Abstract
Background
Trypanosoma brucei rhodesiense is the causative agent of acute human African trypanosomiasis. Identification of T. b. rhodesiense in tsetse populations is essential for understanding transmission dynamics, assessng human disease risk, and monitoring spatiotemporal trends and impact of control interventions. Accurate detection and characterisation of trypanosomes in vectors relies on molecular techniques. For the first time in Malawi, a molecular technique has been used to detect trypanosomes in tsetse flies in Nkhotakota Wildlife Reserve.
Methods
A polymerase chain reaction (PCR) technique was used to identify the serum resistance associated (SRA) gene of T. b. rhodesiense in tsetse flies. Of 257 tsetse flies that were randomly caught, 42 flies were dissected for microscopic examination. The midguts of 206 flies were positive and were individually put in eppendorf tubes containing phosphate-buffered saline (PBS buffer) for DNA extraction. Internal transcribed spacer (ITS)-PCR was first used to isolate all trypanosome species from the flies. TBR PCR was then used to isolate the Trypanozoon group. T. brucei-positive samples were further evaluated by SRA PCR for the presence of the SRA gene.
Results
Of 257 flies caught, 185 (72%) were Glossina morsitans morsitans and 72 (28%) were Glossina pallidipes. Three were tenerals and 242 were mature live flies. Of the 242 flies dissected, 206 were positive, representing an 85.1% infection rate. From 206 infected flies, 106 (51.5%) were positive using ITS-PCR, 68 (33.0%) being mixed infections, 18 (8.7%) T. brucei, 9 (4.4%) Trypanosoma vivax, 4 (1.9%) Trypanosoma godfrey, 3 (1.5%) Trypanosoma congolense savanna, 3 (1.5%) Trypanosoma simae, and 1 (0.4%) Trypanosoma simaetsavo. When subjected to TBR PCR, 107(51.9%) were positive for T. brucei. Of the 107 T. brucei-positive samples, 5 (4.7%) were found to have the SRA gene.
Conclusions
These results suggest that wild tsetse flies in Malawi are infected with human-infective trypanosomes that put communities around wildlife reserves at risk of human African trypanosomiasis outbreaks. Further studies need to be done to identify sources of blood meals for the flies and for surveillance of communities around wildlife reserves.
Introduction
Tsetse flies (Glossina spp.) are the vectors for the transmission of both human and animal African trypanosomiasis. Animal trypanosomiasis is called nagana, and it affects domestic animals. The reservoir hosts for both human and animal trypanosomiasis are wild animals—particularly antelopes,1 with domestic animals contributing to a lesser extent.2 The human disease (also known as sleeping sickness) is caused by 2 species of Trypanosoma brucei. These are Trypanosoma brucei rhodesiense, which is confined to East Africa in Malawi and Tanzania, though it is also reported to be present in some parts of Uganda. T. b. rhodesiense is commonly associated with acute infection. Chronic disease is mainly caused by Trypanosoma brucei gambiense, which is currently confined to West Africa, in particular DR Congo, Angola, Guinea, and Côte d'lvoire.3
There are several species of tsetse flies (Glossinidae: Diptera) that feed on both humans and animals. There are specific foci for these flies. The flies that are good vectors for T. b. rhodesiense (namely, Glossina morsitans morsitans, Glossina swynnertinii, Glossina auteni, Glossina longipenis, Glossina pallidipes, Glossina bravepalpis, and Glossina fuscipes fuscipes5–7) are mainly confined to the southwestern part of Africa, while the flies that are good vectors for T. b. gambiense (namely, Glossina fuscipes fuscipes, Glossina tachinoides, and Glossina morsistans8–10) are mainly confined to the western part of the continent. However, there is an overlap of these flies, mainly in northwest and southeast Uganda.4 It is clear that Glossina fuscipes and Glossina morsitans are ubiquitous in their locations, making these species of tsetse flies good vectors for both acute and chronic human African trypanosomiasis.
The agents of sleeping sickness belong to the genus Trypanosoma, subgenus Trypanozoon. There are 3 subspecies of Trypanosoma brucei, namely, Trypanosoma brucei rhodesiense, Trypansoma brucei gambiense, and Trypanosoma brucei brucei; the latter is associated with animal trypanosomiasis and has no effect on human hosts. These species cannot be distinguished morphologically by microscopy. There are molecular techniques that utilise the genetic differences between these parasites that can be used to distinguish them. A commonly used technique involves the serum resistance-associated (SRA) gene, which is lacking in T. b. gambiense but is present in T. b. rhodesiense. The primers for the SRA gene are used in a polymerase chain reaction (PCR) technique to specifically distinguish the 2 forms.11,12 Another molecular method uses the internal transcribed spacer (ITS) regions of the ribosomal DNA (rDNA) of trypanosomes. The ITS region of rDNA is the preferred target for a universal test because of its highly conserved flanking regions and size variability among trypanosomes species and subgroups. Therefore, the ITS1 PCR technique is used to identify almost all trypanosome species.13
The current study was designed to identify if the species of tsetse flies that inhabit Nkhotakota Wildlife Reserve harbour T. b. rhodesiense.
Methods
Two approaches were used to isolate transmissible T. brucei spp. and other trypanosome species: (1) dissection and microscopy, followed by (2) PCR on single midgut samples.
Trapping of tsetse flies
After getting clearance from Nkhotakota Wildlife Reserve officials, and in collaboration with the Malawi Ministry of Agriculture, 5 locations in Nkhotakota Wildlife Reserve were purposefully identified as ideal for trapping tsetse flies. Fly traps were deployed at a distance of 100 m apart. A total of 100 traps were deployed, which included both biconical and Epsilon traps (Figure 1) that were baited with phenols and acetone before being deployed. Tsetse flies were harvested every 24 hours for 2 weeks and there was a break of 3 months during the year. All flies were sorted, according to sex and species, using an ordinary microscope. To distinguish between Glossina morsitans morsitans and Glossina pallidipes, differences in antennal shape and antennal fringe proportions were noted. All tenerals (those that have not yet taken their first blood meal) and dead flies were recorded but were not included in the investigation. Tsetse flies were dissected and each midgut was examined parasitologically (Table 1) using Olympus dissecting and biological microscopes, respectively. Each midgut was then placed in eppendorf tubes containing 200 µL of phosphate-buffered saline (PBS buffer) and were frozen at −200°C for DNA extraction. DNA was extracted from these midguts using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Germantown, MD, USA).
Figure 1.

(A) Biconical and (B) Epsilon traps used to catch tsetse flies at Nkhotakota Wildlife Reserve
Table 1.
Summary showing the number of flies caught and their species, sexes, and infection rates
| Tsetse flies collected | Condition | Males | Females | Total | Infection rate |
| 257 | Infected | 90 | 116 | 206 | 85.1% |
| Non-infected | 19 | 17 | 36 | ||
| 72% G. m. morsitans | Dead | 7 | 5 | 12 | |
| 28% G. pallidipes | Tenerals | 1 | 2 | 3 | |
| Total | 117 | 140 | 257 | ||
PCR steps to identify T. b. rhodesiense
1. ITS1 rDNA PCR to identify all trypanosome species
Midguts of all tsetse flies were tested for the presence of trypanosomes using ITS1 PCR, which can even isolate mixed infections in a sample. The ITS1 rDNA primers used were: The forward 5′-CCGGAAGTTCACCGATATTG-3′ and reverse 5′-TTGCTGCGTTCTTCAACGAA-3′.13 PCR was carried out in 25-µL reaction mixtures containing 5X Phusion HF buffer with a 1.5 mM MgCl2 final reaction concentration (Thermo Fisher Scientific, Waltham, MA, USA), 10 mM of each of the 4 deoxynucleoside triphosphates (dNTPs), primers at 1 µM, 0.25 µL of Taq DNA polymerase (Thermo Fisher Scientific, Waltham, MA, USA), and 3 µL of DNA template. PCR cycles for the primers were: initial step at 98°C for 10 seconds, followed by 35 cycles of 98°C for 30 seconds, 65°C for 30 seconds, 72°C for 30 seconds, and final extension at 72°C for 10 minutes using a Techne Genius thermal cycler (Cole-Parmer, Stone, United Kingdom). Amplification products were visualised in 2 % molecular grade agarose gel (Fisher Biotec, Wembley, Australia) stained with ethidium bromide on an ultraviolet (UV) transluminator (Syngene, Frederick, MD, USA). The PCR product band sizes and species identified are indicated in Table 2 and Figure 2.
Table 2.
Results of various polymerase chain reaction (PCR) methods, species identified, and expected PCR product sizes
| PCR method |
Midguts analysed |
Positive | Species identified | Expected product band size (base pairs) |
Reference |
| ITS1-PCR | 206 | 106 (51.5%) | |||
| 68 (33%) | Mixed infection | ||||
| 18 (8.7%) | T. brucei | 480 | Njiru et al.13 | ||
| 9 (4.4%) | T. vivax | 250 | Njiru et al.13 | ||
| 4 (1.9%) | T. godfrey | 300 | Njiru et al.13 | ||
| 3 (1.5%) | T. simae | 400 | Njiru et al.13 | ||
| 3 (1,5%) | T. congolense savanna | 700 | Njiru et al.13 | ||
| 1 (0.5%) | T. simaetsayo | 370 | Njiru et al.13 | ||
| TBR-PCR | 206 | 107 (51.9%) | T. brucei | 177 | Moser et al.14 |
| SRA-PCR | 107 | 5 (4.7%) | T. b. rhodesiense | 284 | Radwanska et al.12 |
Figure 2.
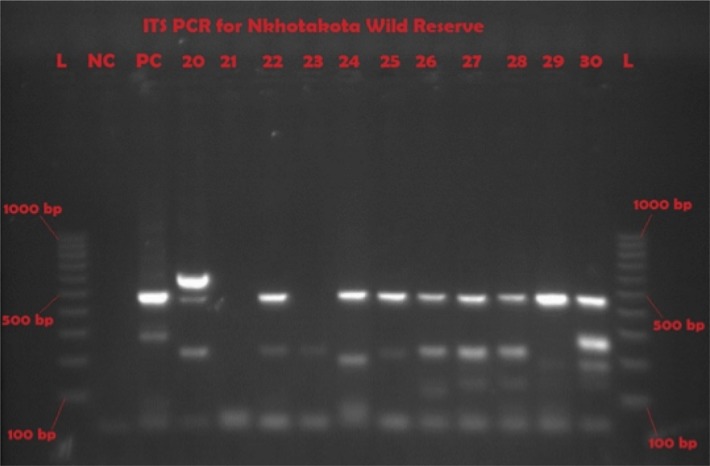
Representative gel of ITS1-PCR showing various trypanosome species and mixed infections
L = molecular ladder; NC = negative control; PC = T. brucei positive control; 20, 22, 24 to 30 are positive samples for different species
2. TBR PCR to identify all trypanozoons
Though ITS1 PCR can identify trypanosomes from the Trypanozoon subgenus, TBR PCR is used as a confirmatory test because it exclusively selects Trypanozoon parasites. The TBR1 and TBR2 PCR primers used were 5′-CGAATGAATATTAAACAATGCGCAG-3′ and 5'-AGAACCATTTATTAGCTTTGTTGC-3',14 respectively. PCR was carried out in 25-µL reaction mixtures containing 5X Phusion HF buffer with a 1.5 mM MgCl2 final reaction concentration (Thermo Fisher Scientific, Waltham, MA, USA), 10 mM of each of the 4 dNTPs, primers at 1 µM, 0.2 µL of Taq DNA polymerase (Thermo Fisher Scientific, Waltham, MA, USA), 7 µL of DNA template for initial PCR, and 5 µL of template for nested PCR. PCR cycles for the primers were: initial step at 98°C for 10 seconds, followed by 35 cycles of 98°C for 30 seconds, 66°C for 30 seconds, 72°C for 30 seconds, and final extension at 72°C for 10 minutes using a Techne Genius thermal cycler (Cole-Parmer, Stone, United Kingdom). Amplification products were visualised in 2% molecular grade agarose (Fisher Biotec, Wembley, Australia) stained with ethidium bromide on a UV transluminator (Syngene, Frederick, MD, USA). For this test the size of the PCR product was 177 bp (Table 2 and Figure 3).
Figure 3.
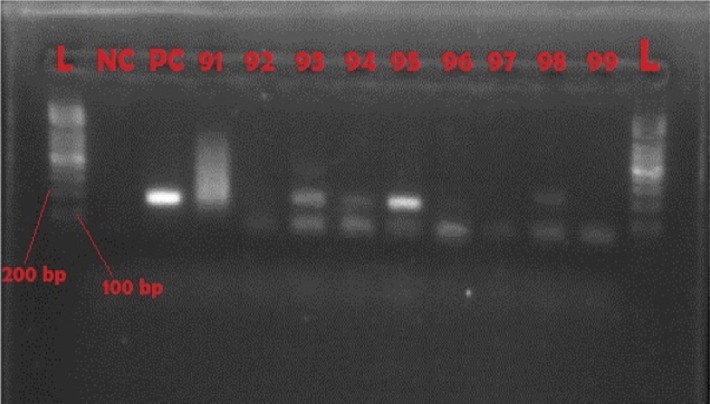
Representative gel of TBR-PCR
NC = negative control; PC = positive control with product size of 177 bp; 93, 94, 95, and 98 are positive samples
3. SRA PCR to identify T. b. rhodesiense
From the Trypanozoon subgenus, our species of interest was T. b. rhodesiense, and SRA PCR was used to differentiate T. b. rhodesiense from the other members of the group. The primers used were: the forward 5′-ATAGTGACAAGATGCGTACTCAACGC-3′ and reverse 5′-AATGTGTTCGAGTACTTCGGTCACGCT-3′.12 PCR was carried out in 20-µL reaction mixtures containing 5X Phusion HF buffer with a 1.5 mM MgCl2 final reaction concentration (Thermo Fisher Scientific, Waltham, MA, USA), 10 mM of each of the 4 dNTPs, primers at 1 µM, 0.2 µL of Taq DNA polymerase (Thermo Fisher Scientific, Waltham, MA, USA), 5 µL of DNA template for initial PCR, and 2 µL of template for nested PCR. PCR cycles for the primers were: initial step at 98°C for 10 seconds, followed by 35 cycles of 98°C for 30 seconds, 66°C for 30 seconds, 72°C for 30 seconds, and final extension at 72°C for 10 minutes using a Techne Genius thermal cycler (Cole-Parmer, Stone, United Kingdom). Amplification products were visualised in 2% molecular grade agarose (Fisher Biotec, Wembley, Australia) stained with ethidium bromide on a UV transluminator (Syngene, Frederick, MD, USA). The size of the PCR product was 284 bp (Table 2 and Figure 4).
Figure 4.
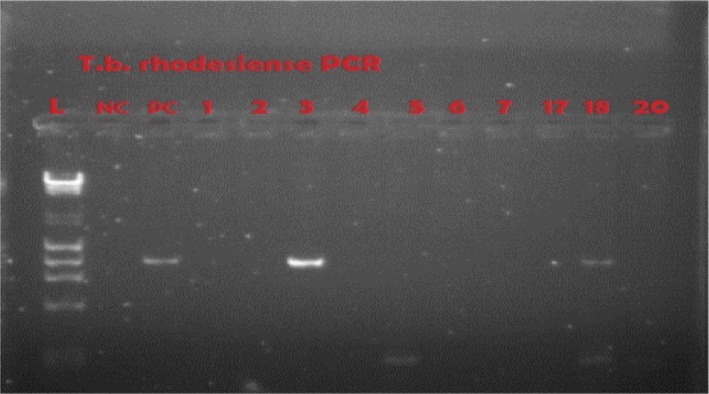
Representative gel of SRA-PCR
NC = negative control; PC = positive control; 3 and 18 are positive at 284 bp
Results
A total of 257 flies were caught (Table 1). Of these, 185 flies (72%) were Glossina morsitan morsitans and 72 (28%) were Glossina pallidipes. Among all flies, 140 (54.5%) were females and 117 (45.5%) were males. Three (1.2%) were tenerals and 12 (4.7%) were dead on the day of sampling. Only mature live flies (242 total, comprising 133 females and 109 males) were dissected for further analysis. In total 206 flies out of the 242 dissected flies were identified with trypanosomes in their midguts, representing an infection rate of 85.1%.
When the subset of live female flies was studied, 116 (87.2%) of 133 were carrying trypanosomes in their midguts. On the other hand, 90 (82.6%) of the 109 live male flies caught were carrying trypanosomes in their midguts. When the 206 infected samples were subjected to ITS rDNA PCR, 106 (51.4 %) were positive (Figure 2), and flies infected wuth more than 1 trypanosome species were observed. Samples from the 206 infected flies were also subjected to TBR PCR (Figure 3), and 107 samples (51.9 %) were positive for T. brucei. This observation showed that tsetse flies in Nkhotakota Wildlife Reserve are highly infected with parasites that could be a risk to human disease.
The 107 DNA samples that were positive for T. brucei subspecies in both ITS rDNA PCR and TBR PCR were subjected to the SRA PCR, and 5 (4.7 %) were positive (Figure 4). This indicated that 4.7 % of the tsetse flies carried T. b. rhodesiense. This was a definitive observation that flies in Nkhotakota Wildlife Reserve carry trypanosomes that are infective to humans.
Discussion
Previous studies have also shown that Glossina morsitans morsitans and Glossina pallidipes are the most prevalent species of tsetse fly in the wildlife reserves of Malawi.15 The present study is in agreement with this observation. However, the prevalence of Glossina morsitans morsitans was higher than that of Glossina pallidipes. It was also noted that the infection rates of both species was not sex dependent, as there was no difference between the proportion of the male (82.6%) and female (87.2%) flies with midgut infections. This shows that the feeding habits of these flies are similar to one another, and that they are not dependent on the egg maturity of female flies. One would have expected a difference in infection rates if blood meals were directly associated with egg maturity. There would also be a difference if each species had a different preferred host for feeding. The proportion of flies infected was also higher than has been observed in other tsetse flies, both in the region and other areas (unpublished observations). This could partly be explained by a common host animal that is preferred for feeding by the flies in Nkhotakota Wildlife Reserve. It is well established that Glossina morsitans morsitans are efficient vectors of trypanosomiasis, as they easily establish infections in their midguts after a blood meal.16 These flies likely feed on antelopes, which are prevalent in the nature reserve and are a known major reservoir host for trypanosomes.1,17
The present study has also established that Trypanosoma brucei species are the most prevalent parasites circulating in the tsetse flies, implying that they are also prevalent in the reservoir hosts. The direct TBR PCR showed that of all infected flies, 107 (51.9%) of 206 were harbouring Trypanosoma brucei infection. The proportion of DNA samples from the same infected flies showed that 106 (51.4 %) of 206 were positive with ITS rDNA PCR. ITS rDNA PCR does not distinguish one trypanosome species from another. The difference in diagnostic sensitivity between ITS rDNA PCR and TBR PCR is partly explained by mixed infections in some of the tsetse flies.13 If every fly was infected by a single parasite, the proportions between TBR PCR and ITS rDNA PCR would have been the same. Additionally, ITS1 has approximately 100 to 200 copies, as opposed to species-specific tests that target satellite DNA with over 10,000 copies, making them more sensitive than universal tests like ITS1.18 As a result of this finding, it became necessary to distinguish the species of Trypanosoma brucei in the DNA of 107 positive tsetse flies that were identified after ITS rDNA PCR and TBR PCR. This was done by nested PCR for the SRA gene. From the 107 samples, 5 were found positive for the SRA gene (Figure 4). SRA gene identification confirmed the existence of Trypanosoma brucei rhodesiense, the causative agent of the acute human African trypanosomiasis found in Malawi and other countries in the southern African region.
From these molecular studies, it is apparent that animal infections and possibly human trypanosomiasis disease can be transmitted in this wildlife reserve. Indeed, over the years, high levels of human infections of trypanosomiasis have been reported at Nkhotakota District Hospital since records of cases began. The level of human infections has been on the decline, while the data for domestic animal infections is scanty.19–22 The present study has also confirmed that both human and animal infections are likely to be caused by the Trypanosoma brucei subspecies in and around Nkhotakota Wildlife Reserve. Cases of trypanosomiasis have been reported in tourists and soldiers that have visited some of Malawi's wildlife reserves.23–25 Moreover, the cases of trypanosomiasis have also been shown to be subacute in some studies, and showing a genetic susceptibility in some people.26,27 Notably, the high infection rates that have been shown in the present study have not translated in a trypanosomiasis epidemic in Malawi. The possible explanation of this is lack of a human-tsetse fly interface.28
Conclusions
The present study has shown that tsetse flies in Nkhotakota Wildlife Reserve have a high infection rate at 85.1%, and more than half of these infections are attributed to Trypanosoma brucei species that are causative agents of both human and animal disease. There is a need to sample blood from both animals and humans to establish whether the risk translates to infections in human beings, as well as cattle, pigs, goats, and wildlife in and around the reserves. In addition, there is a strong need to intensify tsetse trapping to reduce the number of flies in the park. Blood meal analysis would assist in identifying the common sources of infection between flies. This would clarify the observation that tsetse flies seem to prefer a common source of host feeding, as suggested by the same infection rate between species and sexes. Detection of trypanosomes in the vector indicates the presence of potentially infective to both humans and animals in the surrounding areas. It is sensible to suggest that resources should be mobilised to prevent and mitigate potential trypanosomiasis epidemics, both in humans and animals.
Acknowledgements
We are grateful to the Consortium for Development and Application of Xenomonitoring Tools in Human African Trypanosomiasis, spearheaded by Makerere University, with funding from the Bill & Melinda Gates Foundation, for providing resources for this study. We are highly indebted to the Malawi government, particularly the Ministry of Health; the Ministry of Agriculture and Food Security; and the Ministry of Information, Culture and Tourism, for supporting the present study. Above all, we thank all the technical staff for working hard both in the field and laboratory.
Competing interests
All authors declare that they have no competing interests related to this work.
References
- 1.Duke HL. Antelope and their relation to trypanosomiasis. Proceedings B of the Royal Society. 1912;85:577. [Google Scholar]
- 2.Van den Bossche P, Staak C. The importance of cattle as a food source for Glossina morsitans morsitans Katete district, Eastern Province, Zambia. Acta Trop. 1997;65:105–109. doi: 10.1016/s0001-706x(97)00658-x. [DOI] [PubMed] [Google Scholar]
- 3.WHO, author. Human African Trypanosomiasis (sleeping sickness) Fact sheet. 2013. Available: http://www.who.int/trypanosomiasis_african/en/
- 4.Aksoy E, Telleria EL, Echodu R, et al. Analysis of multiple tsetse fly populations in Uganda reveals limited diversity and species-specific gut microbiota. Appl Environ Microbiol. 2014;80(14):4301–4312. doi: 10.1128/AEM.00079-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahama CI, Desquesness M, Dia ML, Losson B, De Deken R, Geerts S. A cross-sectional epidemiological survey of bovine trypanosomiasis and its vectors in the Savelugu and West Mamprusi districts of northern Ghana. Vet Parasitol. 2004;122:1–13. doi: 10.1016/j.vetpar.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Van den Bossche P, Shumba W, Makhambera P. The distribution and epidemiology of bovine trypanosomiasis in Malawi. Vet Parasitol. 2000;88:163–176. doi: 10.1016/s0304-4017(99)00222-8. [DOI] [PubMed] [Google Scholar]
- 7.Sigauque I, van den Bossche P, Motana M, Jamal S, Neves L. The distribution of tsetse (Diptera: Glossinidae) and bovine trypanosomiasis in the Matutuine District, Maputo Province, Mozambique. Onderstepoort J Vet Res. 2000;67:162–172. [PubMed] [Google Scholar]
- 8.Saunders DS. Age determination for female tsetse flies and the age compositions of samples of Glossina pallidipes Aust, G.palpalis fuscipes Newst., and G. brevipalpis Newst. Bullentin of Entomological Research. 1962;53:579–595. [Google Scholar]
- 9.Woolhouse MEJ, Bealby K, Mcnamara JJ, Silutongwe J. Trypanosome infection of the tsetse fly Glossina pallidipes in Luangwa Valley, Zambia. Int J Parasitol. 1994;24:987–983. doi: 10.1016/0020-7519(94)90164-3. [DOI] [PubMed] [Google Scholar]
- 10.Wamwiri FN, Nkwengulila G, Clausen PH. Hosts of Glossina fuscipes fuscipes and G. pallidipes in sleeping sickness endemic areas of western Kenya, as determined by egg-yolk (IgY) enzyme-linked immunosorbent assay. Annals of Trop Med Parasitol. 2007;101:225–232. doi: 10.1179/136485907X156979. [DOI] [PubMed] [Google Scholar]
- 11.Gibson W, Bachouse T, Griffiths A. The human serum resistance associated gene is ubiquitous and conserves in Trypanosoma brucei rhodesiense throughout East Africa. Infect Genet Evol. 2002;1:207–214. doi: 10.1016/s1567-1348(02)00028-x. [DOI] [PubMed] [Google Scholar]
- 12.Radwanska M, Chamekh M, van Hamme L, et al. The serum resistance-associated gene as a diagnostic tool for the detection of Trypanosoma brucei rhodesiense. Am J Trop Med Hyg. 2002;67:684–690. doi: 10.4269/ajtmh.2002.67.684. [DOI] [PubMed] [Google Scholar]
- 13.Njiru ZK, Constatine CC, Guya S, et al. The use of ITS rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res. 2005;95:186–192. doi: 10.1007/s00436-004-1267-5. [DOI] [PubMed] [Google Scholar]
- 14.Moser DR, Cook GA, Ochs DE, Bailey CP, McKane MR, Donelson JE. Detection of Trypanosoma congolense and Trypanosoma brucei subspecies by DNA amplification using the polymerase chain reaction. Parasitol. 1989;99:57–66. doi: 10.1017/s0031182000061023. [DOI] [PubMed] [Google Scholar]
- 15.Mthepheya MWP, Msiska JGM. Report on Tsetse and Trypanosomiasis situation in Malawi. Lilongwe: Department of Animal Health and Industry; 1989. [Google Scholar]
- 16.Mihok S, Olubayo RO, Wesonga DF. Infection rates in Glossina morsitans morsitans fed on waterbuck and Boran cattle infected with Trypanosoma congolense. Acta Trop. 1991;49(3):185–191. doi: 10.1016/0001-706x(91)90037-k. [DOI] [PubMed] [Google Scholar]
- 17.Moloo SK, Orinda GO, Sabwa CL, Minja SH, Masake RA. Study on the sequential tsetse-transmitted Trypanosoma congolense, T. brucei brucei and T. vivax infections to African buffalo, eland, waterbuck, N'Dama and Boran cattle. Vet Parasitol. 1999;80(3):197–213. doi: 10.1016/s0304-4017(98)00209-x. [DOI] [PubMed] [Google Scholar]
- 18.Desquesnes M, McLaughlin G, Zoungrana A, Davila AM. Detection and identification of Trypanosoma of African livestock through a single PCR based on internal transcribed spacer 1 of rDNA. Int J Parasitol. 2001;31:610–614. doi: 10.1016/s0020-7519(01)00161-8. [DOI] [PubMed] [Google Scholar]
- 19.Chisi JE, Nkhoma A, Sternberg JM. An overview of trypanosomiasis in Malawi. Newsletter on Integrated Control of Pathogenic Trypanosomes and their Vectors (ICPTV) 2003;7:9–10. [Google Scholar]
- 20.Chisi J, Misiri H, Zverev Y, Nkhoma A, Sternberg JM. Anaemia in Human African Trypanosomiasis caused by Trypanosoma brucei rhodesiense. East Afr Med J. 2004;81(10):505–508. doi: 10.4314/eamj.v81i10.9232. [DOI] [PubMed] [Google Scholar]
- 21.Chisi J, Nkhoma A, Sternberg J. Presentation of Trypanosomiasis in Nkhotakota. Malawi Medical Journal. 2007;19(4):140–141. doi: 10.4314/mmj.v19i4.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chisi JE, Muula AS, Ngwira B, Kabuluzi S. A retrospective study of Human African Trypanosomiasis in the three Malawi districts. Tanzania Journal of Health Research. 2011;13(1):79–86. doi: 10.4314/thrb.v13i1.61014. [DOI] [PubMed] [Google Scholar]
- 23.Croft AM, Jackson CJ, Friend HM, Minton EJ. African trypanosomiasis in a British soldier. J R Army Med Corps. 2006;152(3):156–160. doi: 10.1136/jramc-152-03-08. [DOI] [PubMed] [Google Scholar]
- 24.Croft AM, Kitson MM, Jackson CJ, Minton EJ, Friend HM. African trypanosomiasis in a British soldier. Mil Med. 2007;172(7):765–769. doi: 10.7205/milmed.172.7.765. [DOI] [PubMed] [Google Scholar]
- 25.Darby JD, Huber MG, Sieling WL, Spelman DW. African trypanosomiasis in two short-term Australian travelers to Malawi. J Travel Med. 2008;15(5):375–377. doi: 10.1111/j.1708-8305.2008.00242.x. [DOI] [PubMed] [Google Scholar]
- 26.MacLean L, Chisi JE, Odiit M, et al. Severity of Human African Trypanosomiasis in East Africa is associated with geographic location, parasite genotype, and host inflammatory cytokine response profile. Infect Immun. 2004;72(12):7040–7044. doi: 10.1128/IAI.72.12.7040-7044.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacLean LM, Odiit M, Chisi JE, Kennedy PGE, Sternberg JM. Focus-Specific Clinical Profiles in Human African Trypanosomiasis caused by Trypanosoma brucei rhodesiense. PLOS Neglected Tropical Diseases. 2010;4(12):e906. doi: 10.1371/journal.pntd.0000906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gondwe N, Marcotty T, Vanwambeke SO, De Pus C, Mulumba M, Van den Bossche P. Distribution and density of tsetse flies (Glossinidae: Diptera) at the game/people/livestock interface of the Nkhotakota Game Reserve human sleeping sickness focus in Malawi. Ecohealth. 2009;6(2):260–265. doi: 10.1007/s10393-009-0252-y. (2009) [DOI] [PubMed] [Google Scholar]


