ABSTRACT
The Study for Monitoring Antimicrobial Resistance Trends (SMART) global surveillance program collected 103,960 isolates of Enterobacteriaceae from 2008 to 2014. From this isolate collection, all ertapenem-nonsusceptible isolates (MIC, ≥1 μg/ml; n = 3,428) and 9,371 isolates of Escherichia coli, Klebsiella pneumoniae, Klebsiella oxytoca, and Proteus mirabilis with an ertapenem-susceptible extended-spectrum-β-lactamase (ESBL)-positive phenotype were assessed for the presence of common carbapenemase genes using a Check-MDR CT101 microarray (Check-Points, Wageningen, the Netherlands) and published multiplex PCR assays. Testing identified 1,493 isolates that harbored a carbapenemase gene (1,485 ertapenem-nonsusceptible isolates and 8 ertapenem-susceptible ESBL-positive isolates) and accounted for 1.4% (1,493/103,960) of all isolates of Enterobacteriaceae. The most frequently identified carbapenemase genes were the KPC (n = 794), OXA-48-like (n = 300), and NDM (n = 290) genes. Carbapenemase genes were most frequently identified in Klebsiella pneumoniae (n = 1,127), Escherichia coli (n = 149), and Enterobacter cloacae (n = 110). Among the carbapenemase-positive isolates, 66.7% (2/3), 37.0% (111/300), 20.0% (8/40), 3.3% (3/92), 2.3% (18/794), and 0% (0/290) of the isolates with genes for GES, OXA-48-like, IMP, VIM, KPC, and NDM, respectively, were susceptible to imipenem (MIC, ≤1 μg/ml). Isolates that tested as susceptible to imipenem were not uncommon among carbapenemase-positive isolates (9.4%, 141/1,493) and most frequently carried OXA-48-like enzymes (78.7%; 111/141); however, overall, these isolates remained rare (0.1%, 141/103,960). The practice of screening clinical isolates of Enterobacteriaceae that test as susceptible to carbapenems in vitro for the presence of carbapenemase genes remains controversial and requires further study.
KEYWORDS: imipenem, carbapenemase, surveillance, global, SMART
INTRODUCTION
Carbapenemase-producing Enterobacteriaceae (CPE) have been identified worldwide. Their propensity for spread and the escalating frequency of their isolation by clinical laboratories have been well documented (1–3). The prevalence of specific carbapenemase enzymes in Enterobacteriaceae continues to demonstrate some geographic dependence (1–3). Currently, the most common carbapenemases detected in Enterobacteriaceae include KPCs, a family of Ambler class A β-lactamases; the zinc-dependent class B metallo-β-lactamases NDM, VIM, and IMP; and OXA-48-like enzymes, members of class D β-lactamases (3). The genes for all of the aforementioned carbapenemases may be plasmid borne, facilitating their spread.
CPE can cause serious local and systemic infections and have been associated with mortality rates as high as 40% in some case series (2, 4). In developed countries, CPE transmission occurs almost exclusively within health care settings, with the main route of spread from patient to patient being via contaminated hands of health care workers (3, 5). Risk factors for infection with CPE include intensive care unit (ICU) stay, mechanical ventilation, indwelling catheters, transplantation, septic shock, exposure to broad-spectrum antimicrobial agents, inadequate initial antimicrobial therapy, and previous colonization with an isolate of CPE (2, 4). Treatment options for carbapenem-resistant Enterobacteriaceae and other Gram-negative bacilli are often limited, as β-lactamase genes are frequently present on the same mobile genetic elements as genes in other classes conferring resistance to antimicrobials (6) and as new antimicrobial agents with novel mechanisms of action have been slow to appear. The ideal therapy for infections attributable to CPE is currently unknown as double-blind, controlled studies have not been performed. Published retrospective observational studies and case reports suggest that combination therapy generates better outcomes and less toxicity than therapy with a single agent such as colistin, tigecycline, amikacin, or fosfomycin, even though in vitro susceptibility testing demonstrates that most pathogens isolated from infected patients are susceptible to these agents (2–4, 7–9). The efficacy of carbapenems in combination with another agent (e.g., colistin, tigecycline, aminoglycosides, fosfomycin) for treating infections due to CPE with low-level carbapenem resistance or susceptibility to carbapenems (e.g., OXA-48-like enzymes) remains undetermined (2, 9, 10). Optimal screening methods to identify patients carrying CPE are also needed, as are strict infection control measures to prevent transmission of CPE from infected individuals.
The detection of the vast majority of isolates of CPE is anticipated to occur using current in vitro antimicrobial susceptibility testing methods in combination with CLSI (11) or EUCAST (12) breakpoints. However, it has been suggested that some CPE may not be identified by clinical laboratory testing methods as they produce MICs that do not exceed susceptible MIC breakpoints for carbapenems (11–13). This potential limitation to antimicrobial susceptibility testing methods may be particularly important for isolates harboring OXA-48-like enzymes (10). Data comparing the prevalence and identities of carbapenemases to the in vitro activity of carbapenems in sizable collections of clinical isolates of Enterobacteriaceae are absent from the published literature. Using data from the Study for Monitoring Antimicrobial Resistance Trends (SMART) global surveillance program from 2008 to 2014, we attempted to identify all isolates that harbored a carbapenemase gene by testing both ertapenem-nonsusceptible isolates (MIC, ≥1 μg/ml) and a random sample of approximately 50% of the available ertapenem-susceptible phenotypically extended-spectrum-β-lactamase (ESBL)-positive isolates of Escherichia coli, Klebsiella pneumoniae, Klebsiella oxytoca, and Proteus mirabilis (>12,000 isolates in total) for the presence of common carbapenemase genes. We then analyzed the in vitro activity of imipenem against these isolates to determine whether imipenem CLSI MIC breakpoints (11) identified all carbapenemase producers and how frequently carbapenemase-positive isolates were imipenem susceptible.
RESULTS
Most (100,495/103,960) isolates of Enterobacteriaceae tested by the SMART global surveillance program from 2008 to 2014 were susceptible to ertapenem (96.7% susceptibility). Of the 3,428 isolates of Enterobacteriaceae that were ertapenem nonsusceptible, 2,338 were collected from intra-abdominal infections (IAI) and 1,090 were collected from urinary tract infections (UTI), comprising 3.4% and 3.1%, respectively, of all isolates from these sources. A total of 1,485 (43.3%) ertapenem-nonsusceptible isolates were carbapenemase positive (i.e., positive for carbapenemase genes by PCR). The prevalence of carbapenemase-positive isolates was highest for isolates of ertapenem-nonsusceptible K. pneumoniae (67.2%; 1,123/1,672), Citrobacter freundii (50.0%; 34/68), E. coli (35.8%; 146/408), and Serratia marcescens (31.0%; 18/58) followed by Enterobacter cloacae (11.6%; 110/951) and Enterobacter aerogenes (9.7%; 11/113) and for 158 isolates (4.6%) of 15 other species. Isolates of E. cloacae (43.3%; n = 841), K. pneumoniae (28.3%; n = 549), E. coli (13.5%; n = 262), E. aerogenes (5.2%; n = 102), Enterobacter asburiae (2.6%; n = 51), and S. marcescens (2.1%; n = 40) and 98 (5.0%) isolates of 20 other species composed the 1,943 ertapenem-nonsusceptible, carbapenemase-negative isolates. Only 8 of 9,371 (0.1%) molecularly characterized ertapenem-susceptible ESBL-positive isolates were carbapenemase positive, with 3 isolates carrying blaVIM (ertapenem MICs of ≤0.03, 0.25, and 0.5 μg/ml), 2 isolates carrying blaOXA-48-like (MICs of 0.12 and 0.25 μg/ml), 1 isolate carrying blaKPC (MIC, 0.5 μg/ml), 1 isolate carrying blaIMP (MIC, 0.5 μg/ml), and 1 isolate carrying blaGES (MIC, 0.5 μg/ml). This resulted in a total of 1,493 carbapenemase-positive Enterobacteriaceae isolates (1.4% of all collected), with the majority (98.3%, 1,468/1,493) harboring a single carbapenemase gene. The proportions of carbapenemase-positive isolates were similar among isolates from IAI (1.4% [930/68,653]) and UTI (1.6% [563/35,307]). A total of 1,943 ertapenem-nonsusceptible isolates did not carry carbapenemases included in the screening algorithm. Elevated ertapenem MICs can result from increased production of ESBLs or AmpC β-lactamases combined with the loss of one or both outer membrane porins (14); though acquired or intrinsic β-lactamases were identified in the vast majority of these isolates, porin profiling was not performed in all cases.
Table 1 depicts the imipenem MIC distributions for all 1,493 carbapenemase-positive isolates. Among the carbapenemase-positive isolates, 9.4% (141/1,493) were imipenem susceptible (8.7% [81/930] of isolates collected from IAI and 10.7% [60/563] of isolates from UTI; data not shown); 78.7% (111/141) and 12.8% (18/141) of these isolates encoded OXA-48-like and KPC enzymes, respectively, with 1 isolate carrying genes for both enzymes. Isolates carrying OXA-48-like enzymes were more likely to be imipenem susceptible than other carbapenemase-positive isolates (P < 0.05): 2.3% of 794 KPC-positive Enterobacteriaceae isolates, 0% of 290 NDM-positive isolates, 3.3% of 92 VIM-positive isolates, and 20.0% of 40 IMP-positive isolates were imipenem susceptible, compared with 37.0% of 300 OXA-48-like-positive isolates. A total of 13.8% (206/1,493) of carbapenemase-positive isolates tested imipenem intermediate (MIC, 2 μg/ml) (11), and the majority of these also carried OXA-48-like (106/206, 51.5%) or KPC (66/206, 32.0%) enzymes. As expected, the 21 isolates that carried both an OXA-48-like carbapenemase and a more efficient carbapenemase (KPC, NDM, or VIM; Table 1) were resistant to imipenem (MICs, 4 to >8 μg/ml), with the exception of the 1 isolate carrying OXA-48 and KPC mentioned above.
TABLE 1.
MIC frequency distributions for imipenem against carbapenemase-positive isolates of Enterobacteriaceae collected by the SMART global surveillance program from 2008 to 2014
| Isolate genotype (no. of isolates) | No. (%) of isolates with indicated imipenem MIC (μg/ml)b |
|||||||
|---|---|---|---|---|---|---|---|---|
| 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | >8 | |
| All carbapenemase-positive isolates (1,493)a | 3 (0.2) | 10 (0.7) | 27 (1.8) | 101 (6.8) | 206 (13.8) | 206 (13.8) | 245 (16.4) | 695 (46.6) |
| KPC (794) | 2 (0.3) | 16 (2.0) | 66 (8.3) | 103 (13.0) | 136 (17.1) | 471 (59.3) | ||
| NDM (290) | 2 (0.7) | 28 (9.7) | 70 (24.1) | 190 (65.5) | ||||
| OXA-48-like (300) | 2 (0.7) | 9 (3.0) | 22 (7.3) | 78 (26.0) | 106 (35.3) | 42 (14.0) | 18 (6.0) | 23 (7.7) |
| VIM (92) | 3 (3.3) | 13 (14.1) | 27 (29.3) | 22 (23.9) | 27 (29.3) | |||
| IMP (40) | 1 (2.5) | 1 (2.5) | 3 (7.5) | 3 (7.5) | 19 (47.5) | 7 (17.5) | 3 (7.5) | 3 (7.5) |
| GES (3) | 2 (66.7) | 1 (33.3) | ||||||
The 1,493 carbapenemase-positive isolates included 1,485 isolates that were ertapenem nonsusceptible and 8 isolates that were ertapenem susceptible and phenotypically ESBL positive. Twenty-four isolates carried two carbapenemases: KPC plus VIM (KPC+VIM) (Greece, 3 isolates); KPC+NDM (Colombia, 1 isolate); KPC+OXA-48-like (Argentina, 1 isolate; Italy, 1 isolate; Morocco, 1 isolate); NDM+OXA-48-like (Egypt, 3 isolates; India, 1 isolate; Jordan, 1 isolate; Romania, 1 isolate; Serbia, 4 isolates; Tunisia, 1 isolate; United Arab Emirates, 2 isolates; Vietnam, 2 isolates); VIM+OXA-48-like (Croatia, 1 isolate; Turkey, 1 isolate). One isolate carried three carbapenemases: OXA-48-like+NDM+VIM (Turkey).
Imipenem MIC breakpoints: susceptible, ≤1μg/ml (gray shading); intermediate, 2 μg/ml; resistant, ≥4 μg/ml (11).
K. pneumoniae was the carbapenemase-positive species of Enterobacteriaceae most commonly identified (Table 2). KPC was the carbapenemase most commonly identified in K. pneumoniae. NDM was more frequently found than KPC or OXA-48-like enzymes in E. coli and E. cloacae. VIM was more common in K. pneumoniae and E. cloacae than in E. coli. Table 2 also depicts imipenem MIC distributions for the three most commonly identified species of Enterobacteriaceae (E. coli, K. pneumoniae, and E. cloacae) that were carbapenemase positive. Among the carbapenemase-positive isolates, the percentages of imipenem-susceptible isolates were 7.2% for K. pneumoniae (81/1,127), 12.7% for E. cloacae (14/110), and 21.5% for E. coli (32/149), with the percentage of E. coli significantly higher than that of K. pneumoniae (P < 0.0001) and approaching statistical significance compared to that of E. cloacae (P = 0.07). We also found a significantly higher proportion of OXA-48-like carbapenemases (P < 0.0001) in carbapenemase-positive E. coli isolates (30.9% [46/149]) than in K. pneumoniae isolates (19.3% [217/1127]) and E. cloacae isolates (14.5% [16/110]).
TABLE 2.
Imipenem MIC frequency distributions against carbapenemase-positive isolates of K. pneumoniae, E. coli, and E. cloacae stratified by carbapenemase type
| Organism or genotypea (no. of isolates) | No. (%) of isolates with indicated imipenem MIC (μg/ml)b |
|||||||
|---|---|---|---|---|---|---|---|---|
| 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | >8 | |
| K. pneumoniae (1,127) | 1 (0.1) | 5 (0.4) | 16 (1.4) | 59 (5.2) | 138 (12.2) | 146 (13.0) | 170 (15.1) | 592 (52.5) |
| KPC (682) | 1 (0.1) | 7 (1.0) | 39 (5.7) | 83 (12.2) | 117 (17.2) | 435 (63.8) | ||
| OXA-48-like (217) | 1 (0.5) | 4 (1.8) | 14 (6.5) | 49 (22.6) | 82 (37.8) | 36 (16.6) | 12 (5.5) | 19 (8.8) |
| NDM (169) | 13 (7.7) | 29 (17.2) | 127 (75.1) | |||||
| VIM (52) | 4 (7.7) | 11 (21.2) | 11 (21.2) | 26 (50.0) | ||||
| IMP (24) | 1 (4.2) | 1 (4.2) | 2 (8.3) | 13 (54.2) | 3 (12.5) | 2 (8.3) | 2 (8.3) | |
| GES (2) | 2 (100) | |||||||
| E. coli (149) | 1 (0.7) | 5 (3.4) | 6 (4.0) | 20 (13.4) | 31 (20.8) | 21 (14.1) | 29 (19.5) | 36 (24.2) |
| KPC (42) | 1 (2.4) | 3 (7.1) | 16 (38.1) | 7 (16.7) | 5 (11.9) | 10 (23.8) | ||
| OXA-48-like (46) | 5 (10.9) | 5 (10.9) | 17 (37.0) | 13 (28.3) | 4 (8.7) | 2 (4.3) | ||
| NDM (57) | 1 (1.8) | 7 (12.3) | 24 (42.1) | 25 (43.9) | ||||
| VIM (2) | 1 (50.0) | 1 (50.0) | ||||||
| IMP (3) | 1 (33.3) | 2 (66.7) | ||||||
| GES (0) | ||||||||
| E. cloacae (110) | 1 (0.9) | 4 (3.6) | 9 (8.2) | 17 (15.5) | 22 (20.0) | 29 (26.4) | 28 (25.5) | |
| KPC (22) | 2 (9.1) | 2 (9.1) | 3 (13.6) | 9 (40.9) | 6 (27.3) | |||
| OXA-48-like (16) | 1 (6.3) | 2 (12.5) | 3 (18.8) | 5 (31.3) | 2 (12.5) | 3 (18.8) | ||
| NDM (40) | 1 (2.5) | 7 (17.5) | 10 (25.0) | 22 (55.0) | ||||
| VIM (28) | 3 (10.7) | 5 (17.9) | 10 (35.7) | 10 (35.7) | ||||
| IMP (9) | 2 (22.2) | 1 (11.1) | 4 (44.4) | 1 (11.1) | 1 (11.1) | |||
| GES (0) | ||||||||
Data include 7 ertapenem-susceptible isolates (K. pneumoniae, 4 isolates; E. coli, 3 isolates).
Imipenem MIC breakpoints: susceptible, ≤1μg/ml (gray shading); intermediate, 2 μg/ml; resistant, ≥4 μg/ml (11).
Table 3 shows the activity of antimicrobial agents against carbapenemase-positive Enterobacteriaceae stratified by susceptibility and nonsusceptibility to imipenem. The activities of all tested antimicrobial agents against carbapenemase-positive isolates were low (<50%), with amikacin showing greater activity (49.8% susceptibility) than all other agents tested (<17% susceptibility). Among carbapenemase-positive isolates, the isolates identified as resistant to imipenem were also more resistant to other agents than the carbapenemase-positive imipenem-susceptible isolates. Table 3 shows that even the imipenem-susceptible carbapenemase producers were >50% resistant to all tested agents except amikacin.
TABLE 3.
Activity of antimicrobial agents against carbapenemase-positive isolates of Enterobacteriaceae collected by the SMART global surveillance program from 2008 to 2014
| Genotype/phenotype (no. of isolates) | Antimicrobial agent | MIC50 (μg/ml) | MIC90 (μg/ml) | MIC range (μg/ml) | % susceptible |
|---|---|---|---|---|---|
| All carbapenemase-positive isolates (1,493) | Ertapenem | >4 | >4 | <0.03 to >4 | 0.5 |
| Imipenem | 8 | >8 | 0.12 to >8 | 9.4 | |
| Cefepime | >32 | >32 | <0.5 to >32 | 5.8 | |
| Ceftriaxone | >32 | >32 | <1 to >32 | 3.1 | |
| Cefotaxime | >128 | >128 | <0.5 to >128 | 2.3 | |
| Ceftazidime | >128 | >128 | <0.5 to >128 | 7.9 | |
| Cefoxitin | >16 | >16 | <2 to >16 | 12.7 | |
| Ampicillin-sulbactam | >16 | >16 | <2 to >16 | 0.1 | |
| Piperacillin-tazobactam | >64 | >64 | <2 to >64 | 1.6 | |
| Ciprofloxacin | >2 | >2 | <0.25 to >2 | 10.0 | |
| Levofloxacin | >4 | >4 | <0.5 to >4 | 16.8 | |
| Amikacin | 32 | >32 | <4 to >32 | 49.8 | |
| Carbapenemase-positive imipenem-nonsusceptible isolates (1,352) | Ertapenem | >4 | >4 | <0.03 to >4 | 0.2 |
| Imipenem | >8 | >8 | 2 to >8 | 0 | |
| Cefepime | >32 | >32 | <0.5 to >32 | 4.0 | |
| Ceftriaxone | >32 | >32 | <1 to >32 | 1.6 | |
| Cefotaxime | >128 | >128 | <0.5 to >128 | 1.1 | |
| Ceftazidime | >128 | >128 | <0.5 to >128 | 6.0 | |
| Cefoxitin | >16 | >16 | <2 to >16 | 9.4 | |
| Ampicillin-sulbactam | >16 | >16 | <2 to >16 | 0.1 | |
| Piperacillin-tazobactam | >64 | >64 | <2 to >64 | 1.0 | |
| Ciprofloxacin | >2 | >2 | <0.25 to >2 | 8.7 | |
| Levofloxacin | >4 | >4 | <0.5 to >4 | 15.0 | |
| Amikacin | 32 | >32 | <4 to >32 | 46.5 | |
| Carbapenemase-positive imipenem-susceptible isolates (141) | Ertapenem | 2 | >4 | <0.03 to >4 | 3.5 |
| Imipenem | 1 | 1 | 0.12 to 1 | 100 | |
| Cefepime | >32 | >32 | <0.5 to >32 | 23.4 | |
| Ceftriaxone | >32 | >32 | <1 to >32 | 17.7 | |
| Cefotaxime | >128 | >128 | <0.5 to >128 | 14.2 | |
| Ceftazidime | 64 | >128 | <0.5 to >128 | 26.2 | |
| Cefoxitin | 16 | >16 | <2 to >16 | 44.7 | |
| Ampicillin-sulbactam | >16 | >16 | 16 to >16 | 0 | |
| Piperacillin-tazobactam | >64 | >64 | <2 to >64 | 7.8 | |
| Ciprofloxacin | >2 | >2 | <0.25 to >2 | 23.4 | |
| Levofloxacin | >4 | >4 | <0.5 to >4 | 34.0 | |
| Amikacin | <4 | >32 | <4 to >32 | 80.9 |
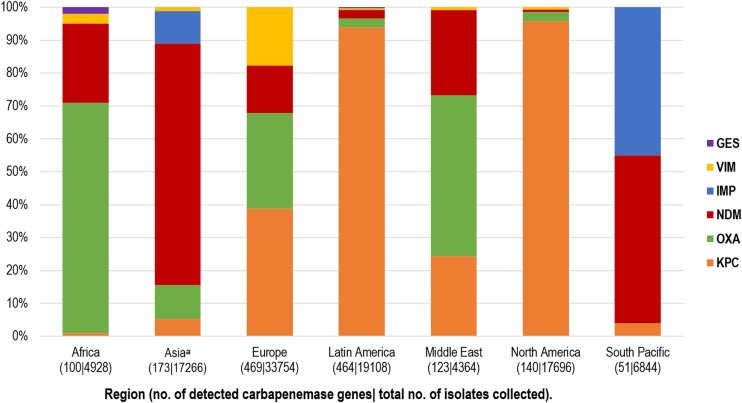
Figure 1 provides an overview of the geographical distribution of the types of carbapenemases detected in characterized Enterobacteriaceae isolates using cumulative data. OXA-48-like enzymes were the carbapenemases most commonly detected in Africa (70.0%, 70/100 detected genes), NDM the carbapenemase most commonly detected in Asia (73.8%, 127/172), KPC (38.8%, 182/469) and OXA-48-like (29.0%, 136/469) the carbapenemases most commonly detected in Europe, KPC (94.0%, 436/464) the carbapenemase most commonly detected in Latin America, OXA-48-like enzymes (48.8%, 60/123) the carbapenemases most commonly detected in the Middle East, KPC (95.7%, 134/140) the carbapenemase most commonly detected in North America, and NDM (51.0%, 26/51) and IMP (45.1%, 23/51) the carbapenemases most commonly detected in the South Pacific region. The distributions of carbapenemase types also differed widely among countries, even within regions, as did the proportions of carbapenemase-positive isolates (Table 4). The countries with the highest carbapenemase-positive rates were in Africa (Egypt, 10.5%), Europe (Greece, 10.0%; Serbia, 7.3%), Latin America (Puerto Rico, 7.5%), and Asia (India, 6.8%) (Table 4). Among the countries with >25 isolates of Enterobacteriaceae that were carbapenemase positive, KPC was the most prevalent carbapenemase in Argentina (90.4% of carbapenemase-positive isolates), Brazil (98.7%), Colombia (98.8%), Puerto Rico (100%), Greece (78.0%), Italy (77.9%), Israel (100%), and the United States (95.6%). OXA-48-like enzymes were the most common carbapenemases in Tunisia (97.0%), Turkey (94.0%), and Saudi Arabia (91.2%). Those three countries also had correspondingly high rates of imipenem-susceptible, carbapenemase-positive isolates. NDM was most commonly identified in Egypt (59.4%), India (90.8%), Vietnam (77.5%), Serbia (86.2%), United Arab Emirates (80.8%), and Philippines (59.5%). VIM was the most prevalent carbapenemase in Spain (63.3%) and accounted for 23.5% and 17.4% of carbapenemase-positive isolates in Greece and Italy, respectively. Among the carbapenemase-positive isolates collected in Philippines, 35.7% carried IMP.
FIG 1.
Relative percentages of carbapenemase types detected, by region. The countries included in each region are listed in Table 4. The data indicated by “Asiaa” exclude isolates from mainland China (all years) and India (most isolates from 2010 and all isolates from 2011 to 2014) that were unavailable for molecular characterization due to export restrictions.
TABLE 4.
Distribution of carbapenemase genes in characterized Enterobacteriaceae isolates by country, 2008 to 2014a
| Country (total no. of isolates collected per country; years of study participation) | No. of sites per country | No. of carbapenemase-positive isolates (% of isolates collected) | No. of isolates carrying specified carbapenemase genesb (% of carbapenemase-positive isolates) |
No. of imipenem-susceptible isolates (% of carbapenemase-positive isolates) | ||||
|---|---|---|---|---|---|---|---|---|
| KPC | OXA-48-like | NDM | IMP | VIM | ||||
| Africa | ||||||||
| Egypt (304; 3) | 2 | 32 (10.5) | 0 (0.0) | 15 (46.9) | 19 (59.4) | 0 (0.0) | 1 (3.1) | 2 (6.3) |
| Kenya (58; 2) | 1 | 1 (1.7) | 0 (0.0) | 0 (0.0) | 1 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Morocco (838; 5) | 2 | 22 (2.6) | 1 (4.5) | 21 (95.5) | 1 (4.5) | 0 (0.0) | 0 (0.0) | 14 (63.6) |
| South Africa (2708; 7)c | 6 | 7 (0.3) | 0 (0.0) | 2 (28.6) | 2 (28.6) | 0 (0.0) | 1 (14.3) | 1 (14.3) |
| Tunisia (1020; 4) | 2 | 33 (3.2) | 0 (0.0) | 32 (97.0) | 1 (3.0) | 0 (0.0) | 1 (3.0) | 8 (24.2) |
| Asia | ||||||||
| Hong Kong (1,482; 7) | 2 | 0 (0.0) | ||||||
| India (1,110; 2)d | 9 | 76 (6.8) | 0 (0.0) | 7 (9.2) | 69 (90.8) | 1 (1.3) | 0 (0.0) | 2 (2.6) |
| Japan (553; 3) | 4 | 7 (1.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 7 (100) | 0 (0.0) | 2 (28.6) |
| Kazakhstan (279; 3) | 1 | 0 (0.0) | ||||||
| Korea, South (1,382; 7) | 4 | 0 (0.0) | ||||||
| Malaysia (1,220; 7) | 2 | 1 (0.1) | 0 (0.0) | 0 (0.0) | 1 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Singapore (1,659; 7) | 2 | 0 (0.0) | ||||||
| Taiwan (5,929; 7) | 9 | 9 (0.2) | 2 (22.2) | 0 (0.0) | 0 (0.0) | 5 (55.6) | 2 (22.2) | 2 (22.2) |
| Thailand (1,050; 7) | 2 | 5 (0.5) | 0 (0.0) | 0 (0.0) | 2 (40.0) | 3 (60.0) | 0 (0.0) | 0 (0.0) |
| Vietnam (2,602; 7) | 4 | 71 (2.7) | 7 (9.9) | 11 (15.5) | 55 (77.5) | 0 (0.0) | 0 (0.0) | 3 (4.2) |
| Europe | ||||||||
| Croatia (461; 4) | 1 | 7 (1.5) | 1 (14.3) | 1 (14.3) | 0 (0.0) | 0 (0.0) | 6 (85.7) | 0 (0.0) |
| Czech Republic (372; 3) | 1 | 1 (0.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (100) | 0 (0.0) |
| Estonia (701; 6) | 2 | 0 (0.0) | ||||||
| France (3252; 7) | 7 | 2 (0.1) | 0 (0.0) | 2 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (100) |
| Germany (3,521; 7) | 7 | 8 (0.2) | 5 (62.5) | 2 (25.0) | 0 (0.0) | 0 (0.0) | 1 (12.5) | 0 (0.0) |
| Greece (1,324; 7) | 3 | 132 (10.0) | 103 (78.0) | 0 (0.0) | 1 (0.8) | 0 (0.0) | 31 (23.5) | 1 (0.8) |
| Hungary (1,270; 5) | 2 | 4 (0.3) | 0 (0.0) | 1 (25.0) | 0 (0.0) | 0 (0.0) | 3 (75.0) | 0 (0.0) |
| Italy (2,687; 7) | 4 | 86 (3.2) | 67 (77.9) | 5 (5.8) | 0 (0.0) | 0 (0.0) | 15 (17.4) | 1 (1.2) |
| Latvia (854; 7) | 1 | 0 (0.0) | ||||||
| Lithuania (967; 7) | 2 | 0 (0.0) | ||||||
| Portugal (2,412; 7) | 3 | 2 (0.1) | 2 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Romania (1,181; 6) | 2 | 17 (1.4) | 0 (0.0) | 9 (52.9) | 6 (35.3) | 0 (0.0) | 3 (17.6) | 3 (17.6) |
| Serbia (894; 4) | 2 | 65 (7.3) | 1 (1.5) | 12 (18.5) | 56 (86.2) | 0 (0.0) | 0 (0.0) | 2 (3.1) |
| Slovenia (532; 4) | 1 | 1 (0.2) | 0 (0.0) | 1 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Spain (9,295; 7) | 12 | 30 (0.3) | 3 (10.0) | 8 (26.7) | 0 (0.0) | 0 (0.0) | 19 (63.3) | 4 (13.3) |
| Switzerland (149; 2) | 2 | 0 (0.0) | ||||||
| Turkey (2,295; 7) | 6 | 100 (4.4) | 0 (0.0) | 94 (94.0) | 5 (5.0) | 0 (0.0) | 4 (4.0) | 39 (39.0) |
| United Kingdom (1,587; 7) | 5 | 1 (0.1) | 0 (0.0) | 1 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Latin America | ||||||||
| Argentina (1,654; 7) | 2 | 83 (5.0) | 75 (90.4) | 9 (10.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 8 (9.6) |
| Brazil (2,776; 7) | 6 | 151 (5.4) | 149 (98.7) | 1 (0.7) | 0 (0.0) | 1 (0.7) | 0 (0.0) | 4 (2.6) |
| Chile (1,718; 7) | 2 | 2 (0.1) | 1 (50.0) | 1 (50.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (50.0) |
| Colombia (2,220; 7) | 6 | 86 (3.9) | 85 (98.8) | 0 (0.0) | 2 (2.3) | 0 (0.0) | 0 (0.0) | 3 (3.5) |
| Dominican Republic (683; 7) | 1 | 0 (0.0) | ||||||
| Ecuador (993; 6) | 2 | 19 (1.9) | 19 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Guatemala (1,565; 7) | 2 | 12 (0.8) | 1 (8.3) | 1 (8.3) | 10 (83.3) | 0 (0.0) | 0 (0.0) | 1 (8.3) |
| Mexico (2,846; 7)e | 4 | 7 (0.2) | 4 (57.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (28.6) | 1 (14.3) |
| Panama (1,256; 7) | 3 | 12 (1.0) | 12 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Puerto Rico (986; 7) | 2 | 74 (7.5) | 74 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 6 (8.1) |
| Venezuela (2,411; 7) | 3 | 16 (0.7) | 16 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Middle East | ||||||||
| Georgia (163; 4) | 1 | 4 (2.5) | 0 (0.0) | 3 (75.0) | 1 (25.0) | 0 (0.0) | 0 (0.0) | 1 (25.0) |
| Israel (1,564; 7) | 3 | 29 (1.9) | 29 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Jordan (655; 6) | 3 | 19 (2.9) | 0 (0.0) | 12 (63.2) | 8 (42.1) | 0 (0.0) | 0 (0.0) | 4 (21.1) |
| Lebanon (596; 4) | 2 | 8 (1.3) | 0 (0.0) | 8 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (37.5) |
| Saudi Arabia (967; 5) | 2 | 34 (3.5) | 0 (0.0) | 31 (91.2) | 2 (5.9) | 0 (0.0) | 1 (2.9) | 11 (32.4) |
| United Arab Emirates (419; 5) | 2 | 26 (6.2) | 1 (3.8) | 6 (23.1) | 21 (80.8) | 0 (0.0) | 0 (0.0) | 2 (7.7) |
| North America | ||||||||
| Canada (4,642; 7) | 12 | 4 (0.1) | 4 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| United States (13,054; 7) | 33 | 136 (1.0) | 130 (95.6) | 4 (2.9) | 1 (0.7) | 0 (0.0) | 1 (0.7) | 6 (4.4) |
| South Pacific | ||||||||
| Australia (2,594; 6) | 5 | 9 (0.3) | 0 (0.0) | 0 (0.0) | 1 (11.1) | 8 (88.9) | 0 (0.0) | 1 (11.1) |
| New Zealand (2,575; 7) | 4 | 0 (0.0) | ||||||
| Philippines (1,675; 7) | 2 | 42 (2.5) | 2 (4.8) | 0 (0.0) | 25 (59.5) | 15 (35.7) | 0 (0.0) | 3 (7.1) |
| Total (103,960) | 217 | 1,493 (1.4) | 794 (53.2) | 300 (20.1) | 290 (19.4) | 40 (2.7) | 92 (6.2) | 141 (9.4) |
Details of countries with a carbapenemase-positive rate of ≥3% are bolded; for those countries, the carbapenemases most commonly detected are indicated with gray shading.
The sum of the isolates carrying different carbapenemase genes is greater than the number of carbapenemase-positive isolates because some isolates carried more than one carbapenemase gene.
Not shown, two isolates carrying GES (28.6% of carbapenemase-positive isolates identified).
Only isolates collected from 2008 to 2010 are included.
Not shown, one isolate carrying GES (14.3% of carbapenemase-positive isolates identified).
DISCUSSION
In the current study of isolates of Enterobacteriaceae from 55 countries, 1.4% (1,493/103,960) of all isolates collected from 2008 to 2014 were carbapenemase positive. We observed that isolates that tested as susceptible to imipenem were not uncommon among the carbapenemase-positive isolates (9.4%, 141/1,493) and that the carbapenemase-positive isolates that tested as susceptible to imipenem tended to carry OXA-48-like enzymes (78.7%; 111/141) (Table 1). Overall, carbapenemase-positive imipenem-susceptible isolates remained uncommon (0.1%, 141/103,960) and the vast majority of isolates of Enterobacteriaceae, in most locales, were susceptible to ertapenem (96.7% of all isolates in the current study) and to other carbapenems (15–17). Consequently, given that carbapenemase-positive imipenem-susceptible isolates do occur, as we observed in the current study, is it important to determine whether any carbapenem-susceptible isolate encountered in the clinical laboratory is a carbapenemase producer? This issue is complex. Carbapenemase-positive imipenem-susceptible isolates may serve as reservoirs of carbapenemase genes that can be transmitted to other pathogens; in addition, these isolates may increase their level of carbapenemase production or acquire porin mutations and become carbapenem resistant. Therapeutic failures have been reported for infections caused by carbapenem-susceptible isolates (18). Furthermore, the carbapenem MIC alone may be an unreliable predictor of therapeutic efficiency against CPE that display an inoculum effect (19). Another important consideration is the prevalence of OXA-48-like enzymes among isolates in a given geographic area. Isolates carrying OXA-48-like β-lactamases have a propensity to demonstrate a limited increase in carbapenem MICs unless they also harbor a permeability defect (20), making it difficult to identify some isolates by phenotypic methods. Meropenem nonsusceptibility is a highly specific phenotypic marker for carbapenemase production but shows poor sensitivity for OXA-48 producers (21). High-level resistance to temocillin is a sensitive and specific marker of OXA-48 production, but temocillin is not included in the automated susceptibility testing methods used by most laboratories (21, 22). Potron and colleagues (23) studied 107 OXA-48 Enterobacteriaceae isolates collected from 2001 to 2011 in Europe and North Africa and reported that 65% of the isolates were susceptible to imipenem and meropenem according to CLSI guidelines. Most of the isolates in that study showed intermediate susceptibility or resistance to ertapenem, suggesting that, in the absence of temocillin susceptibility testing, ertapenem may be the most appropriate carbapenem for use in detection of OXA-48 producers; our study data would appear to confirm this observation.
The clinical relevance of OXA-48-like enzymes remains unclear (13). The efficacy of carbapenems in treatment of infections in humans caused by OXA-48 producers with susceptibility or low-level resistance remains debatable, as imipenem-containing therapy has been reported both to succeed (24) and to fail (25, 26) in treating serious infections attributable to these organisms. In the current study, we observed that, even for imipenem-susceptible carbapenemase producers, >50% of the isolates were resistant to almost all other antimicrobial agents tested (Table 3), leaving few possible treatment options. Knowing this information may be important for empirical therapy considerations as well as for antimicrobial stewardship and infection control efforts to control the spread of carbapenemases. Our observations suggest that it may be worthwhile to identify carbapenemase producers even if they are imipenem susceptible. Controlled trials are needed to determine the true clinical efficacy of carbapenems in treating infections due to OXA-48-like producers such as has been done for KPC producers (4, 8).
The CLSI (11), the FDA, and EUCAST (12) currently recommend reporting antimicrobial susceptibility test results for carbapenems and extended-spectrum cephalosporins without editing the result on the basis of identifying a particular mechanism of resistance. In reference to carbapenems, Livermore and colleagues have suggested that this recommendation is misguided because a significant percentage (50%) of isolates of CPE with carbapenem-susceptible MICs fail therapy, because routine susceptibility testing in clinical laboratories is less precise than the testing performed in research settings and published in peer-reviewed literature, and because there is the potential for laboratories to not look for carbapenemases, or ESBLs, at all if they are not mandated to do so, which would lead to a loss of critical information for infection control practitioners and epidemiologists (13, 27). Kavi and others later suggested that revising carbapenem susceptibility testing results on the basis of available mechanistic or class resistance information would be useful given that altered antimicrobial agent pharmacokinetics may occur in some patients and that a response to antimicrobial therapy may be difficult to judge in the first 24 to 48 h of treatment (28). Livermore and colleagues suggested that it is prudent to seek carbapenemases and ESBLs directly and, when they are identified, generally to avoid substrate antimicrobial agent classes as therapy or, at a minimum, to use only agents from the affected class in combination with another antimicrobial agent with known susceptibility data (13).
Adler et al. reported that infections caused by carbapenemase-producing K. pneumoniae may be more difficult to treat than infections caused by carbapenemase-negative isolates with the same carbapenem MIC because carbapenemase producers exhibit a marked inoculum effect and are more resistant to the bactericidal effect of carbapenems, suggesting that MIC measurements alone may not be sufficient to predict the therapeutic efficacy of carbapenems against CPE (19). The authors suggested that carbapenemase testing, in addition to its epidemiological importance, may also have therapeutic implications, as not all CPE can be detected reliably using current carbapenem MIC breakpoints (11, 12, 19). For the purposes of screening patients for potential carbapenemase producers, EUCAST publishes specific screening MIC breakpoints which detect CPE with high reliability (12). EUCAST screening MIC breakpoints for ertapenem (>0.12 μg/ml), meropenem (>0.12 μg/ml), and imipenem (>1 μg/ml) are 1 dilution step higher than the currently defined epidemiological cutoff (ECOFF) values in order to increase specificity (12). Automated susceptibility testing instruments may not allow for testing at the lower antimicrobial concentrations needed to apply the EUCAST screening MIC breakpoints.
Limitations of the current study include that a small number of carbapenemase producers may have been missed among the carbapenem-susceptible isolates, particularly among the OXA-48-like positive isolates, because our isolate selection protocol for ertapenem-susceptible carbapenemase producers used only a sample (approximately 60%) of ESBL-positive isolates; no ESBL-negative ertapenem-susceptible isolates were tested. However, ertapenem is generally considered the carbapenem with the lowest activity against CPE and, therefore, the most sensitive globally available indicator of carbapenemase activity. Also, the identification of a carbapenemase gene does not guarantee its expression; however, given that many of the screened genes are often plasmid mediated, they are highly likely to be expressed. There is also the possibility that novel carbapenemases or rarer enzymes such as IMI/NMC and SME that were not included in the screening may have been present in the isolates tested. Strengths of the study included that it tested a large global collection of isolates and that its results fill a void in the current literature, as no other large population studies have been performed that provide an estimate of how frequently carbapenemase-positive isolates may be encountered among imipenem-susceptible isolates. However, our study needs to be interpreted in light of the fact that there will be potential location-specific variation depending primarily on the prevalence of OXA-48-like enzymes.
In conclusion, we found that 1.4% (1,493/103,960) of isolates in a recent global collection of clinical isolates of Enterobacteriaceae were carbapenemase positive and that almost all of these isolates (99.5%; 1,485/1,493) were ertapenem-nonsusceptible isolates; only 0.1% (8/9,371) of the characterized ertapenem-susceptible ESBL-positive isolates were carbapenemase positive. Carbapenemase-positive isolates most commonly carried KPC (n = 794), followed by OXA-48-like (n = 300) and NDM (n = 290) enzymes, and K. pneumoniae (n = 1,127), E. coli (n = 149), and E. cloacae (n = 110) were among the species most frequently identified. Among the carbapenemase-positive isolates, 66.7% (2/3), 37.0% (111/300), 20.0% (8/40), 3.3% (3/92), 2.3% (18/794), and 0% (0/290) of isolates carrying GES, OXA-48-like, IMP, VIM, KPC, and NDM enzymes, respectively, tested as susceptible to imipenem (MIC, ≤1 μg/ml). We found that isolates with a carbapenemase gene that tested as susceptible to imipenem were not uncommon (9.4%, 141/1,493) among carbapenemase-positive isolates but remained rare (0.1%, 141/103,960) among our large unselected population of clinical isolates. The practice of screening imipenem-susceptible isolates of Enterobacteriaceae for the presence of carbapenemase genes remains controversial and requires further study.
MATERIALS AND METHODS
Bacterial isolates tested.
The SMART global surveillance program has monitored the in vitro antimicrobial susceptibility patterns of clinical isolates of Gram-negative bacilli collected worldwide from patients with intra-abdominal infections (IAI) since 2002 and from patients with urinary tract infections (UTI) since late 2009. The SMART program collected 103,960 isolates of Enterobacteriaceae from 217 clinical laboratories in 55 countries from 2008 to 2014, of which 68,653 isolates were from IAI and 35,307 from UTI. Isolates from mainland China (all years) and India (most isolates from 2010 and all isolates from 2011 to 2014) were excluded from all aspects of this study because they were not available for molecular characterization due to export restrictions. Specific isolate collection protocols for the SMART program have been described previously (29). All isolates in this study were shipped to International Health Management Associates, Inc. (IHMA, Schaumburg, IL, USA), where their identities were confirmed using RapID One or NF Plus biochemical systems (Remel, Lenexa, KS) (isolates collected in 2008 to 2011) or a Bruker Biotyper matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) spectrometry instrument (Bruker Daltonics, Billerica, MA, USA) (isolates collected in 2012 to 2014), and reference antimicrobial susceptibility testing (11, 30) was performed. The 103,960 isolates of Enterobacteriaceae included 59,151 (56.9%) E. coli, 18,772 (18.1%) K. pneumoniae, 6,041 (5.8%) Enterobacter cloacae, 5,277 (5.1%) P. mirabilis, and 3,142 (3.0%) K. oxytoca isolates and 11,577 (11.1%) isolates of 63 other identified species.
Per SMART program protocol, all ertapenem-nonsusceptible isolates of Enterobacteriaceae as well as a random selection of 50% of E. coli, K. pneumoniae, K. oxytoca, and P. mirabilis isolates positive for ESBL activity by combination clavulanic acid testing (i.e., phenotypically ESBL positive) were to be characterized molecularly. However, 37 of 3,465 ertapenem-nonsusceptible isolates were not available for characterization, resulting in a final analyzed sample of 3,428 isolates of ertapenem-nonsusceptible Enterobacteriaceae. Furthermore, additional ESBL-positive isolates were sometimes characterized for special analyses in support of publications, thereby resulting in an overall final proportion of characterized ESBL-positive isolates of 61% (10,654/17,440). The 3,428 ertapenem-nonsusceptible isolates consisted of 1,672 (48.8%) K. pneumoniae, 951 (27.7%) E. cloacae, 408 (11.9%) E. coli, 113 (3.3%) Enterobacter aerogenes, 68 (2.0%) Citrobacter freundii, and 58 (1.7%) Serratia marcescens isolates and 158 isolates (4.6%) of 26 other species; 1,283 of the E. coli, K. pneumoniae, K. oxytoca, and P. mirabilis isolates were also phenotypically ESBL positive. The 9,371 characterized ertapenem-susceptible phenotypically ESBL-positive isolates consisted of 6,563 E. coli, 2,482 K. pneumoniae, 142 K. oxytoca, and 184 P. mirabilis isolates.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was performed at IHMA using custom dehydrated MicroScan panels (Beckman Coulter, Inc., West Sacramento, CA). All aspects of broth microdilution testing were conducted following CLSI guidelines (11, 30). All isolates were tested against amikacin, ampicillin-sulbactam, cefepime, cefotaxime, cefoxitin, ceftriaxone, ceftazidime, ciprofloxacin, ertapenem, imipenem, levofloxacin, and piperacillin-tazobactam. E. coli, K. pneumoniae, K. oxytoca, and P. mirabilis isolates with a ceftazidime or cefotaxime MIC of >1 μg/ml were further tested for ESBL activity using combination clavulanic acid-based testing, according to CLSI guidelines (11). Isolates were classified as phenotypically ESBL positive if there was an 8-fold reduction at least in ceftazidime or cefotaxime MICs tested in combination with clavulanic acid at 4 μg/ml versus their MICs when tested alone.
Detection of β-lactamase genes.
In total, 12,799 isolates (3,428 ertapenem-nonsusceptible isolates and 9,371 ertapenem-susceptible ESBL-positive isolates) were screened for the presence of genes encoding carbapenemases using a Check-MDR CT101 microarray (Check-Points, Wageningen, the Netherlands) and published multiplex PCR assays, followed by full-gene DNA sequencing (31). Specifically, genes for the IMP, VIM, NDM, and SPM metallo-β-lactamases as well as the KPC, OXA-48-like, and GES serine β-lactamases were screened. For all molecular assays, genomic DNA was extracted from overnight colonies grown on blood agar (Remel, Lenexa, KS) using a QIAamp DNA minikit and a QIAcube instrument (Qiagen, Valencia, CA) according to the manufacturer's instructions. Enzyme variants were identified by comparison to the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov) and the Lahey Clinic website (www.lahey.org/studies).
Statistical analysis.
Fisher's exact test was used to assess the statistical significance of differences in rates of susceptibility to imipenem between carbapenemase types as well as between bacterial species. A P value of <0.05 was considered significant. Analyses were performed with XLSTAT v2015.2.02.18165.
ACKNOWLEDGMENTS
We thank all SMART participants for their contributions to the program.
The SMART global surveillance program is funded by Merck & Co., Inc., Kenilworth, NJ, which also included compensation fees for services in relation to preparing the manuscript.
J.A.K. is a consultant for International Health Management Associates, Inc. (IHMA, Inc.), and an employee of the University of Manitoba and Diagnostic Services Manitoba. S.H.L., K.M.K., R.E.B., and D.F.S. are employees of IHMA, Inc., which receives funding for the SMART surveillance program. J.A.K. and the IHMA authors do not have personal financial interests in the sponsor of this paper (Merck & Co.). K.Y. and M.R.M. are employees of Merck & Co.
Footnotes
For a commentary on this article, see https://doi.org/10.1128/JCM.00430-17.
REFERENCES
- 1.Nordmann P, Poirel P. 2014. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 2.Tängdén T, Giske CG. 2015. Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J Intern Med 277:501–512. doi: 10.1111/joim.12342. [DOI] [PubMed] [Google Scholar]
- 3.Tzouvelekis LS, Markogiannakis A, Psichoglou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev 25:682–707. doi: 10.1128/CMR.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tumbarello M, Viale P, Viscoli C, Trecarichi EM, Tumietto F, Marchese A, Spanu T, Ambretti S, Ginocchio F, Cristini F, Losito AR, Tedeschi S, Cauda R, Bassetti M. 2012. Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. Clin Infect Dis 55:943–950. doi: 10.1093/cid/cis588. [DOI] [PubMed] [Google Scholar]
- 5.Campos AC, Albiero J, Ecker A, Kuroda CM, Meirelles LEF, Polato A, Tognim MCB, Wingeter MA, Teixeira JJV. 2016. Outbreak of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: a systemic review. Am J Infect Cont 44:1374–1380. doi: 10.1016/j.ajic.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 6.Schultsz C, Geerlings S. 2012. Plasmid-mediated resistance in Enterobacteriaceae: changing landscape and implications for therapy. Drugs 72:1–16. doi: 10.2165/11597960-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 7.Falagas ME, Lourida P, Poulikakos P, Rafailidis PI, Tansarli GS. 2014. Antibiotic treatment of infections due to carbapenem-resistant Enterobacteriaceae: systemic evaluation of the available evidence. Antimicrob Agents Chemother 58:654–663. doi: 10.1128/AAC.01222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daikos GL, Tsaousi S, Tzouvelekis LS, Anyfantis I, Psichogiou M, Argyropoulou A, Stefanou I, Sypsa V, Miriagou V, Nepka M, Georgiadou S, Markagiannakis A, Goukos D, Skoutelis A. 2014. Carbapenemase-producing Klebsiella pneumoniae bloodstream infections: lower mortality by antibiotic combination schemes and the role of carbapenems. Antimicrob Agents Chemother 58:2322–2328. doi: 10.1128/AAC.02166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzouvelekis LS, Markogiannakis A, Piperaki E, Souli M, Daikos GL. 2014. Treating infections caused by carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect 20:862–872. doi: 10.1111/1469-0691.12697. [DOI] [PubMed] [Google Scholar]
- 10.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing. Twenty-fifth informational supplement M100-S25. CLSI, Wayne, PA. [Google Scholar]
- 12.EUCAST. 2015. EUCAST breakpoint tables for interpretation of MICs and zone diameters. Version 5.0, January http://www.eucast.org/clinical_breakpoints/.
- 13.Livermore DM, Andrews JM, Hawkey PM, Ho P-L, Keness Y, Doi Y, Paterson D, Woodford N. 2012. Are susceptibility tests enough, or should laboratories seek ESBLs and carbapenemases directly? J Antimicrob Chemother 67:1569–1577. doi: 10.1093/jac/dks088. [DOI] [PubMed] [Google Scholar]
- 14.Jacoby GA, Mills DM, Chow N. 2004. Role of beta-lactamases and porins in resistance to ertapenem and other beta-lactams in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:3203–3206. doi: 10.1128/AAC.48.8.3203-3206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sader HS, Farrell DJ, Flamm RK, Jones RN. 2014. Antimicrobial susceptibility of gram-negative organisms isolated from patients hospitalized in intensive care units in United States and European hospitals (2009–2011). Diagn Microbiol Infect Dis 78:443–446. doi: 10.1016/j.diagmicrobio.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 16.Kehl SC, Dowzicky MJ. 2015. Global assessment of antimicrobial susceptibility among gram-negative organisms collected from pediatric patients between 2004 and 2012: results from the tigecycline evaluation and surveillance trial. J Clin Microbiol 53:1286–1293. doi: 10.1128/JCM.03184-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biedenbach DJ, Kazmierczak K, Bouchillon SK, Sahm DF, Bradford PA. 2015. In vitro activity of aztreonam-avibactam against a global collection of Gram-negative pathogens from 2012 and 2013. Antimicrob Agents Chemother 59:4239–4248. doi: 10.1128/AAC.00206-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weisenberg SA, Morgan DJ, Espinal-Witter R, Larone DH. 2009. Clinical outcomes of patients with KPC-producing Klebsiella pneumoniae following treatment with imipenem or meropenem. Diagn Microbiol Infect Dis 64:233–235. doi: 10.1016/j.diagmicrobio.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adler A, Ma'ayan B-D, Chmelnitsky I, Carmeli Y. 2015. Effect of resistance mechanisms on the inoculum effect of carbapenem in Klebsiella pneumoniae isolates with borderline carbapenem resistance. Antimicrob Agents Chemother 59:5014–5017. doi: 10.1128/AAC.00533-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oueslati S, Nordmann P, Poirel L. 2015. Heterogeneous hydrolytic features of OXA-48-like β-lactamases. J Antimicrob Chemother 70:1059–1063. [DOI] [PubMed] [Google Scholar]
- 21.Huang T-D, Poirel L, Bogaerts P, Berhin C, Nordmann P, Glupczynski Y. 2014. Temocillin and piperacillin/tazobactam resistance by disc diffusion as antimicrobial surrogate markers for the detection of carbapenemase-producing Enterobacteriaceae in geographic areas with a high prevalence of OXA-48 producers. J Antimicrob Chemother 69:445–450. doi: 10.1093/jac/dkt367. [DOI] [PubMed] [Google Scholar]
- 22.Dortet L, Cuzon G, Plesiat P, Naas T. 2016. Prospective evaluation of an algorithm for the phenotypic screening of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother 71:135–140. doi: 10.1093/jac/dkv308. [DOI] [PubMed] [Google Scholar]
- 23.Potron A, Poirel L, Rondinaud E, Nordmann P. 2013. Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill 18:pii=20549. doi: 10.2807/1560-7917.ES2013.18.31.20549. [DOI] [PubMed] [Google Scholar]
- 24.Maherault A-C, Nordmann P, Therby A, Pangon B. 2012. Efficacy of imipenem for the treatment of bacteremia due to an OXA-48-producing Klebsiella pneumoniae isolate. Clin Infect Dis 54:577–578. doi: 10.1093/cid/cir887. [DOI] [PubMed] [Google Scholar]
- 25.Carrër A, Poirel L, Yilmaz M, Akan OA, Feriha C, Cuzon G, Matar G, Honderlick P, Nordmann P. 2010. Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob Agents Chemother 54:1369–1373. doi: 10.1128/AAC.01312-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuzon G, Ouanich J, Gondret R, Naas T, Nordmann P. 2011. Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob Agents Chemother 55:2420–2423. doi: 10.1128/AAC.01452-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomson KS. 2010. Extended-spectrum-β-lactamase, AmpC, and carbapenemase issues. J Clin Microbiol 48:1019–1025. doi: 10.1128/JCM.00219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kavi J, Bhattacharjee D, Macve J, Weinbren MJ. 2013. Comment on: Are susceptibility tests enough, or should laboratories seek ESBLs and carbapenemases directly? J Antimicrob Chemother 68:246. doi: 10.1093/jac/dks378. [DOI] [PubMed] [Google Scholar]
- 29.Biedenbach D, Bouchillon S, Hackel M, Hoban D, Kazmierczak K, Hawser S, Badal R. 2015. Dissemination of NDM metallo-β-lactamase genes among clinical isolates of Enterobacteriaceae collected during the SMART global surveillance study from 2008 to 2012. Antimicrob Agents Chemother 59:826–830. doi: 10.1128/AAC.03938-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clinical and Laboratory Standards Institute. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: Tenth ed: approved standard M07-A10. CLSI, Wayne, PA. [Google Scholar]
- 31.Lob SH, Kazmierczak KM, Badal RE, Hackel MA, Bouchillon SK, Biedenbach DJ, Sahm DF. 2015. Trends in susceptibility of Escherichia coli from intra-abdominal infections to ertapenem and comparators in the United States according to data from the SMART Program, 2009 to 2013. Antimicrob Agents Chemother 59:3606–3610. doi: 10.1128/AAC.05186-14. [DOI] [PMC free article] [PubMed] [Google Scholar]