Abstract
Complete genome sequences in combination with global screening methods allow parallel analysis of multiple mutant loci to determine the requirement for specific genes in different environments. In this paper we describe a high-definition microarray approach for investigating the growth effects of Tn5 insertions in Escherichia coli K-12. Libraries of insertion mutants generated by a unique Tn5 mutagenesis system were grown competitively in defined media. Biotin-labeled runoff RNA transcripts were generated in vitro from transposon insertions in each population of mutants. These transcripts were then hybridized to custom-designed oligonucleotide microarrays to detect the presence of each mutant in the population. By using this approach, the signal associated with 25 auxotrophic insertions in a 50-mutant pool was not detectable following nine generations of growth in glucose M9 minimal medium. It was found that individual insertion sites could be mapped to within 50 bp of their genomic locations, and 340 dispensable regions in the E. coli chromosome were identified. Tn5 insertions were detected in 15 genes for which no previous insertions have been reported. Other applications of this method are discussed.
Defining the role and regulation of gene products has always been a major goal of microbial geneticists. A first step toward understanding the function of a gene involves identifying growth conditions in which the relevant gene product is required for growth. Using conventional methods, workers have investigated single-knockout mutants and the resulting phenotypes. Although effective, this is a slow and often laborious process. Recent advances in genome-wide strategies, particularly those involving transcriptional profiling, have allowed investigations of multiple genes in a single experiment. Often in such studies microarrays are used to take a snapshot of the transcription profile for a given strain or mutant under a specific growth condition relative to a standard condition. The premise in these experiments is that expression correlates with function. When numerous mutants or growth conditions are being investigated, transcriptional profiling can be labor-intensive and expensive. Furthermore, gene expression levels and gene product requirements do not always correlate. In other words, a gene that is highly expressed under a given condition is not necessarily required for growth under that condition (6).
A more direct method for determining gene function is to assess the role of the gene product in the fitness of the organism by characterizing the phenotypes of knockout mutants under various growth conditions. Efforts to study phenotypes caused by transposon mutations have yielded many global techniques, including in vivo expression technology (13), signature-tagged mutagenesis (8), genetic footprinting (14), transposon site hybridization (10), and similar methods (1). This type of high-throughput approach has the potential to address the fitness of individual unknown bacterial mutants in a pool under certain conditions. This provides an initial starting place for choosing interesting genes to study individually in greater detail. Furthermore, this approach could also be designed to use known mutant pools to probe the nutrient make-up of uncharacterized environments.
Two previously described methods (1, 10) involved the use of microarrays for analyzing large libraries of mutants under defined growth conditions. The probes spotted on the arrays in these studies were either complete (10) or partial (1) open reading frames (ORFs) and represented only a portion of the genome. Thus, the microarrays used in these studies were not optimized to detect and monitor transposon insertions. Using a high-density oligonucleotide microarray with full genome coverage could provide higher resolution and could allow detection of transposon insertions throughout the entire genome.
This study was initiated to develop a high-definition parallel approach for studying pools of transposon insertion mutants generated in the model organism Escherichia coli K-12. Although this organism has been extensively studied, ∼13% of the genes in E. coli are still characterized as having unknown functions (11). Furthermore, even with functional assignments, it is not always clear which gene products are required under what conditions. Here we present a straightforward method for analyzing the growth potential of individual mutants within a population. Using this approach, we defined more than 300 independent insertions that identify regions in the E. coli K-12 chromosome that are dispensable for growth in rich lab medium. Compared to the previous global studies that we are aware of, 15 of these insertions define regions in which no previous insertions have been identified, and 12 represent genes which were previously deemed essential.
(This work was presented in an abbreviated form as a poster at the 103rd General Meeting of the American Society for Microbiology, 2003.)
MATERIALS AND METHODS
Media and reagents.
Bacterial cultures were grown in either Luria-Bertani (LB) broth or M9 minimal medium supplemented with 0.2% glucose (and 1.5% agar when appropriate) (9). All cultures were grown at 37°C. SOC medium (9) was used for recovery of cells following electroporations involved in cloning. LB medium was used to recover cells following electroporations involving mutagenesis with transposon-transposase complexes. When necessary, kanamycin (Sigma, St. Louis, Mo.) was added at a concentration of 40 μg/ml. Restriction enzymes and T4 ligase were purchased from New England Biolabs (Beverly, Mass.) and Promega (Madison, Wis.). Native Pfu DNA polymerase was purchased from Stratagene (La Jolla, Calif.). All oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, Iowa). DNA sequencing was performed by using standard dye terminator chemistry. Phase lock gel tubes were purchased from Eppendorf (Westbury, N.Y.). Biotin-16-UTP was purchased from Enzo Diagnostics (Farmingdale, N.Y.). YM10 Microcon concentration columns were purchased from Amicon/Millipore (Billerica, Mass.). An Ampliscribe T7 high-yield in vitro transcription kit, Kan2-T7 transposon template DNA, the EZ::TN pMOD<MCS> transposon construction vector, and a MasterPure DNA isolation kit were generous gifts from Epicentre DNA Technologies Inc. (Madison, Wis.). QIAGEN (Valencia, Calif.) products were used for plasmid purification and DNA gel extraction. Hyperactive EK54/MA56/LP372 transposase was purified as previously described (2).
Bacterial strains and plasmids.
E. coli DH5α was used for all cloning manipulations, and E. coli K-12 strain MG1655 was used for all mutagenesis experiments. Construction of Tn5-KMW1 (Fig. 1A) was performed stepwise with the EZ::TN pMOD<MCS> transposon construction vector. Initially, pKW3 was constructed by PCR amplifying the Kan2-T7 transposon template DNA (Epicentre Inc.) with end-modified primers T7/KAN2-For1 (5′-GTCCAGTCGACCTCTGATGTTACATTGCAC [the underlining indicates an SalI restriction site]) and T7/KAN2-Rev1 (5′-GTCCAGCATGCGTCTCCCTATAGTGAGTCG [the underlining indicates an SphI restriction site]) and cloning the resulting 1.2-kb Kan2-T7-containing fragment into the SalI and SphI sites of pMOD<MCS>. Next, pKW4 was constructed by ligating a 600-bp oriV-containing PCR fragment that was amplified from pJW436 (a generous gift from W. Szybalski) with modified primers oriV-For2 (5′-GTCCAGAGCTCGGCCGCCGGCGTTGTGGATAC [the underlining indicates an SacI restriction site]) and oriV-Rev2 (5′-GTCCAGAATTCCCTATAGTGAGTCGTATTAATCCCCGGGAGGGTTCGAGAAG [the underlining indicates an EcoRI restriction site, and the boldface type indicates the T7 promoter sequence]) into pKW3. Finally, pKW6 was constructed by amplifying the 2.5-kb araC-pBAD-trfA203 fragment from pJW436 with primers araC-For1 (5′-GTCCAGTCGACCGCAATGCTTGCATAATGTGG [the underlining indicates an SalI restriction site]) and araC-Rev1 (5′-GTCCAGGTACCACACTTGCATCGGATGCAGC [the underlining indicates a KpnI restriction site]).
FIG. 1.
(A) Salient features of Tn5-KMW1. The kanamycin resistance gene (Kan), divergent T7 promoters, and 19-bp inverted repeats or Tn5 transposase binding sites (mosaic ends) are shown. (Not shown are oriV and its trfA regulation system under arabinose control. These features were included on the transposon as a backup to allow plasmid libraries to be isolated following mutagenesis. This plasmid library backup system was not needed, and these extra elements on the transposon do not affect the work that is described here.) (B) T7 in vitro transcription labeling scheme. Biotin-16-UTP-labeled transcripts are generated by first harvesting chromosomal DNA from a mixed population of mutants. The DNA is digested and used as a template for runoff T7 RNA polymerase-driven in vitro transcription. DNA is removed by DNase I digestion. RNA is fragmented and hybridized to oligonucleotide microarrays. Tn5 is indicated by a grey box; the divergent arrows represent T7 promoters. The wavy arrows represent in vitro RNA transcripts.
Transposon-transposase complex formation.
Transposon-transposase complexes were formed in vitro as previously described (7). Tn5-KMW1 was released from pKW6 by PvuII digestion, gel purified, and mixed (0.5 μg of DNA per reaction mixture) with purified hyperactive Tn5 transposase (0.1 to 0.5 μg per reaction mixture) in binding buffer (25 mM Tris-acetate [pH 7.5], 100 mM potassium glutamate) and water to obtain a final volume of 40 μl. The reaction mixtures were incubated at 37°C for 1.5 h, checked for shifts by agarose gel electrophoresis, and subsequently dialyzed against 10% glycerol-5 mM Tris-acetate (pH 7) by using 0.05-μm-pore-size filter disks.
Tn5 mutagenesis.
Transposon-transposase complexes (1 μl) were electroporated by using standard procedures (9) into 50 μl of electrocompetent (efficiency, >109 CFU/μg) MG1655 cells, recovered in LB medium at 37°C with shaking at 250 rpm for 2 h, and plated on selective medium. Dilution plating was done to determine the total numbers of transposon insertion mutants recovered. A total of 2,068 randomly selected mutants were picked into 96-well, sterile, flat-bottom, polystyrene dishes and grown overnight in LB medium containing kanamycin. A medium control and a wild-type (MG1655) control were included in every 96-well dish. Mutants were stocked in 15% glycerol at −80°C in polypropylene dishes with foil covers.
Southern hybridization analysis.
Chromosomal DNA of 14 randomly selected mutants was isolated by using a Master Pure DNA isolation kit (Epicentre Inc.). DNA of each mutant was digested with SalI and fractionated by agarose gel electrophoresis. DNA fragments were transferred to a nylon membrane as previously described (9). Probe DNA was generated by PCR amplification of the kanamycin resistance gene from pKW6 (∼1.2 kb) and was subsequently labeled by using a fluorescein-12-dUTP labeling and detection kit obtained from Dupont NEN (Boston, Mass.). The blot was probed and detected as recommended by the manufacturer.
Auxotroph characterization.
Auxotrophs from the reference collection of 2,068 mutants were identified by replica printing in 48- and 96-well formats on glucose M9 minimal agar and LB agar. Mutants that grew on LB agar but failed to grow on glucose M9 minimal agar were classified as auxotrophs. Auxotroph mutants were further characterized by using a series of 11 diagnostic medium pools to identify the probable nutrient requirements, as previously described (4). Individual auxotrophies were verified by supplementing minimal medium with the nutrient(s) predicted by the diagnostic medium studies.
Pilot library construction.
In general, all libraries were constructed by mixing equal volumes of individually grown overnight cultures and either stored immediately after mixing (no further outgrowth) in 15% glycerol at −80°C or used for chromosomal DNA isolation. Specifically for the 50- and 70-mutant pilot libraries, 3-ml overnight cultures started from single colonies were grown at 37°C in LB medium with kanamycin. One-milliliter portions of all the overnight cultures were mixed, and aliquots were frozen in 15% (final concentration) glycerol. To construct the 70-member limit of detection libraries, five overnight cultures were diluted 1:10 and 1:1,000 prior to mixing. Chromosomal DNA was harvested immediately from these libraries. The mutants used in these libraries are available upon request. The upper limit of detection libraries were determined by first replica printing in 96-well format from −80°C stocks into fresh LB medium with kanamycin and growing the cultures overnight at 37°C in a static incubator. Four pools containing 94, 188, 376, and 564 mutants were constructed by pooling bacterial cultures from wells in either plate 1 alone, plates 1 and 2, plates 1 through 4, or plates 1 through 6. Note that each 96-well plate contained one wild-type well and one medium-only control well. Chromosomal DNA was harvested immediately from these mutant pools.
Competitive outgrowth (LB medium versus M9 medium).
To revive the 50-mutant library, 400 μl of a thawed −80°C glycerol stock aliquot was added to 4.5 ml of LB medium without antibiotics and aerated by shaking the preparation for 3 h at 37°C. Following revival of the library, 1:50 dilutions in LB medium and M9 medium supplemented with 0.2% glucose and kanamycin were prepared. Dilution in M9 medium was preceded by three washes in phosphate-buffered saline to eliminate possible nutrient carryover. Cultures were grown with shaking (250 rpm) at 37°C. Optical densities at 600 nm were determined to monitor the cultures through each growth phase. Once the cultures reached the stationary phase, where the optical densities at 600 nm were no longer increasing, cells were harvested for chromosomal DNA isolation and microarray analysis.
Transposon insertion site determination.
Chromosomal insertion locations for a set of 62 mutants were determined by sequencing inverse PCR products. Briefly, genomic DNA was harvested from cells with a MasterPure DNA isolation kit (Epicentre Technologies Inc.), digested with MluI, and self-ligated. By using outward-facing primers homologous to the ends of the Tn5-KMW1 transposon, a PCR was performed with native Pfu DNA polymerase (Stratagene). For each insertion, the primer annealing temperatures were adjusted to ensure adequate product formation, as determined by agarose gel electrophoresis. The resulting PCR products were sequenced, and nucleotide BLAST homology searches against the E. coli K-12 database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/) were performed.
Microarray sample preparation. (i) DNA template.
Chromosomal DNA harvested from the pool of mutants was digested with HincII. The reaction mixtures were then extracted once with phenol-chloroform-isoamyl alcohol (25:24:1) in phase lock gel tubes and ethanol precipitated. The DNA was pelleted, washed once with ice-cold 70% ethanol, and resuspended in diethyl pyrocarbonate (DEPC)-treated water to a concentration of 2 μg/μl.
(ii) In vitro transcription reactions.
An Ampliscribe T7 high-yield in vitro transcription kit (Epicentre Inc.) was used with the following modifications to the recommended protocol. At room temperature, 10 μg of digested chromosomal DNA template was mixed with (final concentrations) 5 mM ATP, 5 mM GTP, 5 mM CTP, 1.67 mM unlabeled UTP, 3.33 mM biotin-16-UTP, 8.3 mM dithiothreitol, 3 μl of T7 RNA polymerase, and enough DEPC-treated water to bring the volume to 30 μl. The reaction mixtures were incubated at 37°C for ∼16 h. Then the reaction mixtures were mixed with 1 to 2 μl of DNase I and incubated at 37°C for 15 min. Free nucleotides were removed, and samples were concentrated by washing them once in 470 μl of DEPC-treated water with YM10 Microcon columns to obtain a final volume of 10 to 20 μl. RNA was quantified by using A260/A280 readings.
Chromosomal DNA end labeling.
Chromosomal DNA was fragmented to obtain ∼200-bp fragments by digesting it with 0.02 U of DNase I per μg of DNA at 37°C for ∼7 min. The DNase I was inactivated by heating the preparation at 95°C for 10 min. Reaction buffer was exchanged for water and concentrated by using YM10 Microcon filtration columns to obtain a volume of ∼20 μl. The fragmented DNA was end labeled by using terminal transferase and biotin-6-ddATP (Promega). YM10 Microcon columns were used to remove free biotin-6-ddATP from the transferase reaction mixtures.
Chip design and synthesis.
Custom oligonucleotide micorarrays were designed (NimbleGen Systems Inc.) to contain 24-mer probes spaced, on average, every 50 bp throughout the E. coli K-12 strain MG1655 genome. Nonunique regions, such as insertion elements, were not included on the arrays to minimize cross-hybridization between these regions. Probes were designed to represent both DNA strands equally (top strand, 5′ to 3′; bottom strand, 3′ to 5′) and to be nonbiased toward open reading frames and/or intergenic regions. Chips were synthesized by using a maskless array technology (12).
Hybridization, staining, and detection.
All microarray hybridization experiments were performed in the Genome Expression Center facility (http://www.gcow.wisc.edu/Gec/index.htm) at the University of Wisconsin-Madison. Chips were prehybridized and hybridized as recommended by NimbleGen Systems Inc. Prehybridizations were done at 45°C for 15 min. Biotin-labeled RNA transcripts were fragmented in 5× fragmentation buffer (200 mM Tris-acetate [pH 8.1], 500 mM potassium acetate, 150 mM magnesium acetate) at 95°C for 10 min. RNA samples (5 to 10 μg per chip) were hybridized to prehybridized NimbleGen arrays for 16 h at 45°C on a rotating wheel in a hybridization oven. Chromosomal DNA hybridizations were done at 42°C rather than 45°C. Following hybridization, samples were removed, and arrays were washed in a series of nonstringent and stringent wash buffers. A streptavidin-Cy3 conjugate was used to stain each microarray for 25 min at room temperature in the dark. The arrays were scanned by using an Axon 4000b scanner with the GenePix Pro software. Signal intensity data were extracted from tiff images by using the NimbleGen extraction software.
Data processing.
Extracted microarray data were analyzed by using custom-designed Perl scripts and GenVision (DNASTAR). All data were stored in a MySQL database. Perl scripts were used to select three or more consecutive probes on the same strand with signal intensities that were at least three times the average empty-feature background signal intensity. The average empty-feature background signal was determined by averaging signal intensities from blank chip features containing no DNA. Additionally, Perl scripts were used to format data files for use in GenVision and Spotfire Pro. Further data analysis was performed by constructing histograms (in genome order) for the E. coli genes, array signal intensities, expected transposon insertion sites, and restriction enzyme cut sites by using the GenVision software. Visual analysis of these plots allowed clear identification of positive signal intensities relative to the overall chip background.
RESULTS
Transposon mutagenesis with a new Tn5 derivative.
A modified version of Tn5 was constructed that contained a kanamycin resistance gene and two divergent T7 promoters flanked by 19-bp inverted repeat mosaic end sequences. T7 promoters were included to permit transcription of neighboring DNA following a transposition event. Transcripts generated from the transposon's left side are homologous to the top strand (5′ to 3′) of DNA, and transcripts generated from the right side are homologous to the bottom strand (3′ to 5′) of DNA, regardless of the transposon's orientation. The pertinent features of the Tn5-KMW1 transposon are shown in Fig. 1A.
By using a previously described Tn5 mutagenesis strategy (7), TN5-KMW1 transposon-transposase complexes were formed and electroporated into wild-type E. coli K-12 strain MG1655. On average, more than 105 viable transposon insertion mutants per ml of transformed cell culture were recovered. From a single mutagenesis, 2,068 mutants were randomly selected, picked into 96-well dishes, and grown overnight in LB broth in static cultures. The mutants were stored in a 96-well format at −80°C in 15% (final concentration) glycerol as a Tn5 reference collection. Replica printing in a 96-well format was used to screen the library of 2,068 mutants for growth on glucose M9 minimal medium and LB medium. Twenty-seven auxotrophs were identified based on their inability to grow on M9 medium while they were viable on LB medium.
Southern hybridization was used to verify that mutagenesis resulted in single transposon insertions. A 1.2-kb PCR fragment of TN5-KMW1 containing the kanamycin resistance gene was used to probe an SalI digest of chromosomal DNA isolated from 14 randomly selected mutants. Fourteen different bands were detected, indicating that each mutant contained a single insertion and that there were no obvious insertion hotspots (data not shown). The genomic locations of the transposon insertions of 66 mutants, including 19 auxotrophs, were determined by inverse PCR and sequencing (data not shown). The transposon insertions were unique, with two exceptions. In two cases, strains contained identical insertions (carB, wecG) and were considered siblings.
Parallel analysis of Tn5 insertion mutants. (i) Transposon pilot libraries.
To begin development of a high-definition microarray approach for analyzing pools of Tn5 insertion mutants, labeling and hybridization techniques were optimized with pilot libraries of insertion mutants. Pilot libraries were generated with mutants selected from the Tn5 reference collection. Mutants with sequenced insertion sites were chosen to allow better interpretation of the microarray hybridization results. To physically generate these pilot libraries, mutants were grown individually overnight in LB medium, pooled, and frozen as described in Materials and Methods.
(ii) Tn5 insertion labeling.
Labeled runoff transcripts of the DNA flanking each transposon insertion mutant in a given Tn5 mutant pool were generated simultaneously by using a modified version of two previously described methods (1, 10). The most significant changes in these methods were that DNA amplification steps were eliminated and the number of DNA-RNA manipulations that were performed between mutagenesis and microarray analysis was limited in an effort to minimize biases in our results. Figure 1B shows a schematic diagram of this modified labeling method. Briefly, chromosomal DNA was harvested from mutant pools and digested with HincII, a six-base blunt end restriction enzyme with an average fragment size of 1,140 bp for the E. coli K-12 genome. Following cleanup and concentration of the digested DNA, biotin-labeled RNA transcripts were generated from the digested pilot library DNA by using T7 RNA polymerase. On average, 30 to 50 μg of labeled RNA was generated in each in vitro transcription reaction. Following removal of the template DNA with DNase I and cleanup of unincorporated nucleotides, the RNA was fragmented with magnesium and heat and hybridized to custom-designed DNA oligonucleotide microarrays (see Materials and Methods).
Microarray design and data analysis.
Initial hybridization results obtained by using both whole-ORF spotted microarrays and commercially available sense strand Affymetrix GeneChip oligonucleotide microarrays demonstrated that only ∼50% of the expected Tn5 insertions could be detected (data not shown). Commercially available microarrays are typically designed for gene expression and are therefore biased toward ORF-containing DNA sequences. When these studies were initiated, microarrays that fully represented the entire genome sequence of E. coli K-12 were not commercially available. To investigate libraries of Tn5 insertions, a high-resolution, custom-designed, full-coverage oligonucleotide microarray was designed and synthesized by NimbleGen Systems Inc. by using a maskless array synthesis technology (12).
The microarrays were designed to contain unique 24-mer probes spaced approximately every 50 bp throughout the genome, representing both the top and bottom strands of the E. coli chromosome, irrespective of ORFs or intergenic regions. To minimize cross-hybridization, probes were designed to be unique and represent nonrepeated sequences of the E. coli K-12 strain MG1655 genome. Mismatch probes were not included on these microarrays, which allowed room for just over 195,000 unique probes in addition to controls. The probe locations were randomized across the array surface.
Preliminary experiments with pilot libraries were performed to verify that the mutants in a pool could all be detected prior to any further outgrowth. The probe signal intensities surrounding each insertion were analyzed by using sequenced transposon insertion locations and local restriction site patterns as a guide. For some insertions, restriction enzyme sites were located very close to the actual transposon insertion site, which severely limited the length of the runoff transcript. In these cases, positive signals were expected for only one side of the insertion. Positive detection of an insertion was initially defined as three or more consecutive probes with signal intensities that were three times the average background signal intensity from at least one side of the transposon insertion. In multiple experiments, approximately 97% of the mutants in each library tested could be detected (data not shown).
Although numerical analysis methods were initially used, analyzing the data graphically with the GeneVision software (Fig. 2, 3, and 4) proved to be reliable for detecting the relative insertion locations. Histogram files of the extracted microarray data were generated by using custom-designed Perl scripts. Specifically, individual probe signals were graphed as bars in which the width represented the oligonucleotide's length (24 nucleotides) and the height corresponded to the extracted signal intensity. The signal intensities for probes representing the bottom strand of DNA were multiplied by −1 for ease of viewing the data when they were graphed. The position of a probe along the x axis corresponded to the position in the E. coli genome. GeneVision permitted the array data, the gene arrangement in the E. coli genome, the restriction sites, and the known transposon insertion sites to be graphed with respect to genome position. Graphing the data in this way allowed rapid identification of the Tn5 insertion locations.
FIG. 2.
LB medium versus M9 medium library outgrowth. (A) Auxotroph analysis. Array data for regions surrounding insertions in pdxB and aroC are shown for outgrowth in both LB medium and M9 medium. Mutants were detected following outgrowth in LB medium and, as expected, were not detected following outgrowth in M9 medium. (B) Nonauxotroph analysis. Array data for regions surrounding the insertion in yhfK are shown. As expected, this mutant was detected following outgrowth in both LB medium and M9 medium.
FIG. 3.
Tn5 insertion mapping. (A) Schematic diagram for determining the genomic locations of Tn5 insertions by using microarray data. The genomic position is determined by taking the difference between probe 2 (P2) and probe 1 (P1). This provides a genomic range for the actual Tn5 insertion site. P1 represents the last positive top-strand probe (5′ to 3′), and P2 represents the first positive bottom-strand probe (3′ to 5′). (B) Mapping unsequenced transposon insertion locations in the ilv region of E. coli. Microarray data for approximately 14 kb of the E. coli genome are shown. Microarray signals for regions surrounding two insertions designated i1 and i2 were expected based on previous sequencing efforts. These insertions correspond to Tn5 insertions in ilvG_1 and ilvD. Additional microarray probe signals in ilvA and ilvC suggest possible insertions designated i3* and i4*. The genome positions of such inserts were mapped by using the method shown in panel A.
FIG. 4.
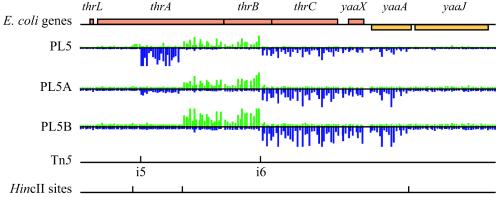
Limit of detection. Probe data for regions surrounding the insertions in thrA (i5) and thrB (i6) are shown. The thrA mutant was diluted 1:10 and 1:1,000 in PL5A and PL5B, respectively, while the thrB mutant was kept constant at a 1:1 ratio in each library tested. The signal from thrA is noticeably decreased in PL5A and completely absent in PL5B.
Variability in the signal intensities for all stretches of positive probes was observed. To determine if this variability was due to hybridization differences between oligonucleotides, chromosomal DNA hybridizations were performed by using wild-type DNA end labeled with biotin. The probe signal intensities from the chromosomal hybridizations showed the same type of variability that was seen with the labeled RNA hybridizations (data not shown). Furthermore, this probe signal variability was reproducible between arrays for both DNA and RNA hybridizations. These data suggest that the observed probe signal variability was due to differences in hybridization efficiency for individual oligonucleotide probes. This variability did not affect the ability to detect positive probe signals surrounding each insertion.
Auxotrophs are lost following competitive growth in M9 minimal medium.
The consequences of growing a pilot library comprised of 50% auxotrophs in both LB and glucose M9 minimal medium were examined. To create this pilot library, 50 mutants from the Tn5 reference collection were selected. Mutants with both sequenced insertion sites (25 nonauxotrophs and 17 auxotrophs) and unsequenced insertion sites (eight auxotrophs) were chosen. Mutants with unsequenced transposon insertion sites were included to determine if new transposon site locations could be determined solely from the microarray hybridization data.
To verify that each mutant could be detected in the library prior to competitive outgrowth, microarray hybridization was performed immediately following mutant pooling. All 42 sequenced insertions were detected in addition to nine other signals, which were presumed to correspond to the eight auxotrophs with uncharacterized insertion locations. For competitive outgrowth, the library was revived from −80°C by growing the organisms in LB medium at 37°C for 3 h. Cells were pelleted, washed with phosphate-buffered saline, and diluted 1:50 in each type of medium. The cultures were grown to the stationary phase, as determined by no further increase in the A600. By using A600 values obtained throughout the outgrowth of these cultures, the total numbers of generations for the cultures were determined to be 13 and 9 for the LB and M9 minimal-medium cultures, respectively. Chromosomal DNA was harvested from cells grown in each type of medium. Biotin-labeled RNA from each sample was generated in vitro, as described above, and hybridized to separate microarrays.
Following competitive outgrowth in M9 minimal medium, none of 25 auxotrophs and all 25 nonauxotrophs were detectable by microarray hybridization. Microarray analysis of the competitive outgrowth in LB medium revealed that 23 of 25 auxotrophs and all 25 nonauxotrophs were detectable. Three insertions, icdA (sequenced), cmr (not sequenced), and yhjB (not sequenced), that were previously detected by microarray hybridization with samples analyzed prior to competitive outgrowth were not detected following outgrowth in LB medium.
Figure 2 shows two representative sections of the E. coli genome that demonstrate the pilot library outgrowth results. Figure 2A shows a region of the E. coli genome in which three previously sequenced insertions are located. Following outgrowth in LB medium, strong probe signals were detected for the insertions in pdxB and aroC, although a distinction between the signals for the neighboring aroC insertions could not be made. The signal was lost for these insertions following outgrowth in glucose M9 minimal medium, as expected based on the inability of the organisms to grow on glucose M9 minimal medium. Correspondingly, Fig. 2B shows the probe data for the genome region surrounding the insertion in yhfK for both LB and M9 medium outgrowth. The insertion in yhfK was detected equally under the two conditions, as expected from the ability of the mutant to grow on both LB and M9 media.
Transposon insertion site mapping.
As mentioned above, eight auxotrophs with previously uncharacterized insertion locations were present in the 50-member pilot library. Additional positive signals were detected in the following nine genes: cmr, yhjB, metC, ilvA, ilvC, argE, serB, pheA, and leuB (Table 1). These findings suggested possible locations for the eight unsequenced insertion mutants plus one additional insertion. This additional insert was likely due to one mutant with two insertions. Insertion locations were mapped by calculating the difference between the genome position of the first positive bottom-strand probe and the genome position of the last positive top-strand probe (Fig. 3A). The average mapping distance for the nine additional signals was 50 bp. The majority of these newly characterized insertions were located in amino acid biosynthetic genes, which was expected based on the glucose M9 auxotroph phenotypes of the mutants chosen. These auxotrophs were further characterized by using diagnostic medium pools (4) to identify specific nutrient requirements. Nutrient requirements were correlated to five of the auxotrophs based on the insertion locations identified (Table 1). Microarray hybridization data for the ilv region of the E. coli genome are shown in Fig. 3B. These data predict locations for potential transposon insertions in ilvA and ilvC and demonstrate that both sequenced (ilvG_1 and ilvD) and unsequenced (ilvA and ilvC) transposon insertion sites can be determined by this method.
TABLE 1.
Auxotroph mapping and characterization
| Gene | b no.a | Top strandb | Bottom strandc | Distance (bp)d | Mutant | Nutritional deficiency |
|---|---|---|---|---|---|---|
| leuB | b0073 | 81170 | 81219 | 49 | 020:G2 | Leucine |
| pheA | b2599 | 2735838 | 2735872 | 34 | 006:A6 | Phenylalanine |
| ilvA | b3772 | 3953883 | 3953926 | 43 | 007:A2 | Isoleucinc-valine |
| ilvC | b3774 | 3956841 | 3956887 | 46 | 007:B1 | Isoleucinc-valine |
| serB | b4388 | 4622451 | 4622498 | 47 | 015:A5 | Serine |
| cmr | b0842 | 883043 | 883107 | 64 | NDe | ND |
| melC | b3008 | 3150839 | 3150859 | 20 | ND | ND |
| yhjB | b3520 | 3669108 | 3669161 | 53 | ND | ND |
| argE | b3957 | 4151699 | 4151793 | 94 | ND | ND |
Number given to the open reading frame of the E. coli K-12 strain MG1655 genome (3).
Nucleotide position of the last positive top-strand probe (5′ to 3′).
Nucleotide position of the first positive bottom-strand probe (3′ to 5′).
Difference between the first positive bottom-strand probe and last positive top-strand probe nucleotide positions.
ND, not determined.
Limit of detection.
To determine the limit of detection for this method (i.e., the point at which outcompeted mutants were no longer detectable), a microarray hybridization analysis of three separate libraries was performed. These libraries were generated by mixing 65 mutants at 1:1 ratios. Five additional mutants were added at a ratio of either 1:1 (PL5), 1:10 (PL5A), or 1:1,000 (PL5B) relative to each of the other 65 mutants. These five mutants were chosen because they were reproducibly detected in our initial hybridization analyses. The diluted mutants had insertions in thrA, caiC, ycaM, aldB, and ilvG_1. Analysis of the signals from the insertions in the 1:10 library showed noticeable decreases in the signals for all five diluted mutants compared to the library with mutants at 1:1 ratios. All other mutants were detected equally in the three libraries. In the 1:1,000 library only aldB was detectable by microarray analysis. In this case (aldB), a decreased level of signal was detected, but this was attributed to a readthrough signal from the neighboring upstream insertion in yiaV. As a representation of these results, the probe data surrounding the insertions in thrA (diluted) and thrB (undiluted) for all three libraries are shown in Fig. 4.
Large library analysis.
To test the upper limit of detection for this method, four pools consisting of 94, 188, 376, and 564 mutants were constructed and tested for detection by microarray hybridization. Following overnight growth in the 96-well format in LB medium with kanamycin, the mutants were pooled and analyzed with no additional competitive outgrowth. The smallest pool (94 mutants) was constructed by mixing the cultures from all wells of plate 1. All subsequent pools were constructed in an additive manner, in which mutants from plates 1 and 2 (188 mutants), plates 1 through 4 (376 mutants), and plates 1 through 6 (564 mutants) were mixed together. No further growth was allowed following pooling of the mutants. Microarray analysis of labeled RNA transcripts generated from chromosomal DNA isolated from each pool was performed as described above.
A summary of the results of the microarray analysis of these four pools is shown in Table 2. Pools consisting of 94, 188, 376, and 564 mutants gave 95, 150, 274, and 333 signals, respectively. A total of 340 different signals were detected in this analysis. With a few exceptions, the insertions detected correlated with nonessential gene designations from collections of systematic gene deletions (http://www.genome.wisc.edu/) and other previously published data sets for E. coli K-12 (5; http://www.shigen.nig.ac.jp/ecoli/pec/index.jsp). Our best approximation was that insertions were detected in 12 genes previously described as essential (Table 3) and 15 insertions were detected in genes for which the collections mentioned above have no current gene classifications (Table 4). These results both support and complement the previous global knockout studies that have been performed.
TABLE 2.
Upper limit library analysis
| No. of mutants pooled | No. of signals detected | % Detected |
|---|---|---|
| 94 | 95 | 101.1a |
| 188 | 150 | 79.8 |
| 376 | 274 | 72.9 |
| 564 | 333 | 59.0 |
The value greater than 100% indicates a potential false positive or double insertion.
TABLE 3.
Previously classified essential genes
| b no. | Gene | Global studya |
|---|---|---|
| b1238 | tdk | A |
| b1269 | yciL | A |
| b1359 | ydaU | A |
| b1463 | nhoA | A |
| b1471 | yddk | A |
| b1765 | ydjA | A |
| b1874 | cutC | A |
| b1961 | dcm | A |
| b2163 | yeiL | A |
| b3134 | agaW | A |
| b3219 | yhcF | A |
| b3863 | polA | B |
Previously described as essential by Gerdes et al. (5) (A) or at http://www.shigen.nig.ac.jp/ecoli/pec/index.jsp (B).
TABLE 4.
New nonessential gene classifications
| b no. | Gene |
|---|---|
| b0363 | yaiP |
| b0382 | yaiB |
| b1044 | ymdA |
| b1725 | yniA |
| b2372 | yfdV |
| b2649 | ypjB |
| b2656 | b2656 |
| b2848 | yqeJ |
| b2851 | ygeG |
| b2852 | ygeH |
| b3046 | yqiG |
| b3050 | yqiJ |
| b3269 | yhdX |
| b3471 | yhhQ |
| b3897 | frvR |
DISCUSSION
It is informative to know an organism's full genome sequence; however, understanding which genes are required for growth and under what conditions they are required is critical to understanding the organism as a whole. While many genetic techniques are used to study the growth effects of individual mutations, the development of high-throughput technologies allows multiple mutations to be analyzed concurrently. In the technique described in this paper DNA microarrays are used to detect individual transposon insertion mutants within a population, and thus this technique can track multiple mutants in parallel. The use of a high-density full genome microarray allows transposon insertions to be mapped to within 50 bp (average) of their genomic locations and avoids all amplification steps associated with sample labeling, reducing the potential for bias.
In vivo Tn5 mutagenesis (7) was used to generate more than 105 E. coli K-12 viable mutants. From this mutagenesis, a reference collection of 2,068 transposon mutants was obtained. This Tn5 mutagenesis strategy eliminated the need for constructing suicide vectors or using more complex phage delivery systems for performing transposon mutagenesis. The mutagenesis resulted in only two observed sibling pairs. These results and further optimization performed during this work suggested that outgrowth following electroporation should be reduced to 1 h to avoid siblings.
As a proof of method, a 50-member mutant pool containing 50% auxotrophs was competitively grown in both a rich medium (LB medium) and a minimal medium (glucose M9 medium). All 25 known M9 minimal medium auxotrophs in the pool were not detectable following nine generations of growth in M9 minimal medium. Three insertions, icdA (sequenced), cmr (not sequenced), and yhjB (not sequenced), were not detectable following 13 generations of outgrowth in LB medium. The failure to detect these three insertions may have been due to freeze-thaw sensitivity or to underrepresentation due to slow growth compared to the other mutants. All 25 nonauxotrophs were detectable following outgrowth in both LB and M9 minimal media, as expected.
Microarray hybridization results further revealed that the mutations of individual insertion mutants could be mapped to within 50 bp (average) of their chromosomal locations, as previously determined by sequencing. To this end, the insertion locations of nine previously uncharacterized insertions were mapped by using mircroarray hybridization results. Eight of these signals corresponded to the eight uncharacterized glucose M9 medium auxotrophs in the 50-member pilot library. The additional insertion likely corresponded to a double insertion in one of the mutants in the pilot library. It is possible that the insertions identified in cmr and yhjB were in the same mutant strain, as both insertions were previously not identified by sequencing and both were not detectable following outgrowth in LB medium. Diagnostic medium pools (4) were used to characterize the nutrient deficiencies of five individual mutants. The nutrient deficiencies identified correlated with the gene knockouts identified by microarray analysis.
Overall, the proof of method results suggest that this technique could be used to define general nutrient compositions of various growth environments. For instance, a pool of representative auxotrophs could be used to determine if an environment is limiting for a given amino acid or cofactor. Applying this technique to a pathogenesis study may be useful in determining whether mutants that fail to grow in a particular animal model are affected by the lack of nutrients in the given environment (i.e., lung, spleen, or bladder) or whether they fail to grow because a virulence factor required for setting up and maintaining the infection has been knocked out. The additional benefits of incorporating this microarray screening technique into global pathogenesis methods, such as signature-tagged mutagenesis (8), include the fact that many of the associated time-consuming steps can be alleviated. Some of these steps include individual transposon construction and gene identification for insertions in mutants that fail to survive the imposed in vivo growth selection.
In an effort to determine the upper and lower limits of detection for this technique, two separate experiments were performed. To study the lower limit of detection, it was determined that mutants diluted 1:1,000 in general were no longer detectable by microarray hybridization. To begin studying the upper limit of detection, it was shown that 95 microarray signals could be detected from a pool of 94 mutants. The additional signal represented one false-positive which may have been a result of one mutant with two insertions. For the largest library (564 mutants), only 333 signals (∼59%) were detected by microarray hybridization.
There are several potential reasons for the decreased number of signals found in the larger pools, including the dilution of T7 promoters that are present for the in vitro transcription reaction. As more mutants are combined, the relative amount of each T7 promoter is decreased, which in turn decreases the number of biotin-labeled RNA transcripts produced. More signals might be detected if the DNA template used in the in vitro transcription reactions could be enriched for DNA fragments containing the T7 promoters (i.e., transposon ends plus flanking chromosomal DNA). Alternatively, amplification methods could be used, with the understanding that biases might be introduced.
In addition to the sensitivity information described above, the 340 signals detected provide information for more than 300 Tn5 insertion mutants in standard LB medium. By comparing our results to other global studies performed with E. coli K-12, insertions were identified in 12 genes that were previously identified as essential (Table 3) and in 15 genes for which no previous insertions have been reported (Table 4). The 12 genes previously described as essential had been classified based on a failure to detect Tn5 insertions in these genes (5). This previous result could simply represent a chance failure to obtain an insertion in these genes or a problem with Tn5 target bias. Therefore, the data presented here are not inconsistent with the data of Gerdes et al. (5) but rather extend the study of these workers.
It is worth noting that the microarray data revealed an insertion in or near ribB, a gene previously defined as essential in all three previous global studies. The results are ambiguous for the exact location of the Tn5 insertion, and therefore it is not clear if the insertion is located just upstream of or just inside the 5′ start for ribB. It is possible that the Kan2/T7 transposon contains putative E. coli-type promoters at the ends and is capable of initiating transcription of neighboring genes. This has been suggested by Gerdes et al. (5) for the EZ::Tn<Kan2> transposon, which is identical to the kanamycin-containing portion of the transposon used in this study. Therefore, it may be that a functional RibB protein is produced regardless of the chromosomal Tn5 insertion location.
The data described above suggest that the high-definition parallel approach can be used to effectively screen libraries of 96 mutants and could be a convenient method for analyzing growth phenotypes of transposon mutant pools. The Tn5 transposition system that was used can be easily modified and allows any DNA feature to be combined into a transposon in vitro and delivered randomly into genomes of any organism that can be made electrocompetent. This alone provides a means for generating libraries of transposon mutants in organisms for which extensive genetic techniques may not be readily available. We anticipate that this parallel strategy will greatly benefit current molecular genetic techniques when high-density oligonucleotide DNA microarrays are used to monitor pools of mutants. This technique has the potential not only to facilitate screening of large numbers of mutants but also to define key nutritional components of various environments and/or media. As new sequences become available, parallel approaches such as the one described here should enable researchers to quickly investigate gene functions in organisms for which little more than the sequence is known.
Acknowledgments
We thank Jeremy Glasner for his help with utilizing GenVision (DNASTAR Inc.) for visualizing microarray data and for countless hours of discussion in the early stages of this project. We thank Todd Richmond (NimbleGen Systems Inc.) for the design of the MG1655 microarray chip. We also thank D. Downs and M. Steiniger-White for their critical reviews of the manuscript. We further thank all members of the Reznikoff lab for their thoughtful insights and discussions of this work. We especially thank Deb Hug for medium preparation.
This work was funded by NSF grant MCB0084089. Microarrays were provided by NimbleGen Systems Inc. under grant NIH-SBIR 2 R44HG002193-02.
REFERENCES
- 1.Badarinarayana, V., P. W. Estep III, J. Shendure, J. Edwards, S. Tavazoie, F. Lam, and G. M. Church. 2001. Selection analyses of insertional mutants using subgenic-resolution arrays. Nat. Biotechnol. 19:1060-1065. [DOI] [PubMed] [Google Scholar]
- 2.Bhasin, A., I. Y. Goryshin, and W. S. Reznikoff. 1999. Hairpin formation in Tn5 transposition. J. Biol. Chem. 274:37021-37029. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Davis, R. W., D. Botstein, and J. R. Roth. 1980. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 5.Gerdes, S. Y., M. D. Scholle, J. W. Campbell, G. Balazsi, E. Ravasz, M. D. Daugherty, A. L. Somera, N. C. Kyrpides, I. Anderson, M. S. Gelfand, A. Bhattacharya, V. Kapatral, M. D'Souza, M. V. Baev, Y. Grechkin, F. Mseeh, M. Y. Fonstein, R. Overbeek, A. L. Barabasi, Z. N. Oltvai, and A. L. Osterman. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185:5673-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 7.Goryshin, I. Y., J. Jendrisak, L. M. Hoffman, R. Meis, and W. S. Reznikoff. 2000. Insertional transposon mutagenesis by electroporation of released TN5 transposition complexes. Nat. Biotechnol. 18:97-100. [DOI] [PubMed] [Google Scholar]
- 8.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 9.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. USA 98:12712-12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serres, M. H., S. Goswami, and M. Riley. 2004. GenProtEC: an updated and improved analysis of functions of Escherichia coli K-12 proteins. Nucleic Acids Res. 32(Database issue):D300-D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh-Gasson, S., R. D. Green, Y. Yue, C. Nelson, F. Blattner, M. R. Sussman, and F. Cerrina. 1999. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 17:974-978. [DOI] [PubMed] [Google Scholar]
- 13.Slauch, J. M., M. J. Mahan, and J. J. Mekalanos. 1994. In vivo expression technology for selection of bacterial genes specifically induced in host tissues. Methods Enzymol. 235:481-492. [DOI] [PubMed] [Google Scholar]
- 14.Smith, V., D. Botstein, and P. O. Brown. 1995. Genetic footprinting: a genomic strategy for determining a gene's function given its sequence. Proc. Natl. Acad. Sci. USA 92:6479-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]