.0.65368-0. [DOI] [PubMed] [Google Scholar]
- 32.Alazard D, Joseph M, Battaglia-Brunet F, Cayol JL, Ollivier B. 2010. Desulfosporosinus acidiphilus sp. nov.: a moderately acidophilic sulfate-reducing bacterium isolated from acid mining drainage sediments. New taxa: Firmicutes (class Clostridia, order Clostridiales, family Peptococcaceae). Extremophiles 14:305–312. doi: 10.1007/s00792-010-0309-4. [DOI] [PubMed] [Google Scholar]
- 33.Stackebrandt E, Sproer C, Rainey FA, Burghardt J, Päuker O, Hippe H. 1997. Phylogenetic analysis of the genus Desulfotomaculum: evidence for the misclassification of Desulfotomaculum guttoideum and description of Desulfotomaculum orientis as Desulfosporosinus orientis gen. nov., comb. nov. Int J Syst Bacteriol 47:1134–1139. doi: 10.1099/00207713-47-4-1134. [DOI] [PubMed] [Google Scholar]
- 34.Yamada T, Sekiguchi Y, Hanada S, Imachi H, Ohashi A, Harada H, Kamagata Y. 2006. Anaerolinea thermolimosa sp. nov., Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new classes Anaerolineae classis nov. and Caldilineae classis nov. in the bacterial phylum Chloroflexi. Int J Syst Evol Microbiol 56:1331–1340. doi: 10.1099/ijs.0.64169-0. [DOI] [PubMed] [Google Scholar]
- 35.Sekiguchi Y, Yamada T, Hanada S, Ohashi A, Harada H, Kamagata Y. 2003. Anaerolinea thermophila gen. nov., sp. nov. and Caldilinea aerophila gen. nov., sp. nov., novel filamentous thermophiles that represent a previously uncultured lineage of the domain Bacteria at the subphylum level. Int J Syst Evol Microbiol 53:1843–1851. doi: 10.1099/ijs.0.02699-0. [DOI] [PubMed] [Google Scholar]
- 36.Emerson D, Fleming EJ, McBeth JM. 2010. Iron-oxidizing bacteria: an environmental and genomic perspective. Annu Rev Microbiol 64:561–583. doi: 10.1146/annurev.micro.112408.134208. [DOI] [PubMed] [Google Scholar]
- 37.Hedrich S, Schlömann M, Johnson DB. 2011. The iron-oxidizing proteobacteria. Microbiology 157:1551–1564. doi: 10.1099/mic.0.045344-0. [DOI] [PubMed] [Google Scholar]
- 38.Johnston CW, Wyatt MA, Li X, Ibrahim A, Shuster J, Southam G, Magarvey NA. 2013. Gold biomineralization by a metallophore from a gold-associated microbe. Nat Chem Biol 9:241–243. doi: 10.1038/nchembio.1179. [DOI] [PubMed] [Google Scholar]
- 39.Hanert H. 1992. The genus Gallionella, p 4082–4088. In Balows A, Trüper H, Dworkin M, Harder W, Schleifer K-H (ed), The prokaryotes. Springer, New York, New York. doi: 10.1007/978-1-4757-2191-1_69. [DOI] [Google Scholar]
- 40.Miao V, Davies J. 2010. Actinobacteria: the good, the bad, and the ugly. Antonie Van Leeuwenhoek 98:143–150. doi: 10.1007/s10482-010-9440-6. [DOI] [PubMed] [Google Scholar]
- 41.Lal S, Tabacchioni S. 2009. Ecology and biotechnological potential of Paenibacillus polymyxa: a minireview. Indian J Microbiol 49:2–10. doi: 10.1007/s12088-009-0008-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wouters K, Moors H, Boven P, Leys N. 2013. Evidence and characteristics of a diverse and metabolically active microbial community in deep subsurface clay borehole water. FEMS Microbiol Ecol 86:458–473. doi: 10.1111/1574-6941.12171. [DOI] [PubMed] [Google Scholar]
- 43.Fierer N, Jackson RB. 2006. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J, Vollrath S, Behrends T, Bodelier PL, Muyzer G, Meima-Franke M, Den Oudsten F, Van Cappellen P, Laanbroek HJ. 2011. Distribution and diversity of Gallionella-like neutrophilic iron oxidizers in a tidal freshwater marsh. Appl Environ Microbiol 77:2337–2344. doi: 10.1128/AEM.02448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fleming EJ, Cetinić I, Chan CS, Whitney King D, Emerson D. 2014. Ecological succession among iron-oxidizing bacteria. ISME J 8:804–815. doi: 10.1038/ismej.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masurat P, Eriksson S, Pedersen K. 2010. Evidence of indigenous sulphate-reducing bacteria in commercial Wyoming bentonite MX-80. Appl Clay Sci 47:51–57. doi: 10.1016/j.clay.2008.07.002. [DOI] [Google Scholar]
- 47.Boivin-Jahns V, Ruimy R, Bianchi A, Daumas S, Christen R. 1996. Bacterial diversity in a deep-subsurface clay environment. Appl Environ Microbiol 62:3405–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barns SM, Takala SL, Kuske CR. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl Environ Microbiol 65:1731–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward NL, Challacombe JF, Janssen PH, Henrissat B, Coutinho PM, Wu M, Xie G, Haft DH, Sait M, Badger J, Barabote RD, Bradley B, Brettin TS, Brinkac LM, Bruce D, Creasy T, Daugherty SC, Davidsen TM, DeBoy RT, Detter JC, Dodson RJ, Durkin AS, Ganapathy A, Gwinn-Giglio M, Han CS, Khouri H, Kiss H, Kothari SP, Madupu R, Nelson KE, Nelson WC, Paulsen I, Penn K, Ren Q, Rosovitz MJ, Selengut JD, Shrivastava S, Sullivan SA, Tapia R, Thompson LS, Watkins KL, Yang Q, Yu C, Zafar N, Zhou L, Kuske CR. 2009. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol 75:2046–2056. doi: 10.1128/AEM.02294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naether A, Foesel BU, Naegele V, Wüst PK, Weinert J, Bonkowski M, Alt F, Oelmann Y, Polle A, Lohaus G, Gockel S, Hemp A, Kalko EK, Linsenmair KE, Pfeiffer S, Renner S, Schöning I, Weisser WW, Wells K, Fischer M, Overmann J, Friedrich MW. 2012. Environmental factors affect acidobacterial communities below the subgroup level in grassland and forest soils. Appl Environ Microbiol 78:7398–7406. doi: 10.1128/AEM.01325-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Castro AP, Quirino BF, Allen H, Williamson LL, Handelsman J, Krüger RH. 2011. Construction and validation of two metagenomic DNA libraries from Cerrado soil with high clay content. Biotechnol Lett 33:2169–2175. doi: 10.1007/s10529-011-0693-6. [DOI] [PubMed] [Google Scholar]
- 52.Muyzer G, Stams AJ. 2008. The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol 6:441–454. doi: 10.1038/nrmicro1892. [DOI] [PubMed] [Google Scholar]
- 53.Weld JT, Gunther A. 1947. The antibacterial properties of sulfur. J Exp Med 85:531–542. doi: 10.1084/jem.85.5.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider T, Baldauf A, Ba LA, Jamier V, Khairan K, Sarakbi MB, Reum N, Schneider M, Röseler A, Becker K, Burkholz T, Winyard PG, Kelkel M, Diederich M, Jacob C. 2011. Selective antimicrobial activity associated with sulfur nanoparticles. J Biomed Nanotechnol 7:395–405. doi: 10.1166/jbn.2011.1293. [DOI] [PubMed] [Google Scholar]
- 55.Eder SH, Gigler AM, Hanzlik M, Winklhofer M. 2014. Sub-micrometer-scale mapping of magnetite crystals and sulfur globules in magnetotactic bacteria using confocal Raman micro-spectrometry. PLoS One 9:e107356. doi: 10.1371/journal.pone.0107356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bertel D, Peck J, Quick TJ, Senko JM. 2012. Iron transformations induced by an acid-tolerant Desulfosporosinus species. Appl Environ Microbiol 78:81–88. doi: 10.1128/AEM.06337-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sokolova TG, González JM, Kostrikina NA, Chernyh NA, Slepova TV, Bonch-Osmolovskaya EA, Robb FT. 2004. Thermosinus carboxydivorans gen. nov., sp. nov., a new anaerobic, thermophilic, carbon-monoxide-oxidizing, hydrogenogenic bacterium from a hot pool of Yellowstone National Park. Int J Syst Evol Microbiol 54:2353–2359. doi: 10.1099/ijs.0.63186-0. [DOI] [PubMed] [Google Scholar]
- 58.Fabisch M, Beulig F, Akob DM, Küsel K. 2013. Surprising abundance of Gallionella-related iron oxidizers in creek sediments at pH 4.4 or at high heavy metal concentrations. Front Microbiol 4:390. doi: 10.3389/fmicb.2013.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner C, Mau M, Schlömann M, Heinicke J, Koch U. 2007. Characterization of the bacterial flora in mineral waters in upstreaming fluids of deep igneous rock aquifers. J Geophys Res 112:2156–2202. doi: 10.1029/2005JG000105. [DOI] [Google Scholar]
- 60.Jones DS, Kohl C, Grettenberger C, Larson LN, Burgos WD, Macaladya JL. 2015. Geochemical niches of iron-oxidizing acidophiles in acidic coal mine drainage. Appl Environ Microbiol 81:1242–1250. doi: 10.1128/AEM.02919-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bruneel O, Duran R, Casiot C, Elbaz-Poulichet F, Personné JC. 2006. Diversity of microorganisms in Fe-As-rich acid mine drainage waters of Carnoules, France. Appl Environ Microbiol 72:551–556. doi: 10.1128/AEM.72.1.551-556.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim JJ, Kim SJ, Lee SS. 2003. Gallionella ferruginea in ochreous precipitates from acid mine drainage in Donghae coal mine area, Korea. Geosci J 7:289–292. doi: 10.1007/BF02919558. [DOI] [Google Scholar]
- 63.Fabisch M, Freyer G, Johnson CA, Büchel G, Akob DM, Neu TR, Küsel K. 2016. Dominance of “Gallionella capsiferriformans” and heavy metal association with Gallionella-like stalks in metal-rich pH 6 mine water discharge. Geobiology 14:68–90. doi: 10.1111/gbi.12162. [DOI] [PubMed] [Google Scholar]
- 64.Hallberg KB, Coupland K, Kimura S, Johnson DB. 2006. Macroscopic streamer growths in acidic, metal-rich mine waters in north Wales consist of novel and remarkably simple bacterial communities. Appl Environ Microbiol 72:2022–2030. doi: 10.1128/AEM.72.3.2022-2030.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katsoyiannis IA, Zouboulis AI. 2004. Application of biological processes for the removal of arsenic from groundwaters. Water Res 38:17–26. doi: 10.1016/j.watres.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 66.Xie Y, Wright S, Shen Y, Du L. 2012. Bioactive natural products from Lysobacter. Nat Prod Rep 29:1277–1287. doi: 10.1039/c2np20064c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parsley LC, Linneman J, Goode AM, Becklund K, George I, Goodman RM, Lopanik NB, Liles MR. 2011. Polyketide synthase pathways identified from a metagenomic library are derived from soil Acidobacteria. FEMS Microbiol Ecol 78:176–187. doi: 10.1111/j.1574-6941.2011.01122.x. [DOI] [PubMed] [Google Scholar]
- 68.Whelan FJ, Verschoor CP, Stearns JC, Rossi L, Luinstra K, Loeb M, Smieja M, Johnstone J, Surette MG, Bowdish DM. 2014. The loss of topography in the microbial communities of the upper respiratory tract in the elderly. Ann Am Thorac Soc 11:513–521. doi: 10.1513/AnnalsATS.201310-351OC. [DOI] [PubMed] [Google Scholar]
- 69.Bartram AK, Lynch MD, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. 2011. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl Environ Microbiol 77:3846–3852. doi: 10.1128/AEM.02772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bachmann BJ. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev 36:525–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Library and read statistics for next-generation sequencing. Download TABLE S1, DOCX file, 0.03 MB (33KB, docx) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mineralogical composition of selected KC core samples. Download TABLE S2, DOCX file, 0.03 MB (36KB, docx) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Acid-digested elemental composition of KC core sample bulk clay. Download TABLE S3, DOCX file, 0.04 MB (43.9KB, docx) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Physicochemical characteristics of KC core samples. Download TABLE S4, DOCX file, 0.03 MB (32.3KB, docx) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Alpha rarefaction for sequencing data. Download FIG S1, PDF file, 0.1 MB (134.6KB, pdf) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Core bacterial community (CBC) OTUs for SA, SB, and D, as well as shared OTUs. Download TABLE S5, DOCX file, 0.05 MB (49KB, docx) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Presence of Gallionellaceae sequences in KC sample amplicons, shown according to PCoA clusters (SA, SB, and D). Download FIG S2, PDF file, 0.2 MB (173.1KB, pdf) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Redox potential, antibacterial activity, and Gallionella sequences. Download FIG S3, PDF file, 0.1 MB (78.4KB, pdf) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Potentially economically or medically interesting bacteria in Kis3 samples. Download FIG S4, PDF file, 0.1 MB (126.2KB, pdf) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental methods. Download TEXT S1, DOCX file, 0.04 MB (47.8KB, docx) .
Copyright © 2017 Svensson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.