Abstract
Accumulation of unfolded proteins in the endoplasmic reticulum (ER) causes a state of cellular stress known as ER stress. The cells respond to ER stress by activating the unfolded protein response (UPR), a signaling network emerging from the ER-anchored receptors IRE1α, PERK and ATF6. The UPR aims at restoring ER protein-folding homeostasis, but turns into a toxic signal when the stress is too severe or prolonged. Recent studies have demonstrated links between the UPR and inflammation. Consequently, small molecule inhibitors of IRE1α and PERK have become attractive tools for the potential therapeutic manipulation of the UPR in inflammatory conditions. TNF is a master pro-inflammatory cytokine that drives inflammation either directly by promoting gene activation, or indirectly by inducing RIPK1 kinase-dependent cell death, in the form of apoptosis or necroptosis. To evaluate the potential contribution of the UPR to TNF-induced cell death, we tested the effects of two commonly used PERK inhibitors, GSK2606414 and GSK2656157. Surprisingly, we observed that both compounds completely repressed TNF-mediated RIPK1 kinase-dependent death, but found that this effect was independent of PERK inactivation. Indeed, these two compounds turned out to be direct RIPK1 inhibitors, with comparable potency to the recently developed RIPK1 inhibitor GSK'963 (about 100 times more potent than NEC-1s). Importantly, these compounds completely inhibited TNF-mediated RIPK1-dependent cell death at a concentration that did not affect PERK activity in cells. In vivo, GSK2656157 administration protected mice from lethal doses of TNF independently of PERK inhibition and as efficiently as GSK'963. Together, our results not only report on new and very potent RIPK1 inhibitors but also highlight the risk of misinterpretation when using these two PERK inhibitors in the context of ER stress, cell death and inflammation.
The endoplasmic reticulum (ER) is a eukaryotic organelle that has a crucial role in many cellular processes. In particular, it controls the synthesis, folding, maturation and trafficking of proteins destined for secretion or cell surface display. It does so with the help of various chaperones and enzymes that respectively promote folding and complex post-translational modification of the nascent proteins. Many physiological and pathological conditions can, however, disturb the ER protein-folding environment, which results in the accumulation of un- or misfolded proteins within the organelle lumen. This situation creates a state of cellular stress commonly called ‘ER stress'. Eukaryotic cells have developed a quality control system, known as the unfolded protein response (UPR), to sense and adapt to ER stress.1, 2 In metazoans, the UPR is activated by inositol-requiring protein 1 (IRE1α), protein kinase RNA-like ER-kinase (PERK) and activating transcription factor 6 (ATF6), three ER-anchored receptors that sense the protein-folding status in the ER lumen and transduce the information in the cytosol. In the absence of ER stress, the chaperone-binding immunoglobulin protein (BIP) maintains these sensors in an inactive state by binding to their ER luminal domain. On BIP release, these receptors activate cytosolic signaling pathways aimed at restoring protein homeostasis by reducing the protein load in the ER. Nevertheless, when the stress is too severe and/or prolonged, the UPR is insufficient to restore ER homeostasis and turns into a toxic signal leading to cell death.2 Recently, the UPR has been shown to have critical functions in immunity and inflammation, and to be linked to a variety of human inflammatory pathologies such as infectious, autoimmune, neurodegenerative and metabolic disorders, as well as cancers.3, 4, 5, 6 At the cellular level, signaling downstream of all three ER sensors has been linked, directly or indirectly, to inflammation. In the case of PERK, its activation was for example reported to activate nuclear factor-κB (NF-κB), a master transcriptional regulator of pro-inflammatory pathways.7 In resting conditions, NF-κB is kept inactive in the cytosol through its association with IκB. Activated PERK indirectly disturbs the IκB-mediated inhibition of NF-κB by inhibiting global protein synthesis via phosphorylation of eukaryotic translation initiator factor 2α (eIF2α). Owing to the short half-life of IκB, the translational break increases the NF-κB to IκB ratio, thereby freeing NF-κB for nuclear translocation. In addition, although inhibiting global protein synthesis, phosphorylated eIF2α promotes the CAP-independent translation of certain mRNAs, including the one encoding activating transcription factor 4 (ATF4), which has an important role in many physiological processes, such as the anti-oxidative stress response, autophagy and inflammation.8, 9, 10, 11 In light of the role of cell death in triggering inflammatory responses, it is also believed that PERK promotes inflammation by inducing cell death. Indeed, CCAAT/enhancer-binding protein homologous protein (CHOP) is a downstream target of ATF4 that has been reported to induce cell death in various ways.2
As a consequence of the link between UPR and inflammation, small molecule inhibitors of the UPR have become attractive tools for the potential therapeutic manipulation of the UPR in inflammatory conditions.6, 12 These inhibitors are also increasingly used both in vitro and in vivo to demonstrate the contribution of the UPR to specific inflammatory pathways. Tumor necrosis factor (TNF) is a master pro-inflammatory cytokine that promotes inflammation by activating the mitogen-activated protein kinases (MAPKs) and NF-κB pathways, which collectively lead to the transcriptional upregulation of pro-inflammatory molecules.13 In addition, it recently became clear that TNF also promotes/exacerbates inflammation by inducing RIPK1 kinase-dependent cell death, in the form of apoptosis or necroptosis.14, 15 As previous studies reported activation of PERK by TNF signaling,16, 17 we decided to evaluate the contribution of PERK signaling to TNF-induced cell death by making use of two commonly used PERK inhibitors, GSK2656157 and GSK2606414. Surprisingly, we identified both compounds as new potent RIPK1 inhibitors, thereby highlighting the risk of misinterpretation when using these inhibitors in the fields of cell death and inflammation.
Results
GSK2606414 and GSK2656157 inhibit TNF-mediated RIPK1 kinase-dependent cell death
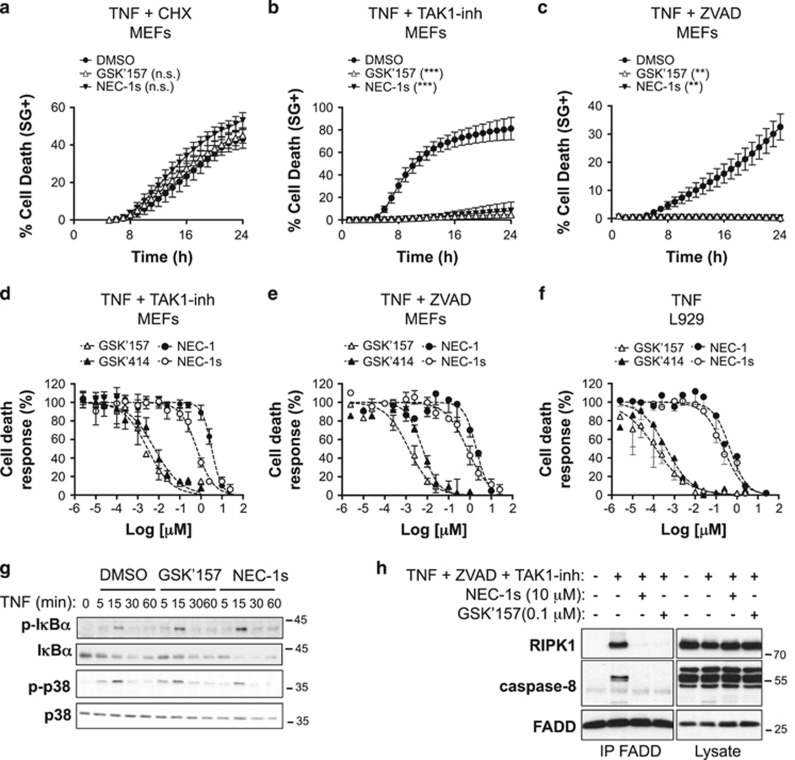
Binding of TNF to TNFR1 can trigger both RIPK1-independent apoptosis and RIPK1 kinase-dependent apoptosis or necroptosis according to the cellular context.14 In mouse embryonic fibroblasts (MEFs), it was previously demonstrated that TNF induces RIPK1-independent apoptosis in the presence of cycloheximide (CHX), RIPK1 kinase-dependent apoptosis when combined to the TAK1 inhibitor (5Z)-7-Oxozeaenol (TAK1-inh) and RIPK1 kinase-dependent necroptosis in the presence of the pan caspase inhibitor zVAD-fmk (ZVAD).18, 19 To evaluate the contribution of PERK signaling to the different cellular death outcomes of TNFR1 engagement in MEFs, we first tested the effect of PERK inhibition by GSK2656157 (GSK'157), an optimized version of the PERK inhibitor GSK2606414 (GSK'414) that was selected for preclinical development.20, 21, 22 NEC-1s, an optimized version of the RIPK1 inhibitor Necrostatin-1 (NEC-1), was used as positive control to monitor RIPK1 kinase dependency in the induced cell-death modalities.23, 24 Although GSK'157 had no effect on TNF-induced RIPK1-independent apoptosis (Figure 1a), it completely prevented RIPK1 kinase-dependent apoptosis (Figure 1b) and necroptosis (Figure 1c). Remarkably, both PERK inhibitors GSK'414 and GSK'157 turned out to be at least 100 times more potent than NEC-1 or NEC-1s in inhibiting TNF-mediated RIPK1 kinase-dependent apoptosis and necroptosis in MEFs (Figures 1d and e). The calculated IC50 values were respectively of 6.3–7.3 μM for GSK'414, 2.9–1.9 μM for GSK'157, 3290–2070 μM for NEC-1 and 650–740 μM for NEC-1s (Table 1). A similar difference in potency was observed when monitoring inhibition of caspase-3 activity following stimulation with TNF and the TAK1 inhibitor (Table 1). The protective effect of these PERK inhibitors was not cell specific as they similarly inhibited TNF-induced RIPK1 kinase-dependent necroptosis in L929 cells (Figure 1f and Table 1).
Figure 1.
The PERK inhibitors GSK2606414 (GSK'414) and GSK2656157 (GSK'157) protect cells from TNF-mediated RIPK1 kinase-dependent cell death. (a–c) Immortalized MEFs were pretreated for 30 min with the indicated compounds (1 μM GSK'157, 10 μM NEC-1s, 250 ng/ml CHX, 1 μM TAK1-inh, 50 μM ZVAD-fmk) and TNF-mediated cell death was measured in function of time by SytoxGreen positivity.40 RIPK1-independent apoptosis was induced by CHX+mTNF (1 ng/ml), RIPK1 kinase-dependent apoptosis by TAK1-inh+hTNF (0.1 ng/ml) and RIPK1 kinase-dependent necroptosis by ZVAD+hTNF (1 ng/ml). The results are presented as mean±S.E.M. of three independent experiments. Statistical significance was determined by two-way ANOVA. Significance between samples is indicated in the figures as follows: NS= P > 0.05; *P<0.05; **P<0.01; ***P<0.001. (d–f) Immortalized MEFs (d and e) and L929 cells (f) were pretreated for 30 min with increasing concentrations of the indicated compounds in presence or absence of TAK1-inh (1 μM) or ZVAD-fmk (50 μM) and TNF-mediated cell death was measured at 24 h post-stimulation by SytoxGreen positivity.40 (g) Immortalized MEFs were pretreated for 30 min with GSK'157 (0.1 μg/ml) or NEC-1s (10 μg/ml), stimulated with hTNF (1 ng/ml) for the indicated time, and the cell lysates were then immunoblotted as indicated. (h) Immortalized MEFs were pretreated for 30 min with ZVAD-fmk (50 μM), TAK1-inh (1 μM) and the indicated compounds and then stimulated for 2 h with hTNF (1 ng/ml). Complex IIb was isolated by FADD immunoprecipitation and RIPK1 and caspase-8 binding revealed by immunoblotting
Table 1. GSK2606414 and GSK2656157 are potent RIPK1 inhibitors.
|
IC50
(μM) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| System | Stimulus | Biological process | Parameter/methodology | GSK'414 | GSK'157 | NEC-1 | NEC-1s | GSK'963 | AMG'44 |
| MEFs | TNF + TAK1-inh | Apoptosis | SYTOX Green+ | 0.0063 | 0.0029 | 3.29 | 0.65 | 0.0018 | NI |
| MEFs | TNF + TAK1-inh | Caspase-3 activity | C3 DEVD-AMC | 0.0068 | 0.0040 | 3.35 | 1.34 | ND | ND |
| MEFs | TNF + zVAD | Necroptosis | SYTOX Green+ | 0.0073 | 0.0019 | 2.07 | 0.74 | ND | NI |
| L929s | TNF | Necroptosis | SYTOX Green+ | 0.0004 | 0.0001 | 0.47 | 0.23 | ND | ND |
| MEFs | Tm | PERK- phosphorylation | Electrophoretic mobility shift | ND | 0.1977 | ND | ND | NI | 0.2178 |
| MEFs | Tm | ATF4 induction | WB | ND | 0.2079 | ND | ND | N.D | 0.7392 |
| GST-hRIPK | – | Kinase activity | ADP-Glo assay | 0.0182 | 0.0691 | ND | 0.8655 | 0.0178 | NI |
| GST-hPERK | – | Kinase activity | ADP-Glo assay | 0.0038 | 0.0097 | ND | NI | NI | 0.1812 |
ND, not determined; NI, no inhibition until 10 μM.
Engagement of TNFR1 by TNF results in the sequential assembly of a membrane-bound primary signaling complex (TNFR1 complex I or TNFR1-SC) that drives gene activation and of a secondary cytoplasmic complex (TNFR1 complex IIb) composed of RIPK1, FADD and caspase-8, that mediates RIPK1 kinase-dependent apoptosis. In the scenario of caspase-8 inhibition, RIPK1 then associates with RIPK3 to form the necrosome and to initiate necroptosis.14 Interestingly, we found that GSK'157 treatment of MEFs did not alter TNFR1 signaling to MAPK or NF-κB, as monitored by phosphorylation of p38 and IκBα, but completely prevented complex IIb formation, similarly as NEC-1s (Figures 1g and h). Of note, GSK'157 was able to inhibit complex IIb assembly at a concentration 100 times lower than the concentration commonly used for NEC-1s.19 Together, our results indicate that GSK'414 and GSK'157 specifically inhibit TNF-mediated RIPK1 kinase-dependent death by preventing complex IIb formation, about 100 times more potently than NEC-1s.
GSK2656157 protects from TNF-mediated RIPK1 kinase-dependent cell death independently of PERK inhibition
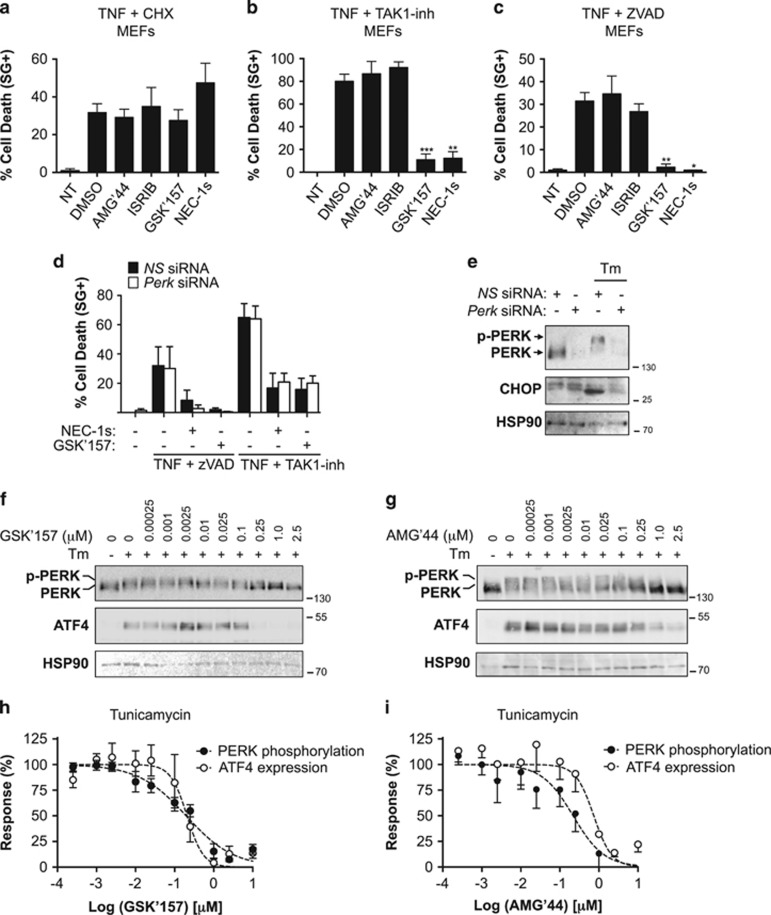
To test for potential off-target effects of GSK'414 and GSK'157, we next evaluated the effect of AMG'44, a structurally distinct PERK inhibitor,25 and of ISRIB, a potent inhibitor of PERK signaling that reverses the effects of eIF2α phosphorylation,26 on TNF-induced cell death in MEFs. Surprisingly, and in contrast to the results obtained with GSK'414 and GSK'157, we found that none of these two additional inhibitors protected the cells from TNF-induced death (Figures 2a–c). The PERK-independent protection obtained by GSK'157 was further illustrated by siRNA-mediated repression of PERK. Indeed, the transient repression of PERK did not protect the cells from RIPK1 kinase-dependent necroptosis or apoptosis triggered by TNF in conjugation with ZVAD-fmk or TAK1-inh, respectively (Figure 2d). In contrast, GSK'157 or NEC-1s still provided protection to the MEFs repressing, or not, PERK expression (Figure 2d). The functionality of the PERK knockdown was confirmed by exposing the MEFs transfected with control siRNA (NS siRNA) and Perk siRNA to the ER stress inducer tunicamycin (Tm). We observed that Tm-induced PERK auto-phosphorylation and CHOP induction was absent in the Perk siRNA-treated cells, confirming functional PERK knockdown (Figure 2e).
Figure 2.
GSK2656157 protects cells from TNF-mediated RIPK1 kinase-dependent death independently of PERK inhibition. (a–c) Immortalized MEFs were pretreated for 30 min with the indicated compounds (2.5 μM AMG'44, 1 μM ISRIB, 1 μM GSK'157, 10 μM NEC-1, 10 μM NEC-1s, 250 ng/ml CHX, 50 μM ZVAD-fmk, 1 μM TAK1-inh) and TNF-mediated cell death was measured 24 h post-stimulation by SytoxGreen positivity.40 RIPK1-independent apoptosis was induced by CHX+mTNF (1 ng/ml; a), RIPK1 kinase-dependent apoptosis by TAK1-inh+hTNF (0.1 ng/ml; b) and RIPK1 kinase-dependent necroptosis by ZVAD+hTNF (1 ng/ml; c). The results are presented as mean±S.D. of three independent experiments. Statistical significance was determined by Student's t-test. (d and e) Immortalized MEFs were transfected with control (NS) or PERK siRNAs for 48 h and then (d) analyzed for TNF-mediated RIPK1 kinase-dependent apoptosis and necroptosis as in b and c, or e exposed to Tm (1 μg/ml) for 4 h and the cell lysates immunoblotted as indicated. (f and g) Immortalized MEFs were pretreated for 30 min with increasing concentrations of GSK'157 or AMG'44, stimulated with Tm (1 μg/ml) for 4 h and the cell lysates were then immunoblotted as indicated. (h and i) Quantification of the results obtained in f and g. The results are presented as mean±S.E.M. of three independent experiments
Having established that GSK'157 protects cells from TNF-mediated RIPK1 kinase-dependent cell death independent of PERK inhibition, we next compared the efficiency of GSK'157 in inhibiting PERK signaling versus TNF-induced cell death in MEFs. As cellular readouts for PERK activity, we monitored Tm-induced PERK auto-phosphorylation and ATF4 induction (Figures 2f–i). Pretreatment of MEFs with GSK'157 resulted in a total inhibition of ATF4 induction and greatly repressed PERK auto-phosphorylation when this compound was used at a concentration above 0.25 μM, with calculated IC50 value of 0.2 μM (Figures 2f and h and Table 1). This concentration is two orders of magnitude higher than the calculated IC50 value for the inhibition of TNF-mediated apoptosis/necroptosis induction (Figures 1d–f and Table 1). Of note, the calculated IC50 value for cellular PERK inhibition by AMG'44 was between 0.22 and 0.74 μM (Figures 2g and i and Table 1). Together, these results indicate that GSK'157 protects cells from TNF-mediated death independent of PERK inhibition, and that this compound is a much better cellular inhibitor of RIPK1 kinase-dependent cell death than of PERK signaling.
GSK2606414 and GSK2656157 are direct RIPK1 inhibitors
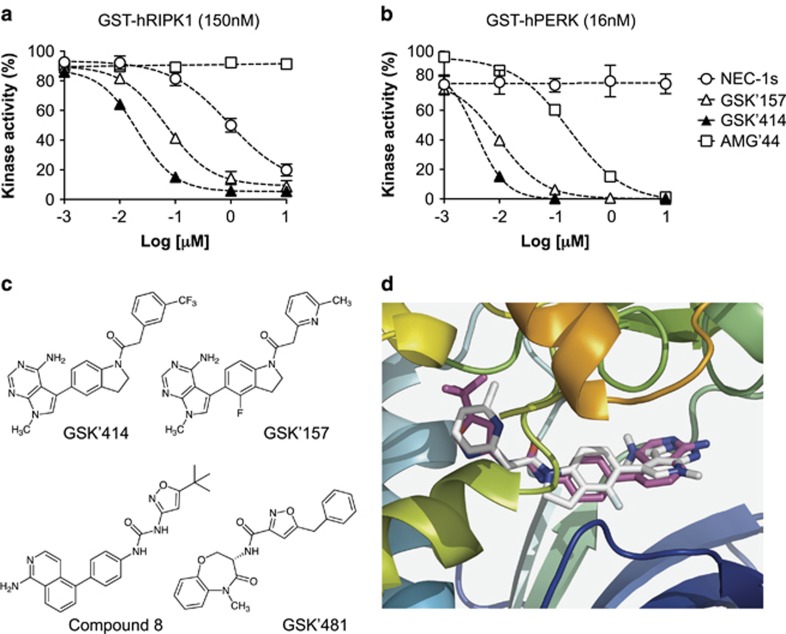
The fact that GSK'414 and GSK'157 could protect cells from TNF-mediated death independent of PERK inhibition, and only in conditions where cell death induction was requiring RIPK1 enzymatic activity, let us hypothesize that RIPK1 could be a substrate directly targeted by these compounds. To test this hypothesis, we performed in vitro ADP-Glo kinase assays using recombinant human RIPK1. Remarkably, we found that both GSK'414 and GSK'157 directly inhibited RIPK1 enzymatic activity with a potency that was at least 10 times higher than NEC-1s (Figure 3a and Table 1). In contrast, and in line with our cellular results (Figures 2b and c), AMG'44 had no effect on RIPK1 kinase activity (Figure 3a). Of note, we observed that while GSK'414, GSK'157 and AMG'44 repressed PERK activity in the ADP-Glo kinase assays with different potencies, the enzymatic activity of PERK was not affected by NEC-1s (Figure 3b).
Figure 3.
GSK2606414 and GSK2656157 are potent inhibitors of RIPK1. (a and b) Quantification of ADP-Glo kinase assays performed with recombinant hRIPK1 (a) or hPERK (b) in presence of increasing concentrations of NEC-1s, GSK'414, GSK'157 and AMG'44. (c) Chemical structures of GSK2606414, GSK2656157, the ligand in pdb-entry 4NEU (compound 8)27 and in pdb-entry 5HX6 (GSK'481).30 (d) Zoom into the ATP-site of RIPK1 crystal structure 4NEU (cartoon, from N-blue to C-red) in complex with the aminoisoquinoline-inhibitor compound 8 (sticks, cyan). The best-docked pose of GSK'157 (sticks, white) occupies the ATP-site in a similar manner as compound 8. Figure prepared with PyMOL
Modeling of the RIPK1–GSK2656157 complex
To predict how GSK'157 interacts with RIPK1, we examined the previously published crystal structures of the RIPK1 kinase domain bound to different inhibitors. The crystal structure pdb-entry 4NEU shows an inactive ‘Asp-Leu-Gly (DLG)-out' conformation of RIPK1 in complex with an aminoisoquinolinyl-phenylurea derivative (compound 8),27 binding into the ATP-pocket. This inhibitor has an overall resemblance to the indolyl-aminopyrimidine derivatives GSK'414 and GSK'157 (Figure 3c). Automated docking with AutoDock-Vina28 of GSK'157 into the ATP-pocket of 4NEU gave, as first predicted docking pose, an active site occupancy that is remarkably similar to that of compound 8: the respective isoquinoline/pyridine rings, central phenyl/indole rings, and tert. butyl isoxazole/methylpyridyl groups, all respectively and similarly are occupying RIPK1's hinge binder zone, the central zone near DLG-out and the allosteric hydrophobic pocket along the C-helix (Figure 3d). RIPK1 structure 4NEU shows a distinct inactive conformation from pdb-entries 4ITI, 4ITJ, 4ITH (bound to necrostatins)29 or the recently published 5HX6.30 In the latter, RIPK1 is in complex with the benzoxazepinone derivative GSK'481, an inhibitor that spans the same three zones occupied by compound 8 in 4NEU. In structure 5HX6, Phe162 points to and partly blocks the central zone for proper docking with GSK'157. However, when Phe162 is set as a flexible residue, GSK'157 again adopts a pose similar to compound 8 in 4NEU (not shown). Together, these results predict a model in which GSK'157 binds RIPK1 in a very similar way as compared with the type II class inhibitor compound 8,27 occupying the ATP-binding pocket with the kinase adopting a DLG-out conformation.
GSK2606157 is a potent inhibitor of TNF-induced lethal shock
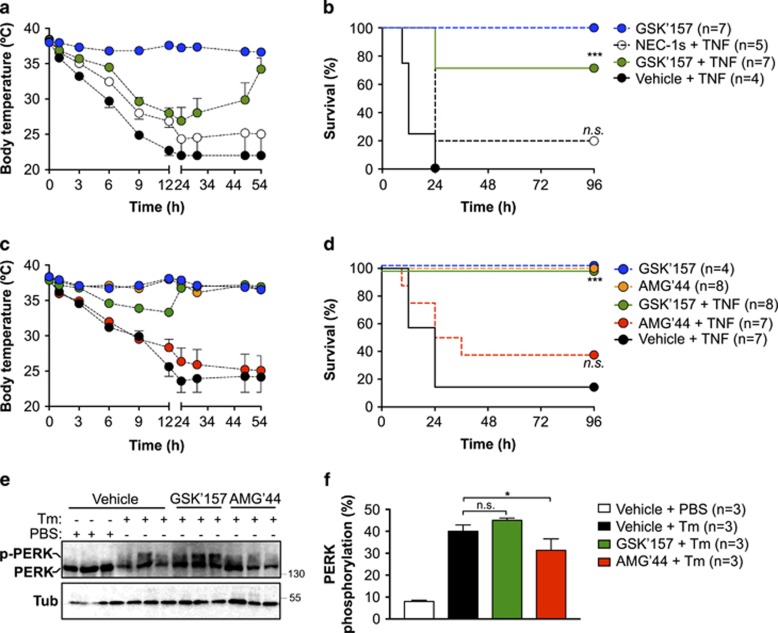
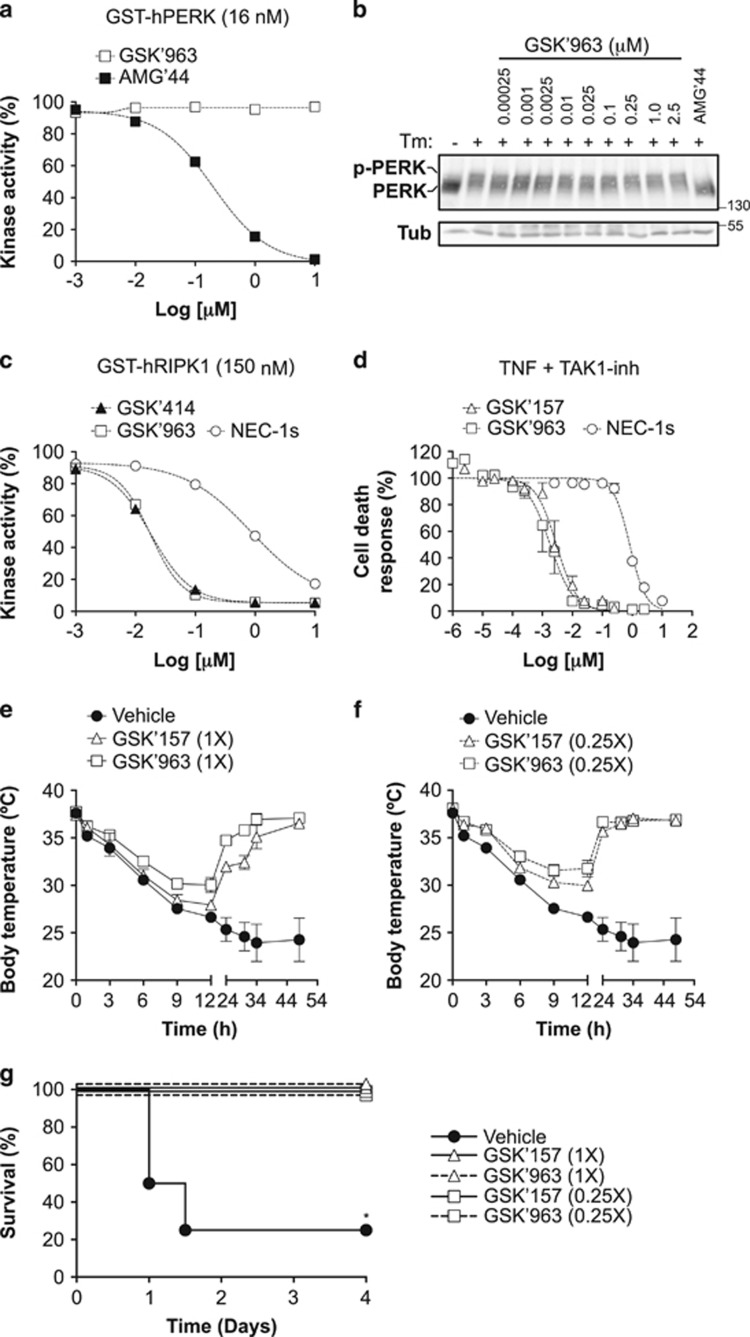
RIPK1 kinase-dependent death has been reported to drive lethal hypothermia in an acute model of sterile shock caused by TNF injection.31, 32 The severity of the shock and the resulting lethality rate depend on the injected dose of TNF, and pharmacological inhibition of RIPK1 by NEC-1s was shown to either fully or partially protect the mice from death.33, 34 In cells, we found that GSK'157 was at least 100 times more potent than NEC-1s in inhibiting TNF-mediated RIPK1 kinase-dependent death (Figures 1d–f). As NEC-1s and GSK'157 show similar pharmacokinetic profiles (T1/2=1 h for NEC-1s24 and =1.25 h for GSK'15722), we tested the potential protective effect of GSK'157 in the model of TNF-induced lethal shock by using it at a concentration 10 times lower than the one commonly used for NEC-1s.33, 34 The mice were injected with a single intravenous dose of vehicle, NEC-1s (6 mg/kg=21.6 μmol/kg) or GSK'157 (0.9 mg/kg=2.16 μmol/kg) 15 min before intravenous injection of TNF. At the high dose of TNF used, 75% of the mice succumbed within 12 h, and all mice were dead by 24 h post injection (Figures 4a and b). Administration of NEC-1s provided partial protection during the first 24 h, but only 25% of the animals survived over a period of 4 days. In clear contrast, 75% of the mice receiving GSK'157 survived the 4-day period and the surviving animals had completely regained their normal body temperature (Figures 4a and b,Table 2). Of note, no toxic effects were observed in control animals administered with GSK'157 alone. To test the potential contribution of PERK inhibition to the protection obtained with GSK'157, we repeated the experiment with AMG'44, a PERK inhibitor that did not inhibit RIPK1 (Figures 2a–c) or protect cells from TNF-induced death (Figures 3a and b). AMG'44 was used at a concentration 10 times higher than GSK'157 (12 mg/kg=21.4 μmol/kg). As shown in Figure 4, the administration of AMG'44 did not provide significant protection against TNF-induced hypothermia and lethality, whereas GSK'157 fully protected the mice in that experiment (Figures 4c and d,Table 2). Importantly, at the concentration used, the administration of AMG'44, and not of GSK'157, significantly repressed PERK auto-phosphorylation in the liver of mice injected with tunicamycin (Tm; Figures 4e and f). Taken together, these results demonstrate that GSK'157 is a much better inhibitor than NEC-1s in the model of TNF-induced lethal shock and that the protection obtained by GSK'157 is independent of PERK inhibition.
Figure 4.
GSK2656157 protects mice from TNF-induced lethal shock independent of PERK inhibition. (a–d) C57BL/6 J female mice were pre-injected intravenously (i.v.) with the indicated compounds 15 min before i.v. injection of mTNF (15 μg/20 g). (a and c) Body temperature as a function of time. (b and d) Cumulative survival rates. Survival curves were compared using Kaplan–Meier and log-rank Mantel–Cox test. (e) C57BL/6 J female mice were pre-injected intravenously (i.v.) with the indicated compounds 15 min before i.p. injection of Tm (2 μg/g). PERK auto-phosphorylation in the liver was analyzed by immunoblot 6 h post injection. Each lane represents a different animal. (f) Quantification of the results obtained in e. The results are presented as mean±S.E.M. Statistical significance was determined by Student's t-test. Significance between samples is indicated in the figures as follows: NS = P > 0.05; *P<0.05; **P<0.01; ***P<0.001
Table 2. Protection conferred by the inhibitors in the in vivo model of TNF-induced lethal shock.
|
Dose (mice) |
||||
|---|---|---|---|---|
| Compound | Factor | μmol/kg | mg/kg | Protection |
| GSK'157 | 1X | 2.16 | 0.9 | +++ |
| 0.25X | 0.54 | 0.226 | +++ | |
| GSK'963 | 1X | 2.16 | 0.496 | +++ |
| 0.25X | 0.54 | 0.124 | +++ | |
| NEC-1s | 10X | 21.6 | 6 | + |
| AMG'44 | 10X | 21.4 | 12 | − |
GSK2606157 has comparable potency as GSK'963, a recently developed potent inhibitor of RIPK1
Having demonstrated the potential use of GSK'157 as a new tool for the in vitro and in vivo study of RIPK1, we next decided to compare its activity to GSK'963, a recently developed and commercially available RIPK1 inhibitor shown to exhibit improved efficacy relative to NEC-1s.35 We first evaluated the effect of GSK'963 on PERK and found that similar to NEC-1s, but in contrast to GSK'414 and GSK'157, GSK'963 had no inhibitory activity on PERK (Figures 5a and b). At the level of RIPK1, GSK'414 and GSK'157 remarkably showed comparable inhibitory activity as GSK'963, respectively, analyzed by in vitro ADP-Glo kinase assay using recombinant human RIPK1 and in cells following TNF-induced RIPK1-dependent apoptosis (Figures 5c and d). This comparable efficacy in inhibiting RIPK1 was also observed in vivo where both GSK'963 and GSK'157 provided full protection to TNF-induced hypothermia and lethality when used at 2.16 μmol/kg (1X) or 0.54 μmol/kg (0.25X; Figures 5e–g,Table 2), concentrations at which GSK'157 does not inhibit PERK (Figures 4e and f).
Figure 5.
GSK2606157 and GSK'963 have comparable inhibitory activities on RIPK1. (a and c) Quantification of ADP-Glo kinase assays performed with recombinant hPERK (a) or hRIP1 (c) in the presence of increasing concentrations of NEC-1s, GSK'414, AMG'44 and GSK'963. (b) Immortalized MEFs were pretreated for 30 min with increasing concentrations of GSK'963, stimulated with Tm (1 μg/ml) for 4 h and the cell lysates were then immunoblotted as indicated. AMG'44 (2.5 μM) was used as a positive control. (d) Immortalized MEFs were pretreated for 30 min with increasing concentrations of the indicated compounds in the presence or absence of TAK1-inh (1 μM) and TNF-mediated cell death was measured 24 h post-stimulation by SytoxGreen positivity.40 (e–g) C57BL/6 J female mice were pre-injected intravenously (i.v.) with the indicated compounds 15 min before i.v. injection of mTNF (15 μg/20 g). (e and f) Body temperature as a function of time. (g) Cumulative survival rates. 1X=2.16 μmol/kg; 0.25X=0.54 μmol/kg
Discussion
The unfolded protein response (UPR), which is activated by IRE1α, PERK and ATF6, is emerging as a signaling network having important roles in cell death, immunity and inflammation.2, 3, 4, 5 Consequently, there is a growing interest on the potential use of small molecule inhibitors of the UPR in the treatment of inflammatory conditions.6, 12 Among these molecules, selective PERK inhibitors have been reported and these compounds are currently commonly used to demonstrate the contribution of PERK signaling in various disease models.20, 21, 22, 36 TNF is a master pro-inflammatory cytokine that promotes inflammation either through gene activation or by inducing RIPK1-dependent cell death.14, 15 In light of the reported activation of PERK by TNF,16, 17 we evaluated the potential role of PERK signaling in regulating TNF-mediated death. Our in vitro and in vivo results allow us to conclude that PERK signaling does not regulate TNF-induced cell death, as PERK inactivation was neither protecting nor sensitizing cells to death induced by TNF. Nevertheless, we identified two reportedly potent and selective PERK inhibitors, GSK2606414 and its optimized analog GSK2656157, to actually be much more potent RIPK1 inhibitors. These compounds consequently provided protection to TNF-mediated RIPK1 kinase-dependent cell death independently of PERK inhibition. The fact that these inhibitors completely inhibited TNF-mediated RIPK1-dependent cell death at concentrations that did not affect PERK activity makes them better (about 100 times) cellular inhibitors of RIPK1 than of PERK, at least in MEFs. The cellular IC50 values that we calculated for GSK2606414 and GSK2656157 in MEFs are higher than the ones initially reported for PERK inhibition in other cell types.20, 21, 22 Nevertheless, our RIPK1- and PERK-dependent functional readouts were monitored in the same cells, demonstrating the higher potency of these inhibitors in targeting RIPK1 than PERK in a given cell type. These findings have important consequences considering that these compounds are commonly used as specific PERK inhibitors. In this study, we focused on the role of RIPK1 downstream of TNFR1, but RIPK1-dependent functions extend this particular pathway or even cell death induction in general. RIPK1 has for example also been involved in other biological processes also influenced by PERK signaling, such as autophagy.37, 38 It is, therefore, highly probable that some of the phenotypes obtained with GSK2606414 or GSK2656157 reflect a consequence of RIPK1 inhibition rather than PERK inhibition. In contrast to GSK2606414 and GSK2656157, we found that AMG'44 inhibits PERK activity without interfering with RIPK1. We would, therefore, recommend using AMG'44 for the pharmacological inhibition of PERK in biological processes that might be influenced by RIPK1.
We found that GSK2656157 displayed about 800-fold increased activity over NEC-1 and 200-fold over NEC-1s in inhibiting RIPK1 in our cell death assays. We also found that GSK2656157 was a much better RIPK1 inhibitor than NEC-1s in the in vivo model of TNF-induced lethal shock. Remarkably, the in vitro and in vivo inhibitory activity of GSK2656157 on RIPK1 was comparable to the one of GSK'963, a recently developed RIPK1 inhibitor exhibiting improved efficacy and in vivo availability relative to NEC-1s.35 This makes GSK2656157 one of the most potent RIPK1 inhibitors currently available. These findings therefore open the doors for a broader use of this inhibitor in various cellular and in vivo models involving RIPK1 kinase-dependent functions. In this study, we focused on RIPK1 but we cannot exclude the possibility that GSK2656157 also efficiently targets additional kinases other than RIPK1 and PERK. GSK2656157 was developed to improve the physical properties and pharmacokinetics of its analog GSK2606414.21, 22 Despite its high selectivity for PERK, GSK2656157 was reported to lead to 80% inhibition of 15 other kinases (out of a panel of 300 kinases that did not include RIPK1) when used in vitro at 10 μM. It would therefore be important to re-evaluate the kinase selectivity profile of GSK2656157 on the whole human kinome when using it at a much lower concentration shown to be sufficient to inhibit 100% of RIPK1 activity. In light of the general interest in generating small molecules specifically targeting RIPK1 or PERK in pathological conditions, it would also be of importance to generate derivatives of GSK2656157 in the hope to segregate the specificity for each of the two kinases.
In conclusion, our study led to the identification of GSK2606414 and GSK2656157 as two new potent RIPK1 inhibitors working in the nanomolar range. Our work also highlights the risk of misinterpretation when using these two compounds as ‘selective' PERK inhibitors.
Materials and methods
Antibodies and reagents
Antibodies and reagents were purchased from the following companies: anti-p-IκBα (Ser32/36; Cell Signaling Technology, Danvers, MA, USA; no. 9246), anti-p-p38 (Thr180/Tyr182; Cell Signaling Technology; no. 9211), anti-p38 (Cell Signaling Technology; no. 9212), anti-CHOP (Cell Signaling Technology; no. 2895), anti-PERK (Cell Signaling Technology; no. 3192), anti-IκBα (Santa Cruz Biotechnology; Dallas, TX, USA; no. sc-371), anti-FADD (Santa Cruz Biotechnology; no. sc-6036), anti-RIPK1 (BD Transduction Laboratories, San Jose, CA, USA; no. 610166), anti-Caspase-8 (Abnova, Taipei city, Taiwan; no. MAB3429), anti-FADD (Enzo Life Sciences, Farmingdale, NY, USA; no. ADI-AAM-212-E), anti-HSP90 (Abcam, Cambridge, UK; no. 13480), anti-ATF4 (Proteintech group, Manchester, UK; no. 10835-1), Tunicamicyn (Tm; Sigma-Aldrich, St Louis, MO, USA; no. T7765), Cycloheximide (CHX; Sigma-Aldrich; no. C-7698), 9-Epimer-11,12-dihydro-(5Z)-7-Oxozeanol ((5Z)-7-Oxozeaenol or TAK1-inh; AnalytiCon Discovery GmbH, Potsdam, Germany; no. NP-0009245), ZVAD-fmk (Bachem, Bubendorf, Switzerland; no. N-1510), AMG'44 (Tocris Bioscience, Bristol, UK; no. 5517), GSK2606414 (Toronto Research Chemicals, Toronto, ON, Canada; no. G797800), GSK2656157 (ApexBio, Boston, MA, USA; no. B2175), NEC-1 (Calbiochem, San Diego, CA, USA; no. 480065), NEC-1s (Laboratory of Medicinal Chemistry, University of Antwerp, Belgium), GSK'963 (Aobius, Gloucester, MA, USA; no. 9775). Recombinant human and mouse TNF-α are produced and purified to at least 99% homogeneity in our laboratory and have a respective specific biological activity of 6.8 × 107 IU/mg and 6.12 × 109 IU/mg (4.15 × 109 IU/mg for the one used in vivo).
Cell culture
SV40 large T-immortalized Ripk1+/+ MEFs and L929sAhFas cells (referred to as L929 cells) have been previously described.19, 39 The MEFs and L929 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and l-glutamine (2 mM). For siRNA-mediated repression, MEFs were seeded at 2 × 105 cell/well in six-well plates and transfected 24 h later with ON-TARGETplus smart-pool siRNAs (Dharmacon, Lafayette, CO, USA; PERK: L-044901-00-0010, NS: D-001810-10-2) using Dharmafect Transfection Reagents (Dharmacon, no. T-2001-01). Dharmafect was used at 4 μl for a final concentration of 30 nM siRNA/well. The cells were analyzed for cell death induction 48 h post transfection.
Cell-death analysis
The cell-death measurements were carried out using a Fluostar Omega fluorescence plate reader (BMG Labtech, Ortenberg, Germany) with temperature- and atmosphere-controlled settings, as previously reported.40 In brief, the cells were seeded in triplicate at 10 000 cells per well in a 96-well adherent plate. The next day, the cells were treated with the indicated compounds in the presence of 5 μM SytoxGreen (Invitrogen, Waltham, MA, USA; no. S7020) and 10 μM Ac-DEVD-AMC (Pepta Nova, Sandhausen, Germany; no. 3171-V). SytoxGreen intensity was measured in function of time at intervals of 1 h with an excitation filter of 485 nm, emission filter of 520 nm, gains set at 1100. We used 20 flashes per well and orbital averaging with a diameter of 3 mm. The cell death was calculated by subtracting the background fluorescence from the induced SytoxGreen fluorescence and by dividing the obtained result by the maximal fluorescence (minus the background fluorescence) obtained by permeabilization of the cells by using Triton X-100 at a final concentration of 0.1%. Caspase-3 activity (cleaved DEVD-AMC) was obtained using an excitation filter of 355 nm and an emission filter of 460 nm with the same time intervals and gains of 1000 with 20 flashes per well and 3 mm orbital averaging.
In vitro kinase assays
In vitro kinase assays were performed by ADP-Glo luminescence assays (Promega; Madison, WI, USA; no. V9101) using recombinant GST-hRIPK11–479 (produced in our laboratory) or purchased recombinant GST-hPERK (Invitrogen; no. PV5106), and following the manufacturer's protocol. The primary reaction (150 nM GST-RIPK1 or 16 nM GST-PERK, 50 μM ATP, 50 mM HEPES pH 7.5, 50 mM NaCl, 30 mM MgCl2, 4 mM DTT, 0.5 mg/ml BSA and 0.02% CHAPS, in the presence of increasing concentrations of the indicated compounds) was carried out for 4 h at room temperature. We used a 2 : 2 : 1 ratio of kinase reaction volume to ADP-Glo reagent volume to kinase detection reagent volume.
Immunoprecipitation
MEFs were seeded at 2 × 106 cells in 60 cm2 culture plates. The next day, the cells were pretreated with the indicated compounds for 30 min before stimulation with hTNF (1 ng/ml) for 2 h. Then, the cells were washed two times in ice-cold PBS before lysis in 1 ml of NP-40 lysis buffer containing protease (Roche Applied Science, Penzberg, Germany; no. 11873580001) and phosphate (Roche Applied Science; no. 04906837001) inhibitors. The cell lysates were cleared by centrifugation for 15 min at 4 °C and the supernatant incubated overnight with protein A Sepharose CL-4B (GE Healthcare, Diegem, Belgium; no. 17-0780-01) containing 8 μl of anti-FADD (M-19) antibody (Santa Cruz Biotechnology; no. sc-6036). The next day, the beads were washed three times in NP-40 lysis buffer. The immune complexes were then eluted in 2 × Laemmli buffer and analyzed by western blotting.
TNF-induced lethal shock
C57BL/6 J female mice of 8–11 weeks of age were purchased from Janvier Labs and housed for 2 weeks at the specific-pathogen free animal facility of the IRC at VIB (Ghent, Belgium) before the TNF-induced shock experiment. All the experiments on mice were conducted according to institutional, national and European animal regulations. DMSO stock solutions of NEC-1s, GSK'157, GSK'963 and AMG'44 were diluted in endotoxin-free DPBS (Sigma-Aldrich) and injected intravenously (i.v.) in a volume of 200 μl (6% DMSO final) at 6 mg/kg (21.6 μmol/kg; 10X) for NEC-1s, 0.9 and 0.226 mg/kg (2.16 and 0.54 μmol/kg; 1X and 0.25X, respectively) for GSK'157, 12 mg/kg (21.4 μmol/kg; 10X) for AMG'44 and 0.496 and 0.124 mg/kg (2.16 and 0.54 μmol/kg; 1X and 0.25X, respectively), 15 min before i.v. injection of mTNF (15 μg/20 g in 200 μl endotoxin-free PBS (pH 6.8)). Control mice received an equal amount (i.v.) of DMSO (6%) dissolved in DPBS 15 min before the mTNF challenge (vehicle). Mortality and body temperature were monitored until 4 days after mTNF injection. Rectal body temperature was recorded with an industrial electric thermometer (Comark Electronics, Norwich, UK; model 2001). The dead mice were considered to be at 22 °C.
In vivo analysis of PERK activation/inhibition
C57BL/6 J female mice of 9 weeks of age were i.v. injected with vehicle (DMSO), GSK'157 (2.16 μmol/kg; 1X) or AMG'44 (21.4 μmol/kg; 10X) 15 min before i.p. injection with 2 μg/g of Tm diluted in 150 mM dextrose. After 6 h, the livers were collected and homogenized in lysis buffer (1% NP-40, 200 mM NaCl, 10 mM Tris-HCl pH 7.5, 5 mM EDTA and 10% glycerol) containing protease (Roche Applied Science, Penzberg, Germany; no. 11873580001) and phosphate (Roche Applied Science; no. 04906837001) inhibitors using the Tissuelyser II (Qiagen, Hilden, Germany). PERK activation (auto-phosphorylation) was monitored by western blot analysis using 20 μg of proteins/lane. Each lane represents a different animal. The immunoblot signal was obtained with the Amersham Imager 600 (GE Healthcare Life Sciences, Chicago, IL, USA) and the quantification of the intensity of the bands corresponding to the non-phosphorylated (a) and auto-phosphorylated (b) forms of PERK was performed using ImageJ (https://imagej.nih.gov/ij/). PERK phosphorylation (%)=b/(a+b) × 100.
Docking settings
All waters and ligands of crystal structures 4NEU and 5HX6 were manually deleted from the pdb-text file. The emptied structures were subjected to a local minimization with the GROMOS96 (43B1 parameter set) implementation within the Swiss-PdbViewer,41 and polar hydrogens were added. Ligands were 3D-drawn with Avogadro 1.1.142 and minimized with the built-in general AMBER force field.43 The AutoDockTools 1.5.4 suite44 was used for pdbqt-format preparation of the protein and ligands. Dockings were with AutoDock-Vina 1.1.028 with exhaustiveness set at 32. Grid-box sizes were x, y and z=18 centered at x=28.4, y=61.7 and z=50.9 for 4NEU; x, y and z=18 centered at x=20.6, y=−6.2 and z=56.8 for 5HX6 where Phe162 was set as flexible residue. Visualization was with PyMOL (DeLano Scientific LLC, San Carlos, CA, USA).
Statistical analysis
Statistical analysis was performed with GraphPad Prism V6 software (GraphPad, La Jolla, CA, USA). Statistical significance was determined using two-way ANOVA, Student's test or Kaplan–Meier and log-rank Mantel–Cox test. Significance between samples is indicated in the figures as follows: NS=P>0.05; *P<0.05; **P<0.01; ***P<0.001.
Acknowledgments
Research in the groups of MJMB and PV is supported by grants from the Vlaams Instituut voor Biotechnologie (VIB), grants from Ghent University (MRP, GROUP-ID consortium), grants from the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO)(G017212N, G013715N, G078713N), from the Flemish Government (Methusalem BOF09/01M00709 and BOF16/MET_V/007) and from the Belgian science policy office (BELSPO)(IAP 7/32). RR is paid by VIB grant. TD is financed by FWO grant G0A5413N. The European Research Council (ERC) grant ‘glycotarget' and a BOF grant from Ghent University provide funding for WN.
Footnotes
Edited by P Salomoni
The authors declare no conflict of interest.
References
- Cao SS, Kaufman RJ. Unfolded protein response. Curr Biol 2012; 22: R622–R626. [DOI] [PubMed] [Google Scholar]
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 2012; 13: 89–102. [DOI] [PubMed] [Google Scholar]
- Grootjans J, Kaser A, Kaufman RJ, Blumberg RS. The unfolded protein response in immunity and inflammation. Nat Rev Immunol 2016; 16: 469–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao SS, Luo KL, Shi L. Endoplasmic reticulum stress interacts with inflammation in human diseases. J Cell Physiol 2016; 231: 288–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg AD, Kaczmarek A, Krysko O, Vandenabeele P, Krysko DV, Agostinis P. ER stress-induced inflammation: does it aid or impede disease progression? Trends Mol Med 2012; 18: 589–598. [DOI] [PubMed] [Google Scholar]
- Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer 2014; 14: 581–597. [DOI] [PubMed] [Google Scholar]
- Tam AB, Mercado EL, Hoffmann A, Niwa M. ER stress activates NF-kappaB by integrating functions of basal IKK activity, IRE1 and PERK. PLoS ONE 2012; 7: e45078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Kaufman RJ. All roads lead to ATF4. Dev Cell 2003; 4: 442–444. [DOI] [PubMed] [Google Scholar]
- Zhang C, Bai N, Chang A, Zhang Z, Yin J, Shen W et al. ATF4 is directly recruited by TLR4 signaling and positively regulates TLR4-trigged cytokine production in human monocytes. Cell Mol Immunol 2013; 10: 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Osualdo A, Anania VG, Yu K, Lill JR, Kaufman RJ, Matsuzawa S et al. Transcription factor ATF4 induces NLRP1 inflammasome expression during endoplasmic reticulum stress. PLoS ONE 2015; 10: e0130635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep 2016; 17: 1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Chevet E, Harding HP. Targeting the unfolded protein response in disease. Nat Rev Drug Discov 2013; 12: 703–719. [DOI] [PubMed] [Google Scholar]
- Kalliolias GD, Ivashkiv LB. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat Rev Rheumatol 2016; 12: 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting AT, Bertrand MJ. More to life than NF-kappaB in TNFR1 signaling. Trends Immunol 2016; 37: 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature 2015; 517: 311–320. [DOI] [PubMed] [Google Scholar]
- Xue X, Piao JH, Nakajima A, Sakon-Komazawa S, Kojima Y, Mori K et al. Tumor necrosis factor alpha (TNFalpha) induces the unfolded protein response (UPR) in a reactive oxygen species (ROS)-dependent fashion, and the UPR counteracts ROS accumulation by TNFalpha. J Biol Chem 2005; 280: 33917–33925. [DOI] [PubMed] [Google Scholar]
- Masuda M, Miyazaki-Anzai S, Levi M, Ting TC, Miyazaki M. PERK-eIF2alpha-ATF4-CHOP signaling contributes to TNFalpha-induced vascular calcification. J Am Heart Assoc 2013; 2: e000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell 2008; 133: 693–703. [DOI] [PubMed] [Google Scholar]
- Dondelinger Y, Aguileta MA, Goossens V, Dubuisson C, Grootjans S, Dejardin E et al. RIPK3 contributes to TNFR1-mediated RIPK1 kinase-dependent apoptosis in conditions of cIAP1/2 depletion or TAK1 kinase inhibition. Cell Death Differ 2013; 20: 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axten JM, Medina JR, Feng Y, Shu A, Romeril SP, Grant SW et al. Discovery of 7-methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-p yrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). J Med Chem 2012; 55: 7193–7207. [DOI] [PubMed] [Google Scholar]
- Atkins C, Liu Q, Minthorn E, Zhang SY, Figueroa DJ, Moss K et al. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res 2013; 73: 1993–2002. [DOI] [PubMed] [Google Scholar]
- Axten JM, Romeril SP, Shu A, Ralph J, Medina JR, Feng Y et al. Discovery of GSK2656157: an optimized PERK inhibitor selected for preclinical development. ACS Med Chem Lett 2013; 4: 964–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 2008; 4: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A, Maki JL, Yuan J. Activity and specificity of necrostatin-1, small-molecule inhibitor of RIP1 kinase. Cell Death Differ 2013; 20: 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AL, Andrews KL, Beckmann H, Bellon SF, Beltran PJ, Booker S et al. Discovery of 1H-pyrazol-3(2H)-ones as potent and selective inhibitors of protein kinase R-like endoplasmic reticulum kinase (PERK). J Med Chem 2015; 58: 1426–1441. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, Li H et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife 2013; 2: e00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PA, Bandyopadhyay D, Berger SB, Campobasso N, Capriotti CA, Cox JA et al. Discovery of small molecule RIP1 kinase inhibitors for the treatment of pathologies associated with necroptosis. ACS Med Chem Lett 2013; 4: 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010; 31: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Peng W, Liu Y, Yan C, Maki J, Degterev A et al. Structural basis of RIP1 inhibition by necrostatins. Structure 2013; 21: 493–499. [DOI] [PubMed] [Google Scholar]
- Harris PA, King BW, Bandyopadhyay D, Berger SB, Campobasso N, Capriotti CA et al. DNA-encoded library screening identifies benzo[b][1,4]oxazepin-4-ones as highly potent and monoselective receptor interacting protein 1 kinase inhibitors. J Med Chem 2016; 59: 2163–2178. [DOI] [PubMed] [Google Scholar]
- Duprez L, Takahashi N, Van Hauwermeiren F, Vandendriessche B, Goossens V, Vanden Berghe T et al. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity 2011; 35: 908–918. [DOI] [PubMed] [Google Scholar]
- Berger SB, Kasparcova V, Hoffman S, Swift B, Dare L, Schaeffer M et al. Cutting Edge: RIP1 kinase activity is dispensable for normal development but is a key regulator of inflammation in SHARPIN-deficient mice. J Immunol 2014; 192: 5476–5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Duprez L, Grootjans S, Cauwels A, Nerinckx W, DuHadaway JB et al. Necrostatin-1 analogues: critical issues on the specificity, activity and in vivo use in experimental disease models. Cell Death Dis 2012; 3: e437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondelinger Y, Jouan-Lanhouet S, Divert T, Theatre E, Bertin J, Gough PJ et al. NF-kappaB-independent role of IKKalpha/IKKbeta in preventing RIPK1 kinase-dependent apoptotic and necroptotic cell death during TNF signaling. Mol Cell 2015; 60: 63–76. [DOI] [PubMed] [Google Scholar]
- Berger SB, Harris P, Nagilla R, Kasparcova V, Hoffman S, Swift B et al. Characterization of GSK'963: a structurally distinct, potent and selective inhibitor of RIP1 kinase. Cell Death Discov 2015; 1: 15009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JA, Halliday M, Molloy C, Radford H, Verity N, Axten JM et al. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci Transl Med 2013; 5: 206ra138. [DOI] [PubMed] [Google Scholar]
- Yonekawa T, Gamez G, Kim J, Tan AC, Thorburn J, Gump J et al. RIP1 negatively regulates basal autophagic flux through TFEB to control sensitivity to apoptosis. EMBO Rep 2015; 16: 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Q, Jin L, Jiang CC, Tay KH, Lai F, Liu XY et al. RIPK1 regulates survival of human melanoma cells upon endoplasmic reticulum stress through autophagy. Autophagy 2015; 11: 975–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlangenakker N, Bertrand MJ, Bogaert P, Vandenabeele P, Vanden Berghe T. TNF-induced necroptosis in L929 cells is tightly regulated by multiple TNFR1 complex I and II members. Cell Death Dis 2011; 2: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grootjans S, Hassannia B, Delrue I, Goossens V, Wiernicki B, Dondelinger Y et al. A real-time fluorometric method for the simultaneous detection of cell death type and rate. Nat Protoc 2016; 11: 1444–1454. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 1997; 18: 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminform 2012; 4: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem 2004; 25: 1157–1174. [DOI] [PubMed] [Google Scholar]
- Sanner MF. Python: a programming language for software integration and development. J Mol Graph Model 1999; 17: 57–61. [PubMed] [Google Scholar]