Abstract
Matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) has been used to identify bacteria based upon protein signatures. This research shows that while some different proteins are produced by vegetative bacteria when they are cultured in different growth media, positive identification with MALDI-TOF MS is still possible with the protocol established at the Pacific Northwest National Laboratory (K. H. Jarman, S. T. Cebula, A. J. Saenz, C. E. Petersen, N. B. Valentine, M. T. Kingsley, and K. L. Wahl, Anal. Chem. 72:1217-1223, 2000). A core set of small proteins remain constant under at least four different culture media conditions and blood agar plates, including minimal medium M9, rich media, tryptic soy broth (TSB) or Luria-Bertani (LB) broth, and blood agar plates, such that analysis of the intact cells by matrix-assisted laser desorption/ionization mass spectrometry allows for consistent identification.
There is a growing need for rapid microorganism identification capabilities in cases of terrorist attacks, in food processing plants, in hospitals, and in health care facilities. Bacterial identification by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) has been widely studied (7, 14). The approach chosen at Pacific Northwest National Laboratory (PNNL) was to develop a library of reproducible spectral signature peaks characteristic of different bacterial genera and species for identification of unknown samples (11). The ability of MALDI-TOF MS to identify bacteria to the species level in pure cultures and simple microbial mixtures under carefully controlled culture and analysis conditions has been established already (20). Also established was an automated statistically based algorithm for objective analysis and evaluation of the MALDI-TOF MS data. Questions have arisen as to whether microorganisms can be identified by this method if they are grown under different culturing conditions which may turn on different proteins. A study done by Longo et al. (16) indicated that different nutrients might affect the pattern of protease production in bacteria. Arnold et al. (1) also showed visible differences in the MALDI-TOF MS profiles for Escherichia coli obtained throughout the growth curve. However, Bernardo et al. (2) recently showed that growth on two different media (Mueller-Hinton agar and Columbia blood agar) did not significantly alter the MALDI-TOF MS profiles. Thus, the question remains how applicable will this technology be with automated identification for organisms grown under conditions different from the signature conditions? In particular, can the bacterial signatures generated by MALDI-TOF MS on clean samples cultured in rich media in the laboratory be used to identify the same organisms cultured under different growth conditions, such as minimal media or blood agar plates? The effect of various culture media on MALDI-TOF MS identification is examined with the media selection based on previous reports of inducing physiological changes in the organisms of interest.
According to previous reports, use of the minimal medium chosen for this study has induced changes in protein expression and cell envelope composition for both E. coli and Bacillus subtilis (4). Two-dimensional electrophoresis has been used to visualize the protein expression differences for an E. coli strain grown on both minimal (M9) and rich media. B. subtilis has been extensively studied with regard to the effects of growth media on cell physiology and composition, and several strains have been shown to produce variations in the cell wall polymer teichoic and teichuronic acids as well as phage binding under nutrient (phosphate in particular) limitation (5, 8, 13, 21). In particular, levels of galactose and galactosamine varied in strains of B. subtilis cultured on nutrient broth rich media versus phosphate-amended semidefined minimal medium (similar to M9), demonstrating the variability in cellular composition (8). More recently, numerous genes whose expression is altered by growth on a medium with a minimal salts or carbon source versus a rich LB medium have been identified by transcriptome and proteome analyses (17).
Sheep blood agar (SBA) has also been used as a nutrient-rich medium because of its utility in differentiating Bacillus anthracis strains from the closely related members of the B. cereus strains. Strains of B. cereus and B. thuringiensis exhibit hemolysis, whereas B. anthracis strains do not (21). Laboratories growing and characterizing strains of B. anthracis will use blood agar as an indicator medium, thus making this medium relevant for use in developing a signature for Bacillus species. Blood agar (SSI [Statens Serum Institute] enteric medium, 5% sheep's blood) has also been reported as a suitable medium to culture Yersinia spp. in a clinical setting; thus, blood agar is a relevant medium for culturing Yersinia spp. (3).
The organisms used in this study were chosen as representatives of genera containing agents used as biological weapons (BW). B. subtilis, for example, is selected as a simulant for B. anthracis. B. subtilis var. niger (formerly B. subtilis var. globigii) has a long history of use as a simulant for B. anthracis (9). While the differences between the simulant and the BW agent B. anthracis need to be noted (such as susceptibility of B. subtilis strains to enzymatic lysis by lysozyme [21]), the characterization of the genetics and physiology of B. subtilis (6) make it well suited for this study. Yersinia enterocolitica and E. coli are included as gram-negative representatives to study effects of media and growth conditions. The Y. enterocolitica species was chosen as a gram-negative biological warfare simulant not only because it is a pathogen in its own right but also because of its relatedness to the BW agent Y. pestis, also known as the plague (3, 15).
The patented automated peak extraction and analysis algorithms (11, 12) developed at PNNL were used to compare the signatures of the microorganisms to those in the signature library. The algorithms have been published previously (11, 12), and the code has been made available upon request to government agencies and universities. An extensive library construction process has been utilized at PNNL to capture the variability in both the organism and analytical process. The microorganisms are cultured on three separate days, and duplicate aliquots of each sample are washed and spotted onto a MALDI plate. Sixty replicate spectra are collected over the 3-day period to account for day-to-day variability. The ions present in at least 70% of the spectra are included into a signature library for that microorganism (11). Unknown microorganisms can be analyzed and compared to the signature library to determine a statistical degree of association to the known microorganisms. While this identification system has worked well with individual bacteria cultured on a given medium and even with simple mixtures of bacteria (20), it remains to be seen if microorganisms cultured on different media can still be identified. In each case, a correct identification was made based upon a constant set of proteins in the signature. This parallels the findings of Arnold et al. (1), who examined different stages of growth by using MALDI-MS. The results of MALDI-TOF MS signatures and automated data analysis and identification are described here for E. coli, B. subtilis, and Y. enterocolitica cultured in four different media.
MATERIALS AND METHODS
Supplies.
The cultures used in the study included Y. enterocolitica (ATCC 51871), B. subtilis (ATCC 15841), and E. coli W3110 (ATCC 27325). They were obtained from the American Type Culture Collection, Manassas, Va. Media included M9 minimal salts, purchased from Sigma (St. Louis, Mo.), tryptic soy broth without dextrose (TSB), purchased from Fisher Scientific (Pittsburgh, Pa.), Difco LB, purchased from Fisher Scientific, and tryptic soy agar/sheep blood agar plates (TSA_BA) and Columbia agar with 5% sheep blood (CAB), obtained from PML Microbiologicals (Portland, Oreg.). Protein standards, horse heart cytochrome c, and angiotensin I were purchased from Sigma. Ferulic acid and trifluoroacetic acid (TFA) were purchased from Aldrich (Milwaukee, Wis.). Acetonitrile and ammonium chloride were obtained from J. T. Baker (Phillipsburg, N.J.). The water was from a Milli-Q Plus purification system (Millipore Corp., Bedford, Mass.).
Safety precautions.
Trifluoroacetic acid is corrosive and causes severe burns. Suitable protective clothing, including gloves, laboratory coat, and eye and face protection, should be worn when working with the concentrated solution. Y. enterocolitica is a biosafety level 2 (BSL 2) microorganism and thus was handled accordingly in a BSL 2 laboratory. The organism (50 μl) was rendered nonviable (which was verified by culturing onto TSA plates) with 70 μl of 0.1% TFA and 30 μl of acetonitrile prior to MALDI-TOF MS analysis.
Laboratory methods.
Initial reference MALDI-TOF MS signatures of these organisms were previously generated at PNNL for B. subtilis cultured in TSB and E. coli and Y. enterocolitica cultured in LB broth (20). Additional samples of Y. enterocolitica, B. subtilis, and E. coli were cultured in liquid M9 minimal salts medium. E. coli and Y. enterocolitica were also cultured in TSB without dextrose, which is a rich medium, using 30 g/liter of Milli-Q water. The cultures were incubated in a shaker incubator at ∼150 rpm at 26°C for Y. enterocolitica, 30°C for B. subtilis, and 37°C for E. coli. M9 minimal medium was prepared using M9 minimal salts with magnesium sulfate, calcium chloride, and glucose, with a final pH of 7.0. The cultures were incubated in a shaker incubator at ∼150 rpm at 26°C for Y. enterocolitica, 30°C for B. subtilis, and 37°C for E. coli. The M9 cultures were incubated for 24 to 36 h, and the TSB cultures were incubated for ∼14 h to get similar concentrations for preliminary MALDI-TOF MS analyses, as indicated by the growth curves generated for each type of media (data not shown). Y. enterocolitica did not grow well in the M9 medium and required longer incubation times. The liquid cultures were divided in half, centrifuged, and washed twice with 2% ammonium chloride (NH4Cl). The pellets were reconstituted in 150 to 550 μl of 2% NH4Cl based on optical density readings taken at 600 nm. All three microorganisms were also cultured on tryptic soy/sheep blood agar (TSA_BA) plates and Columbia agar/sheep blood agar (CAB) plates and incubated for ∼22 h at 26°C for Y. enterocolitica, 30°C for B. subtilis, and 37°C for E. coli. After incubation, several colonies were gently scraped from the agar with a sterile inoculating loop, placed into 1 ml of 2% NH4Cl, centrifuged for 2 min at 850 × g (Brinkman Eppendorf model 5415C), and washed four times with 2% ammonium chloride (NH4Cl) to remove any trace of the agar. The pellet was reconstituted in 50 to 150 μl of 2% NH4Cl, depending on the visible concentration. Y. enterocolitica, which is a BSL 2 microorganism, was reconstituted in only 50 μl of 2% ammonium chloride, because 70 μl of 0.1% TFA and 30 μl of acetonitrile were added to the samples to make them nonviable prior to analysis. Rendering the cells nonviable allowed for MALDI analysis without concern for instrument contamination. Viability was checked by plating onto TSA plates.
MALDI-TOF MS analysis.
A 0.5-μl droplet of cell preparation was spotted onto a MALDI stainless steel plate and air dried. Ferulic acid matrix solution was prepared at a concentration of 10 mg/ml in 0.1% TFA (70%) and acetonitrile (30%). Three microliters of a 1-mg/ml concentration of cytochrome c and 0.5 μl of a 1-mg/ml concentration of angiotensin were added as internal calibration standards to 197.5 μl of matrix. The matrix solution (0.5 μl) with internal standard was spotted over the dried sample. Some of the samples cultured on blood agar plates required a second layer of matrix for better signal strength. MALDI analysis was performed on a PerSeptive Biosystems Voyager-DE RP MALDI time-of-flight mass spectrometer with a nitrogen laser (337 nm) operated in the linear, delayed extraction mode with an accelerating voltage of 25 kV. Each spectrum was collected in the positive ion mode as the average of 128 laser shots. Ten replicate spectra from duplicate cultures were collected over a 3-day period for a total of 60 spectra for each combination of organism and growth condition to allow for direct comparison with the signatures in the PNNL reference signature library (11, 20). Each spectrum was internally calibrated with the protonated monomer ion of equine cytochrome c (m/z, 12,361) and the protonated monomer ion of angiotensin I (m/z, 1,297). The raw data files were then transferred to the data analyst for automated peak extraction and analysis.
The PNNL reference signature library contains B. subtilis ATCC 15841 grown in TSB. Signatures for E. coli and Y. enterocolitica were from cultures in LB medium. Each reference signature was constructed by using 10 replicate spectra from duplicate cultures collected over a 3-day period for a total of 60 spectra. The spectra were analyzed by PNNL's automated peak extraction and analysis algorithms (11, 12) for identification.
RESULTS
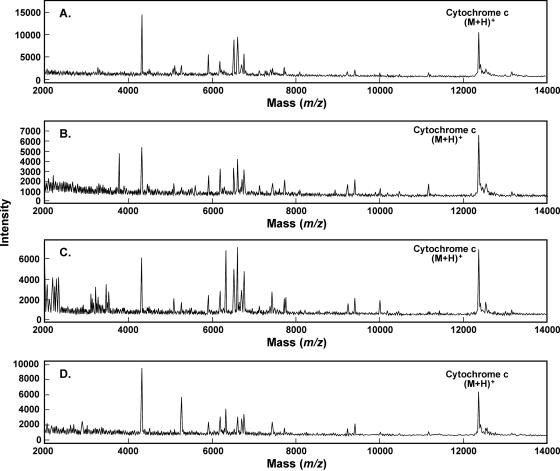
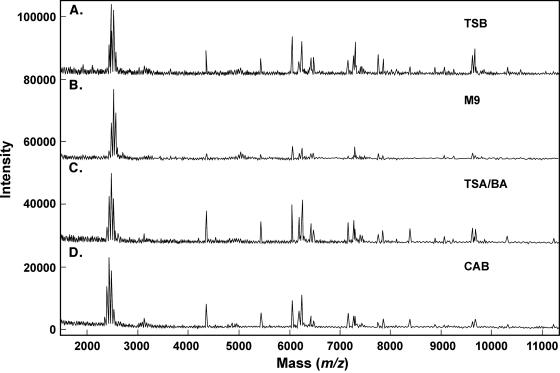
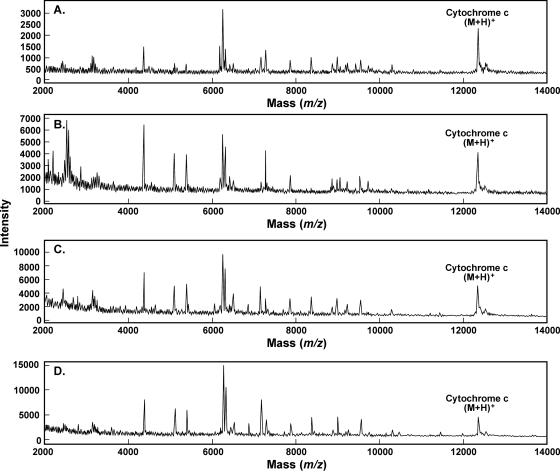
Representative MALDI spectra for each organism, B. subtilis, E. coli, and Y. enterocolitica, cultured with each of four different culture media, are shown in Fig. 1 to 3, respectively. The standard medium refers to what was used in the original signatures of the organisms: TSB for B. subtilis and LB for E. coli and Y. enterocolitica. As noted previously by other researchers (16), there is a visible difference in the MALDI mass spectra of a given organism cultured under different conditions. However, there is also a series of ions that appears reproducibly in all of the conditions tested here for a given organism. Bernardo et al. (2) also determined that most ions were the same between two different media. One major advantage of our automated data analysis approach is that visible differences, primarily in the relative intensity of the ions, are not taken into account. Rather, the reproducible appearance of ions at the correct m/z values is used for our comparison and identification approach. Each of 60 individual mass spectra for each organism under each culture condition collected over multiple days is submitted to our patented automated peak extraction algorithms (12) to obtain peak tables. The 60 replicate peak tables for each condition are then compiled, and m/z values of each peak are aligned within a standard deviation typically less than two m/z wide. An average m/z value is calculated from these aligned peaks, and any that occur in greater than 70% of the replicates (42 out of 60 replicates) are included in the signature and in Tables 1 to 3. The comparison and identification process is described in more detail elsewhere (11). Tables 1 to 3 contain the reproducible signature ions obtained by our signature protocol for each of the three organisms, B. subtilis, E. coli, and Y. enterocolitica, for each of four culture conditions. Peaks that are common to all four culture conditions for each organism are shown in bold in the tables. One column contains the average m/z value, and the other column contains the frequency of appearance of that ion within the 60-replicate spectra. From previous experience, the majority of ions observed for vegetative bacterial samples are between m/z 2,000 and 20,000; thus, we have tabulated ions only within that mass range. Table 4 summarizes the similarity and uniqueness of the MALDI-MS signatures for each culture condition and organism. For B. subtilis, there are 9 peaks common to all four culture conditions; for E. coli, there are 11 peaks in common; for Y. enterocolitica, there are 8 peaks in common. Also worth noting, there are several peaks unique to each culture condition that may prove useful for identifying culture conditions of unknown samples for forensic applications.
FIG. 1.
Representative MALDI spectra (m/z 2,000 to 14,000) for B. subtilis 15841 with each growth medium. (A) Tryptic soy broth; (B) M9; (C) tryptic soy agar/blood agar plate; (D) Columbia blood agar plate.
FIG. 3.
Representative MALDI spectra (m/z 2,000 to 14,000) for Y. enterocolitica 51871 with each growth medium. (A) Tryptic soy broth; (B) M9; (C) tryptic soy agar/blood agar plate; (D) Columbia blood agar plate.
TABLE 1.
MALDI-MS signatures of B. subtilis 15841 using different growth mediaa
| Standard
|
M9
|
TSA_BA
|
CAB
|
||||
|---|---|---|---|---|---|---|---|
| m/z | Freq. | m/z | Freq. | m/z | Freq. | m/z | Freq. |
| 2,045 | 0.70 | 3,755 | 0.98 | 2,003 | 0.77 | 2,041 | 0.92 |
| 2,086 | 0.78 | 4,306 | 0.98 | 2,170 | 0.77 | 2,170 | 0.73 |
| 2,512 | 0.73 | 5,065 | 0.95 | 2,227 | 0.75 | 2,227 | 0.80 |
| 3,254 | 0.80 | 5,254 | 0.78 | 2,257 | 0.87 | 2,257 | 0.93 |
| 3,299 | 0.92 | 5,494 | 0.82 | 2,314 | 0.90 | 2,314 | 0.95 |
| 3,377 | 0.72 | 5,901 | 0.98 | 3,087 | 0.77 | 3,087 | 0.77 |
| 4,306 | 0.98 | 6,507 | 0.98 | 3,117 | 0.78 | 3,200 | 0.92 |
| 4,482 | 0.85 | 6,598 | 0.97 | 3,200 | 0.85 | 3,256 | 0.83 |
| 5,093 | 1.00 | 6,699 | 0.93 | 3,256 | 0.83 | 3,456 | 0.75 |
| 5,254 | 0.87 | 6,755 | 0.97 | 3,456 | 0.87 | 4,304 | 0.80 |
| 5,901 | 1.00 | 7,115 | 0.97 | 3,513 | 0.80 | 5,253 | 0.77 |
| 6,061 | 0.95 | 7,714 | 0.93 | 4,304 | 0.95 | 5,900 | 0.95 |
| 6,320 | 0.73 | 9,224 | 0.95 | 5,253 | 0.77 | 6,312 | 0.87 |
| 6,507 | 1.00 | 9,397 | 0.97 | 5,900 | 0.98 | 6,506 | 0.97 |
| 6,598 | 1.00 | 10,003 | 0.83 | 6,311 | 0.90 | 6,597 | 0.83 |
| 6,700 | 1.00 | 11,156 | 0.92 | 6,507 | 0.93 | 6,699 | 1.00 |
| 6,755 | 1.00 | 6,597 | 0.97 | 6,754 | 0.92 | ||
| 7,115 | 0.90 | 6,699 | 0.90 | 7,426 | 0.78 | ||
| 7,294 | 0.98 | 6,755 | 0.90 | 7,713 | 0.88 | ||
| 7,385 | 0.98 | 7,114 | 0.75 | 9,222 | 0.97 | ||
| 7,441 | 0.75 | 9,224 | 0.88 | 9,395 | 0.92 | ||
| 7,542 | 0.93 | 9,396 | 0.82 | 11,155 | 0.87 | ||
| 7,714 | 0.95 | 10,002 | 0.73 | ||||
| 8,083 | 0.88 | 11,156 | 0.83 | ||||
| 9,224 | 0.95 | ||||||
| 9,397 | 0.97 | ||||||
| 10,004 | 0.88 | ||||||
| 13,148 | 0.93 | ||||||
Boldface ions are common across all growth media. The standard medium is TSB. freq., frequency.
TABLE 3.
MALDI-MS signatures of Y. enterocolitica 51871 using different growth mediaa
| Standard
|
M9
|
TSA BA
|
CAB
|
||||
|---|---|---|---|---|---|---|---|
| m/z | Freq. | m/z | Freq. | m/z | Freq. | m/z | Freq. |
| 2,389 | 0.85 | 2,430 | 0.93 | 2,388 | 0.98 | 2,346 | 0.93 |
| 2,431 | 0.97 | 2,472 | 1.00 | 2,430 | 0.98 | 2,388 | 0.98 |
| 2,473 | 1.00 | 2,514 | 1.00 | 2,472 | 0.98 | 2,430 | 0.98 |
| 2,515 | 0.98 | 2,556 | 1.00 | 2,614 | 1.00 | 2,472 | 0.98 |
| 4,826 | 0.78 | 2,597 | 0.98 | 2,606 | 0.85 | 2,514 | 0.98 |
| 6,045 | 0.77 | 4,350 | 1.00 | 2,649 | 0.73 | 2,606 | 0.90 |
| 6,241 | 0.70 | 5,029 | 0.75 | 2,826 | 0.73 | 2,649 | 0.75 |
| 6,415 | 0.87 | 5,071 | 0.78 | 3,121 | 0.98 | 3,080 | 0.77 |
| 7,262 | 0.87 | 5,428 | 0.98 | 4,350 | 0.98 | 3,121 | 0.98 |
| 7,288 | 0.98 | 6,046 | 1.00 | 5,428 | 0.98 | 3,163 | 0.80 |
| 7,745 | 1.00 | 6,417 | 0.95 | 6,046 | 0.98 | 3,645 | 0.72 |
| 7,841 | 0.93 | 6,474 | 1.00 | 6,242 | 0.98 | 4,350 | 0.98 |
| 9,607 | 0.88 | 7,150 | 0.95 | 6,416 | 0.98 | 5,428 | 0.98 |
| 9,651 | 1.00 | 7,289 | 0.85 | 6,474 | 0.95 | 6,046 | 0.82 |
| 7,384 | 1.00 | 7,150 | 0.98 | 6,242 | 0.98 | ||
| 7,746 | 0.97 | 7,289 | 1.00 | 6,417 | 1.00 | ||
| 7,842 | 0.90 | 7,384 | 0.82 | 6,474 | 0.93 | ||
| 8,371 | 0.93 | 7,746 | 0.92 | 7,150 | 0.95 | ||
| 9,052 | 0.72 | 7,842 | 0.98 | 7,289 | 0.98 | ||
| 9,608 | 0.73 | 8,370 | 0.98 | 7,385 | 0.72 | ||
| 8,865 | 0.88 | 7,746 | 0.98 | ||||
| 9,052 | 0.98 | 7,842 | 0.98 | ||||
| 9,239 | 0.90 | 8,370 | 0.98 | ||||
| 9,608 | 0.82 | 8,865 | 0.75 | ||||
| 9,652 | 0.77 | 9,051 | 0.98 | ||||
| 9,846 | 0.80 | 9,239 | 0.70 | ||||
| 10,300 | 0.98 | 11,214 | 0.88 | ||||
| 11,214 | 0.85 | 12,537 | 0.77 | ||||
| 11,476 | 0.75 | 13,601 | 0.85 | ||||
| 12,537 | 0.82 | ||||||
| 13,601 | 0.82 | ||||||
Boldface ions are common across all growth media. freq., frequency.
TABLE 4.
Commonalities and uniqueness of ions between signatures for different growth media
| No. of peaks in common |
B. subtilis 15841
|
E. coli 33694
|
Y. enterocolitica 51871
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Standard | M9 | TSA_BA | CAB | Standard | M9 | TSA_BA | CAB | Standard | M9 | TSA_BA | CAB | |
| 4 | 9 | 9 | 9 | 9 | 11 | 11 | 11 | 11 | 8 | 8 | 8 | 8 |
| 3 | 5 | 4 | 4 | 5 | 5 | 2 | 7 | 7 | 3 | 8 | 10 | 9 |
| 2 | 1 | 8 | 0 | 7 | 5 | 1 | 8 | 6 | 1 | 0 | 9 | 8 |
| Unique | 13 | 1 | 3 | 3 | 6 | 5 | 2 | 5 | 2 | 4 | 3 | 4 |
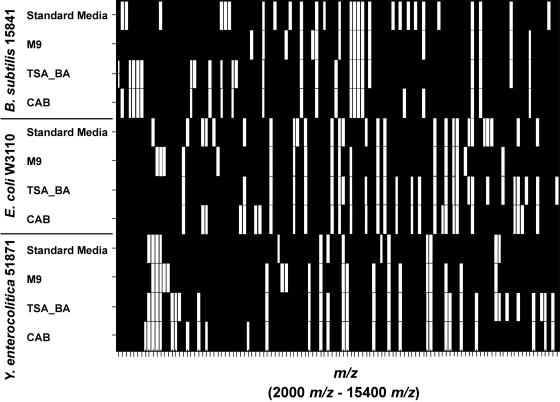
To test the ability of our automated bacterial identification method, the spectra from the samples grown with the three new media (not used for the initial signature) were compared to the signatures of the same organism grown with the original media in the reference library. Specifically, each set of 60 spectra were divided into 12 groups of 5 spectra. These 12 sets of spectra were then compared to the reference signatures following the process outlined by Jarman et al. (11). Using this approach, peaks corresponding to any biomarkers in a given signature are extracted from a given set of five test spectra. A probability score, called the degree of association, is computed based on (i) the collection signature peaks found in the test spectra and (ii) the relative importance of those extracted peaks. The relative importance of each peak is its reproducibility (frequency of appearance) measured from replicate spectra used to construct the reference signature. Using a combination of statistical guidance and empirical experience, a given reference signature is positively identified in a set of test spectra if the degree of association is greater than 0.15. Details of this approach are provided in reference 11. Despite the differences, all of the microorganisms were identified in all media (12 out of 12 samples were positive for each medium tested). It is recognized that this is based upon a small sample size and that the library would have to be expanded significantly to determine if identification is still possible with a much larger database. The limited scope of this experiment only allows for speculation, but it would be significant if unique ions exist for different culture conditions. A visual comparison of the signatures for all three organisms under all four growth conditions is provided in Fig. 4. The similarities within one organism under different conditions and the differences between organisms are more evident in this presentation of the MALDI-MS signatures for the organisms.
FIG. 4.
Image plot of overlapping ions across B. subtilis 15841, E. coli 33694, and Y. enterocolitica 51871 signatures with different growth media. Ions are represented by white bars.
DISCUSSION
Despite being cultured in different media, whole-cell bacteria are identifiable by MALDI-TOF MS and PNNL's automated statistically based data analysis algorithms. A significant number of ions are common to each bacterium in all four growth media used in this study. The presence of proteins common to all growth conditions is not totally unexpected, as many genes are known to be constitutively expressed and perform housekeeping functions in the cell (18, 19). These functions are required despite changing metabolic needs that different culturing conditions present. Thus, it is not surprising that a specific set of low-molecular-weight, constitutively expressed proteins form ions in the MALDI-MS signature regardless of culture conditions. Expansion of this study to include further organisms and more varied growth conditions, including different growth phases, temperature, and pH, needs to be completed for more extensive understanding of the capabilities and limitations of this technology for bacterial identification. One approach the laboratory has taken is to analyze samples grown in chemostats. Specific culture conditions have a decided effect on phenotypic properties at different growth temperatures (D. S. Wunschel, E. A. Hill, J. S. McLean, K. Jarman, N. B. Valentine, Y. A. Gorby, and K. L. Wahl, submitted for publication). This is well documented for members of the genus Yersinia, where the tests used to differentiate strains of this genus must be performed after growth at both 25 and 37°C (10).
It is possible to identify microorganisms grown under different growth conditions by using MALDI mass spectrometry and PNNL's patented algorithms. This technology is also well suited for high-throughput analysis, which is necessary as a screening tool for biological warfare. Furthermore, additional unique ions present in each medium may be useful in forensic applications for providing information on the specific culturing conditions.
FIG. 2.
Representative MALDI spectra (m/z 2,000 to 14,000) for E. coli 33694 with each growth medium. (A) Tryptic soy broth; (B) M9; (C) tryptic soy agar/blood agar plate; (D) Columbia blood agar plate.
TABLE 2.
MALDI-MS signatures of E. coli 33694 using different growth mediaa
| Standard
|
M9
|
TSA BA
|
CAB
|
||||
|---|---|---|---|---|---|---|---|
| m/z | freq. | m/z | freq. | m/z | freq. | m/z | freq. |
| 2,431 | 0.74 | 2,471 | 0.70 | 2,835 | 0.83 | 2,835 | 0.93 |
| 3,081 | 0.86 | 2,513 | 1.00 | 3,638 | 0.90 | 3,127 | 0.72 |
| 3,125 | 0.98 | 2,554 | 1.00 | 4,365 | 1.00 | 3,161 | 0.77 |
| 3,165 | 0.97 | 2,835 | 0.83 | 5,097 | 1.00 | 3,580 | 0.72 |
| 3,207 | 0.91 | 3,247 | 0.93 | 5,381 | 0.98 | 3,638 | 0.72 |
| 3,638 | 0.74 | 4,366 | 1.00 | 6,255 | 1.00 | 3,933 | 0.70 |
| 4,366 | 1.00 | 5,097 | 1.00 | 6,316 | 1.00 | 4,185 | 0.78 |
| 5,097 | 0.97 | 5,382 | 0.98 | 6,412 | 0.92 | 4,365 | 1.00 |
| 5,150 | 0.74 | 6,256 | 1.00 | 6,508 | 1.00 | 5,097 | 1.00 |
| 5,382 | 0.83 | 6,317 | 1.00 | 6,857 | 0.98 | 5,382 | 1.00 |
| 6,256 | 1.00 | 6,509 | 1.00 | 7,159 | 1.00 | 6,255 | 1.00 |
| 6,316 | 0.98 | 7,159 | 0.92 | 7,274 | 0.98 | 6,316 | 1.00 |
| 6,412 | 0.93 | 7,274 | 0.95 | 7,333 | 0.87 | 6,509 | 1.00 |
| 6,857 | 0.79 | 7,872 | 1.00 | 7,450 | 0.77 | 6,857 | 1.00 |
| 7,159 | 0.98 | 8,370 | 0.72 | 7,706 | 0.77 | 7,159 | 1.00 |
| 7,274 | 1.00 | 8,876 | 0.77 | 7,870 | 1.00 | 7,274 | 1.00 |
| 7,871 | 1.00 | 8,994 | 0.93 | 8,369 | 1.00 | 7,334 | 0.97 |
| 8,370 | 0.98 | 9,064 | 0.93 | 8,876 | 0.80 | 7,450 | 0.70 |
| 8,877 | 0.91 | 9,740 | 0.82 | 8,994 | 1.00 | 7,706 | 0.97 |
| 8,994 | 0.98 | 9,190 | 0.80 | 7,870 | 1.00 | ||
| 9,190 | 0.90 | 9,226 | 0.97 | 8,119 | 0.72 | ||
| 9,226 | 0.84 | 9,535 | 0.75 | 8,370 | 1.00 | ||
| 9,428 | 0.95 | 9,738 | 0.97 | 8,876 | 0.93 | ||
| 9,535 | 0.79 | 10,138 | 0.88 | 8,994 | 1.00 | ||
| 9,554 | 0.79 | 10,299 | 1.00 | 9,226 | 0.98 | ||
| 10,299 | 1.00 | 10,693 | 0.70 | 10,138 | 0.92 | ||
| 11,450 | 0.79 | 11,450 | 0.98 | 10,299 | 0.98 | ||
| 15,411 | 0.70 | 10,468 | 0.80 | ||||
| 11,450 | 0.98 | ||||||
Boldface ions are common across all growth media freq., frequency.
Acknowledgments
This work was supported by the United States Department of Energy. Pacific Northwest National Laboratory is operated for the U.S. Department of Energy by Battelle Memorial Institute under contract DE-AC06-76-RLO 1830.
REFERENCES
- 1.Arnold, R., J. Karty, A. Ellington, and J. Reilly. 1999. Monitoring the growth of a bacteria culture by MALDI-MS of whole cells. Anal. Chem. 71:1990-1996. [DOI] [PubMed] [Google Scholar]
- 2.Bernardo, K., N. Pakulat, M. Macht, O. Krut, H. Seifert, S. Fleer, F. Hünger, and M. Krönke. 2002. Identification and discrimination of Staphylococcus aureus strains using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics 2:747-753. [DOI] [PubMed] [Google Scholar]
- 3.Blom, M., A. Meyer, P. Gerner-Smidt, K. Gaarslev, and F. Espersen. 1999. Evaluation of Statens Serum Institut enteric medium for detection of enteric pathogens. J. Clin. Microbiol. 37:2312-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choe, L. H., W. Chen, and K. H. Lee. 1999. Proteome analysis of factor for inversion stimulation (Fis) overproduction in Escherichia coli. Electrophoresis 20:798-805. [DOI] [PubMed] [Google Scholar]
- 5.Ellwood, D. C., and D. W. Tempest. 1972. Effects of environment on bacterial wall content and composition. Adv. Microb. Physiol. 7:83-117. [Google Scholar]
- 6.Errington, J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 57:1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenselau, C., and P. A. Demirev. 2001. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom. Rev. 20:157-171. [DOI] [PubMed] [Google Scholar]
- 8.Fox, K. F., D. S. Wunschel, A. Fox, and G. C. Stewart. 1998. Complementarity of GC-MS and LC-MS analyses for determination of carbohydrate profiles of vegetative cells and spores of bacilli. J. Microbiol. Methods 33:1-11. [Google Scholar]
- 9.Hawley, R. J., and E. M. Eitzen, Jr. 2001. Biological weapons—a primer for microbiologists. Annu. Rev. Microbiol. 55:235-253. [DOI] [PubMed] [Google Scholar]
- 10.Holt, J. G., N. R. Krieg, P. H. A. Sneath, J. T. Staley, and S. T. Williams (ed.). 1994. Bergey's manual of determinative bacteriology, 9th ed. Williams & Wilkins, Baltimore, Md.
- 11.Jarman, K. H., S. T. Cebula, A. J. Saenz, C. E. Petersen, N. B. Valentine, M. T. Kingsley, and K. L. Wahl. 2000. An algorithm for automated bacterial identification using matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 72:1217-1223. [DOI] [PubMed] [Google Scholar]
- 12.Jarman, K. H., D. S. Daly, K. K. Anderson, and K. L. Wahl. 2003. A new approach to automated peak detection. Chemom. Intell. Lab. Syst. 69:61-76. [Google Scholar]
- 13.Lang, W. K., K. Glassey, and A. R. Archibald. 1982. Influences of phosphate supply on teichoic acid and teichuronic acid content of Bacillus subtilis cell walls. J. Bacteriol. 151:367-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lay, J. O., Jr. 2001. MALDI-TOF mass spectrometry of bacteria. Mass Spectrom. Rev. 20:172-194. [DOI] [PubMed] [Google Scholar]
- 15.Leclercq, A., A. Guiyoule, M. El Lioui, E. Carniel, and J. Decallonne. 2000. High homogeneity of the Yersinia pestis fatty acid composition. J. Clin. Microbiol. 38:1545-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longo, M. A., I. S. Novella, L. A. Garcia, and M. Diaz. 1999. Comparison of Bacillus subtilis and Serratia marcescens as protease producers under different operating conditions. J. Biosci. Bioeng. 88:35-40. [DOI] [PubMed] [Google Scholar]
- 17.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Völker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savli, H., A. Karadenizli, F. Kolayli, S. Gundes, U. Ozbek, and H. Vahaboglu. 2003. Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J. Med. Microbiol. 52:403-408. [DOI] [PubMed] [Google Scholar]
- 19.Tobisch, S., D. Zühlke, J. Bernhardt, J. Stülke, and M. Hecker. 1999. Role of CcpA in regulation of the central pathways of carbon catabolism in Bacillus subtilis. J. Bacteriol. 181:6996-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wahl, K. L., S. C. Wunschel, K. H. Jarman, N. B. Valentine, C. E. Petersen, M. T. Kingsley, K. A. Zartolas, and A. J. Saenz. 2002. Analysis of microbial mixtures by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 74:6191-6199. [DOI] [PubMed] [Google Scholar]
- 21.Wunschel, D. S., K. F. Fox, G. E. Black, and A. Fox. 1994. Discriminations among the B. cereus group, in comparison to B. subtilis, by structural carbohydrate profiles and ribosomal RNA spacer region PCR. Syst. Appl. Microbiol. 17:625-635. [Google Scholar]