Abstract
Legionella species are frequently detected in aquatic environments, but their occurrence in extreme, acidic, geothermal habitats has not been explored with cultivation-independent methods. We investigated a predominately eukaryotic algal mat community in a pH 2.7 geothermal stream in Yellowstone National Park for the presence of Legionella and potential host amoebae. Our analyses, using PCR amplification with Legionella-specific primers targeting 16S rRNA genes, detected four known Legionella species, as well as Legionella sequences from species that are not represented in sequence databases, in mat samples and cultivated isolates. The nonrandom occurrence of sequences detected at lower (30°C) and higher (35 to 38°C) temperatures suggests that natural thermal gradients in the stream influence Legionella species distributions in this mat community. We detected only one sequence, Legionella micdadei, from cultivated isolates. We cultured and sequenced partial 18S rRNA gene regions from two potential hosts, Acanthamoeba and Euglena species.
Legionella species are facultative intracellular gram-negative bacilli, well known for their role as the agent of Legionnaires' disease, a severe and sometimes deadly pneumonia caused by Legionella pneumophila (3, 4, 21, 35). In the United States, there may be as many as 25,000 to 100,000 cases of Legionnaires' disease each year. The mortality rate for nosocomial infections can be as high as 50%, a considerable health concern (23, 34). Since person-to-person contact does not spread the disease, prevention of the illness necessitates avoiding contact with contaminated water supplies.
The optimum growth temperature for L. pneumophila in culture is ∼37°C (12, 20), but the bacterium has been isolated from environmental samples at temperatures ranging from 10 to 42°C (14, 26). Heated water in spas, water heaters and pipes, showerheads, and cooling towers provides an ideal habitat for extensive growth and may increase the risk of Legionnaires' disease (35), which usually occurs in outbreaks associated with water systems altered by humans (12, 15). When people inhale water or aerosolized drops contaminated with Legionella species, macrophages in the lung engulf the bacteria. In immunocompromised individuals, these opportunistic bacteria avoid the cell-mediated immune defenses of macrophages and proliferate within these phagocytic cells.
Because a focus of disease prevention is on avoiding contact with contaminated water, molecular methods for sensitive and rapid detection of Legionella spp. in natural consortia has gained new interest (34) and can provide information about the ecology and persistence of Legionella spp. in the environment. Most studies of Legionella spp. are based on cultivated isolates from surface waters, ponds, streams, soils, and other moist areas (4, 12, 34, 35, 38). In these environments, phagocytic amoebae such as Naegleria, Acanthamoeba, and Hartmanella ingest the bacteria while grazing, a process similar to ingestion by human lung macrophages. The legionellae evade amoebal defense mechanisms, avoid digestion, and replicate within vacuoles, eventually lysing the host cells and returning to the environment (35).
Adaptations for survival in amoebal hosts may include traits that allow opportunistic pathogenic legionellae to parasitize and exploit mammalian cells (35). The bacteria can persist and flourish in the relatively protected nutrient-rich microenvironments within amoebae, and bacterial virulence, antibiotic resistance, and survival can be enhanced (10, 27, 30, 34). Within the protective environment of amoebal cysts, Legionella species can withstand desiccation, biocides such as chlorine, and temperature extremes. Cysts also provide a mechanism for dispersal into new habitats (12, 35, 38). Legionella species also survive as free-living cells within microbial biofilm communities (7, 12, 17, 35).
Cultivation-independent methods for surveying microbial diversity in hot springs in Yellowstone National Park (YNP), Wyoming, revealed major phylogenetic groups and new species undetected by traditional culture-based methods (5, 11, 18, 31, 32). In our previous studies at Nymph Creek, a geothermal (temperature ranges from 25 to 60°C), extremely acidic (pH 2.7) stream located in YNP, we used PCR with general bacterial and eukaryotic small subunit rRNA gene primers to characterize an algal mat community that proliferates as an extensive, predominately eukaryotic biofilm in the streambed (31, 32). We detected one sequence that suggested the presence of a vahlkampfiid amoeba, a potential host for Legionella spp. We examined mat samples microscopically and confirmed that low numbers of amoebae and Euglena species were present in addition to the predominant green (Chlorella-like) and red (Cyanidium-like) algal species (8; M. J. Ferris, unpublished data). Using PCR primers that specifically target Naegleria spp., we identified two uncultivated Naegleria sequences that may also be potential hosts for Legionella species (31, 32).
Other Legionella isolates do not grow at pH 2.7 (26), and previous field studies have not identified Legionella spp. in low-pH environments (14). However, we reasoned that Naegleria or Euglena species in Nymph Creek might provide a favorable pH microenvironment for intracellular parasitic bacteria. Our objective in this study was to determine if the acidic biofilm community in Nymph Creek harbors Legionella species. We used PCR amplification with primer sets that target Legionella 16S rRNA and traditional culture methods to identify five Legionella species. We also cultivated and sequenced 18S rRNA gene fragments of two potential hosts, Acanthamoeba and Euglena species, from the Nymph Creek mat.
MATERIALS AND METHODS
Sample collection.
Samples were collected from Nymph Creek, located near Norris Geyser Basin, YNP. The 1- to 10-cm-deep acid-sulfate-chloride stream originates from multiple hot springs and contains an extensive 2- to 3-mm-thick algal mat. Temperature gradients range from 60°C at the source springs to ∼25°C downstream (∼150 m), where the stream flows into Nymph Lake. The pH of the bulk water is 2.7 to 3.3 and contains high levels of sulfur, arsenic, manganese, and iron, characteristic of extreme acidic environments in geothermal areas (8, 11, 13, 31, 32).
Mat samples for DNA extraction were collected, using clean weighing spatulas, along a transect in the stream at sites ∼10 m apart with temperatures of 30, 35, 38, and 47°C. The samples were placed into sterile plastic tubes and stored frozen, as previously described (31). Mat samples for cultivation were similarly collected at a 35°C site, placed in sterile tubes with a small volume of stream water, stored at ambient temperature, and plated within 24 h of collection.
Culturing.
BCYEα agar (buffered charcoal-yeast extract agar containing 0.1% alpha-ketoglutarate) supplemented with 0.3% glycine was prepared for culturing Legionella spp. Negative-control medium was identical except that it also contained the antimicrobial agents polymyxin B (100 U/ml), anisomycin (80 μg/ml), and vancomycin (5 μg/ml) and lacked l-cysteine, in accordance with the protocol of Gorman et al. (16). Mat samples from Nymph Creek were plated directly onto selective medium at a range of pHs from 3 to 7 and incubated at 37°C in a candle jar or plated onto SAG medium (Cyanidium medium 17, pH 2.9; Sammlung von AlgenKulturen Gottingen Culture Collection of Algae [http://www.epsag.uni-goettingen.de/html/culturemedia.html#BM17]) at 25°C for culturing of amoebae.
Nucleic acid extraction and amplification.
DNA was extracted by using a bead-beating method as described previously (31). PCR amplification was performed by using an AmpliTaq Gold DNA polymerase kit (Applied Biosystems, Foster City, Calif.) and deoxynucleoside triphosphate mix (Promega Corporation, Fitchburg, Wis.) according to the manufacturers' instructions. Genomic DNA from L. pneumophila subsp. pneumophila (ATCC 33152D; American Type Culture Collection) was used as a positive control.
An L. pneumophila-specific primer pair, Leg-1 (5′-AGGGTTGATAGGTTAAGAGC-3′) and Leg-2 (5′-CCAACAGCTAGTTGACATCG-3′), which amplifies a 386-bp sequence within the bacterial 16S rRNA gene (19, 28) was used in this study. Reaction conditions were 95°C for 5 min followed by 40 cycles of 95°C for 1 min, 57°C for 1 min 30 s, and 72°C for 1 min, followed by an extension step at 72°C for 10 min.
A second primer set for Legionella species was also utilized. LEG 225 (5′-AAGATTAGCCTGCGTCCGA-3′) and LEG 858 (5′-GTCAACTTATCGCGTTTGCT-3′) amplify a 654-bp fragment of the 16S rRNA gene (24). Reaction conditions were 95°C for 5 min followed by 30 cycles of 95°C for 1 min, 64°C for 1 min, and 74°C for 1 min, followed by an extension step at 74°C for 10 min.
Primers for monocultures of amoebae were NS1 (5′-GTAGTCATATGCTTGTCTTC-3′) and NS2 (5′-GGCTGCTGGCACCAGACTTG-3′), which amplify a segment of ∼555 bp that is proximal to the 5′ end of the 18S rRNA gene (37). Reaction conditions were 95°C for 5 min followed by 30 cycles of 95°C for 15 s, 55°C for 1 min 30 s, and 72°C for 1 min 30 s, followed by an extension step at 72°C for 10 min.
Purified DNA from monocultures of Euglena was amplified with primers 18S F (5′-AA(C/T)TGGTTGATCCTGCCAG(C/T)-3′) and 18S R (5′-TGATCCT(G/C)TGCAGGTTCACC-3′) according to the method described by Sittenfeld et al. (33) by using a GeneAmp XL PCR kit (Perkin-Elmer).
Amoeba and euglenoid species DNAs were amplified with the Leg-1-Leg-2 and LEG 225-LEG 858 primer pairs to test for the presence of Legionella spp. in these potential host organisms.
PCR products were purified with a QIAquick PCR cleanup kit (QIAGEN, Inc., Valencia, Calif.) and sequenced directly by cycle sequencing with Big Dye Terminators (according to the manufacturer's directions) using an ABI Prism 310 DNA sequencer (Applied Biosystems). PCR-amplified DNA fragments were electrophoresed before and after cleanup on 1% agarose gels in 1× TBE (50 mM Tris-HCl, 50 mM boric acid, 1 mM EDTA) containing ethidium bromide and visualized with UV light. A 100-bp DNA ladder (Promega Corporation) was used as a size marker.
Clone library construction.
PCR amplicons were cloned with a TA cloning kit using pCR2.1-TOPO and Escherichia coli TOP10 competent cells (Invitrogen, Carlsbad, Calif.) to produce libraries (36). Individual colonies were selected, and their plasmids were extracted by using the Promega Plus SV miniprep DNA purification system.
Sequencing and phylogenetic analysis.
Plasmid inserts were sequenced with M13 forward and M13 reverse primers by using Big Dye Terminators and an ABI 310 DNA sequencer (Applied Biosystems) as described above. Cloned sequences were compared to those in GenBank (6) by using BLAST (2). Sequences were aligned by using Sequencher 3.1.1 (Gene Codes Corporation, Ann Arbor, Mich.).
DNA extracted from Legionella pure culture isolates was PCR amplified as described above. The products were purified with a QIAquick PCR purification kit (QIAGEN, Inc.) and sequenced with the Leg-1 and Leg-2 primers.
Nucleotide sequence accession numbers.
All rRNA sequences were deposited in GenBank under accession numbers AY682851 to AY682873.
RESULTS AND DISCUSSION
Legionella 16S rRNA gene sequences were obtained by direct PCR amplification of bulk DNA extracted from samples at three sites in the algal mat where temperatures were 38, 35, and 30°C. PCR amplification of DNA isolated from 47°C mat samples did not yield a product with either primer set, despite other culture-based studies that show that L. pneumophila can tolerate temperatures of 50°C (20, 26). Samples from the 38 and 30°C sites were collected on the same day and were amplified with the same primer set (Leg-1 and Leg-2) in order to examine the influence of temperature on population distribution in the mat. We sampled a 35°C site using an independent Legionella-specific primer set approximately 1 year later.
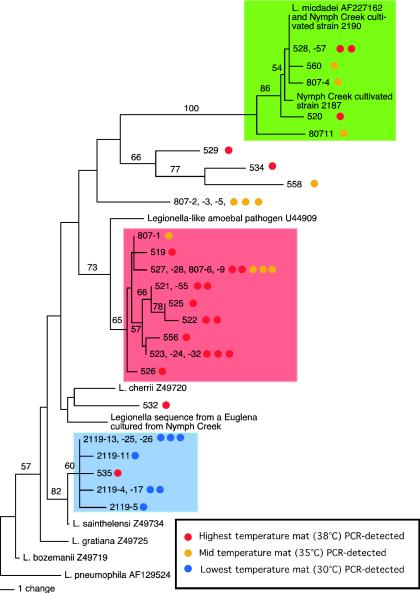
All seven sequences retrieved from the low-temperature (30°C) sample formed a well-supported clade (boot strap value of 82%) (indicated by the blue circles in the blue-shaded area of Fig. 1). Sequences in this clade were nearly identical (>99% sequence similarity) to that of Legionella sainthelensi. Only 1 of 31 total sequences from the higher-temperature sites was L. sainthelensi (red circle in the blue-shaded box in Fig. 1). Thus, it appears that L. sainthelensi is prominent at 30°C but rare at temperatures ≥35°C. Detection of L. sainthelensi in a volcanic basin in YNP is consistent with its original identification near another volcanically active site (9). In the Campbell et al. study (9), L. sainthelensi and other species were cultivated from neutral to slightly alkaline pH (7.4 to 8.2) water near Mt. St. Helens (Washington). However, the water temperature ranged from 10 to 17°C at these sites.
FIG. 1.
Neighbor-joining dendrogram based on total character differences using 346 bp of the Legionella 16S rRNA gene corresponding to positions 457 to 805 (E. coli, GenBank accession number Z83205), which includes two regions of high variability. Bulk DNA, extracted from mat samples collected from Nymph Creek, was PCR amplified, cloned, and sequenced. Sequences retrieved from different temperatures are indicated by blue, yellow, and red circles. The same primer set was used to amplify sequences indicated by red and blue circles, and a different primer set was used to obtain sequences indicated by yellow circles. Sequences of Legionella and Euglena cultivated from mat samples (35°C) are indicated. GenBank accession numbers are included with the named species.
A second clade (bootstrap value of 73%) (indicated by the red-shaded area in Fig. 1) contained the most common sequence detected in Nymph Creek, representing nearly half of the sequences retrieved from the mat by PCR amplification and over two-thirds of the sequences retrieved from the highest-temperature (38°C) sample. Sequences in this clade were most similar (>98%) to that of an unnamed Legionella-like amoebal pathogen (LLAP) (Fig. 1). This prominent LLAP-like sequence was detected by using two different PCR primer sets in samples collected at different times (2002 and 2003) and temperatures (35 and 38°C), suggesting that the prevalence of the sequence is not transient, an artifact of primer bias, or the result of single-sample-point sampling. No LLAP sequences were detected in the low-temperature sample (30°C), suggesting that the species is more abundant at warmer temperatures. LLAPs, unlike legionellae, grow poorly on BCYE α medium. However, LLAPs can often be cocultured with amoebal hosts such as Hartmanella veriformis and Acanthamoeba polyphaga (1). We did not detect the Nymph Creek LLAP sequence among the Legionella isolates cultivated directly from a 35°C mat sample or in our cultures of Euglena species (95% 18S rRNA gene sequence similarity to the sequence in GenBank, accession number EMU532403) or Acanthamoeba sp. (95% 18S rRNA gene sequence similarity to the sequence in GenBank, accession number AY026245.1). Neither 18S rRNA sequence from the pure culture isolates was related to the uncultivated Naegleria-like sequence we previously reported (31, 32).
A third clade (indicated by the green-shaded area in Fig. 1) contained the only sequence we obtained from Legionella culture isolates. This clade contained a minority of the sequences retrieved from the mat by direct PCR detection. Sequences were identical (Nymph Creek isolate 2190) or nearly identical (>99% similarity) to that of L. micdadei. Another sequence detected by PCR was most closely related (>97% similarity) to that of Legionella cherrii, as was a sequence we amplified from the pure-culture Euglena. The sequence detected from the Acanthamoeba isolate was most similar to that of Legionella, but species could not be determined because analysis was based on a short segment of 16S rRNA. Other Nymph Creek Legionella sequences detected directly from the mat by PCR (for example, no. 558 in Fig. 1) were 96% similar to Legionella sequences in public databases. None of the sequences appeared to be chimeric (22).
Previously isolated Legionella species do not grow at a low pH (≤2.7) (14, 26). Our attempts to cultivate Legionella on low-pH medium were unsuccessful (data not shown). We suspect that the Nymph Creek species proliferate in higher-pH microenvironments within host cells. The detection of Legionella sequences by PCR in cultured isolates of Acanthamoeba and Euglena grown on SAG medium at pH 2.9 provides preliminary evidence that these organisms harbor the bacteria. Our previous studies identified other uncultivated potential hosts, Naegleria spp., in Nymph Creek (31, 32). Rogers and Keevil observed microcolonies of Legionella in biofilms (29), and it is possible that microenvironments with favorable pHs exist in interstitial spaces within the Nymph Creek mat. Similar hypotheses were proposed to explain dissimilatory sulfate reduction by uncultivated sulfate-reducing bacteria in Nymph Creek (11, 13). It is also possible that some Legionella spp. can grow and/or remain viable under extremely acidic conditions.
To our knowledge, this is the first report of Legionella species in a hot spring in Yellowstone National Park. Our results revealed at least four known Legionella species and one potentially new species in a low-pH algal mat. The Legionella species are nonrandomly distributed between areas that differ by 5 to 8°C along the stream channel. The distribution is more consistent with well-established, thriving populations of microbes, whereas a random distribution would suggest that nonviable cells washed into the mat via surface runoff. We suggest that Legionella species and host organisms flourish in the mat community in Nymph Creek and that this extreme physical and chemical environment provides a reservoir for potentially pathogenic legionellae.
Acknowledgments
We thank the National Science Foundation (Microbial Observatory grant 9977922); the National Park Service, Department of the Interior; and the Thermal Biology Institute at Montana State University for financial support.
We are grateful to all who assisted in the field and laboratory including Jen Fagg, Tai Takenaka, Emily Kuhn, and Dean Snow. Special thanks to Mary Bateson for sequencing. We thank John Varley, Christie Hendrix, and Liz Cleveland for their assistance.
This project was conducted under the direction of the Yellowstone Center for Resources following the guidelines for research in Yellowstone National Park.
REFERENCES
- 1.Adeleke, A., J. Pruckler, R. Benson, T. Rowbotham, M. Halablab, and B. Fields. 1996. Legionella-like amebal pathogens—phylogenetic status and possible role in respiratory disease. Emerg. Infect. Dis. 2:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atlas, R. M. 1999. Legionella: from environmental habitats to disease pathology, detection and control. Environ. Microbiol. 1:283-293. [DOI] [PubMed] [Google Scholar]
- 4.Barker, J., and M. R. W. Brown. 1994. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology 140:1253-1259. [DOI] [PubMed] [Google Scholar]
- 5.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, D. A., M. S. Boguski, D. J. Lipman, J. Ostell, B. F. Ouellette, B. A. Rapp, and D. L. Wheeler. 1999. GenBank. Nucleic Acids Res. 27:12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brock, T. D. 1978. Thermophilic microorganisms and life at high temperatures. Springer-Verlag, New York, N.Y.
- 8.Brown, M. R. W., and J. Barker. 1999. Unexplored reservoirs of pathogenic bacteria: protozoa and biofilms. Trends Microbiol. 7:46-50. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, J., W. F. Bibb, M. A. Lambert, S. Eng, A. G. Steigerwalt, J. Allard, C. W. Moss, and D. J. Brenner. 1984. Legionella sainthelensi: a new species of Legionella isolated from water near Mt. St. Helens. Appl. Environ. Microbiol. 47:369-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cirillo, J. D. 1999. Exploring a novel perspective on pathogenic relationships. Trends Microbiol. 7:96-98. [DOI] [PubMed] [Google Scholar]
- 11.Ferris, M. J., T. S. Magnuson, J. A. Fagg, R. Thar, M. Kühl, K. B. Sheehan, and J. M. Henson. 2003. Microbially mediated sulphide production in a thermal, acidic algal mat community in Yellowstone National Park. Environ. Microbiol. 5:954-960. [DOI] [PubMed] [Google Scholar]
- 12.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fishbane, S., J. G. Dillon, H. L. Gough, and D. A. Stahl. 2003. Linkage of high rates of sulfate reduction in Yellowstone hot springs to unique sequence types in the dissimilatory sulfate respiration pathway. Appl. Environ. Microbiol. 69:3663-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fliermans, C. B., W. B. Cherry, L. H. Orrison, S. J. Smith, D. L. Tison, and D. H. Pope. 1981. Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 41:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Fulgueiras, A., C. Navarro, D. Fenoll, J. Garcia, P. Gonzales-Diego, T. Jimenez-Bunuelas, M. Rodriguez, R. Lopez, F. Pacheco, J. Ruiz, M. Segovia, B. Balandron, and C. Pelaz. 2003. Legionnaires' disease outbreak in Murcia, Spain. Emerg. Infect. Dis. 8:915-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorman, G. W., J. M. Barbaree, and J. C. Feeley. 1994. Procedures for the recovery of Legionella from the environment. Public Health Service, U.S. Department of Health and Human Services, Centers for Disease Control, Atlanta, Ga.
- 17.Harb, O. S., L-Y. Gao, and Y. A. Kwaik. 2000. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ. Microbiol. 2:251-265. [DOI] [PubMed] [Google Scholar]
- 18.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jonas, D., A. Rosenbaum, S. Weyrich, and S. Bhakdi. 1995. Enzyme-linked immunoassay for detection of PCR-amplified DNA of Legionella in bronchoalveolar fluid. J. Clin. Microbiol. 33:1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusnetsov, J. M., E. Ottoila, and P. J. Martikainen. 1996. Growth, respiration and survival of Legionella pneumophila at high temperatures. J. Appl. Bacteriol. 81:341-347. [DOI] [PubMed] [Google Scholar]
- 21.Kwaik, Y. A., L-Y. Gao, B. J. Stone, C. Venkataraman, and O. S. Harb. 1998. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl. Environ. Microbiol. 64:3127-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maidak, B. L., G. J. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1997. The RDP (Ribosomal Database Project). Nucleic Acids Res. 25:109-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marston, B. J., J. F. Plouffle, T. M. File, B. A. Hackman, S. J. Salstrom, H. B. Lipman, M. S. Kolczak, and R. F. Breiman. 1997. Incidence of community-acquired pneumonia requiring hospitalization—results of a population-based active surveillance study in Ohio. Arch. Intern. Med. 157:1709-1718. [PubMed] [Google Scholar]
- 24.Miyamoto, H., H. Yamamoto, K. Arima, J. Fujii, K. Maruta, K. Izu, T. Shiomori, and S.-I. Yoshida. 1997. Development of a seminested PCR method for detection of Legionella species and its application to surveillance of legionellae in hospital cooling tower water. Appl. Environ. Microbiol. 63:2489-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nwachuku, N., and C. P. Gerba. 2004. Health effects of Acanthamoeba spp. and its potential for waterborne transmission. Rev. Environ. Contam. Toxicol. 180:93-131. [DOI] [PubMed] [Google Scholar]
- 26.Ohno, A., N. Kato, K. Yamad, and K. Yamaguchi. 2003. Factors influencing survival of Legionella pneumophila serotype 1 in hot spring water and tap water. Appl. Environ. Microbiol. 69:2540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pope, D. H., R. J. Soracco, H. K. Gill, and C. B. Fliermans. 1982. Growth of Legionella pneumophila in two-membered cultures with green algae and cyanobacteria. Curr. Microbiol. 7:319-322. [Google Scholar]
- 28.Reischl, U., H.-J. Linde, N. Lehn, O. Landt, K. Barratt, and N. Wellinghausen. 2002. Direct detection and differentiation of Legionella spp. and Legionella pneumophila in clinical specimens by dual-color real-time PCR and melting curve analysis. J. Clin. Microbiol. 40:3814-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogers, J., and C. W. Keevil. 1992. Immunogold and fluorescein immunolabelling of Legionella pneumophila within an aquatic biofilm visualized by using episcopic differential interference contrast microscopy. Appl. Environ. Microbiol. 58:2326-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheehan, K. B., J. A. Fagg, M. J. Ferris, and J. M. Henson. 2003. PCR detection and analysis of the free-living amoeba Naegleria in hot springs in Yellowstone and Grand Teton National Parks. Appl. Environ. Microbiol. 69:5914-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheehan, K. B., M. J. Ferris, and J. M. Henson. 2003. Detection of Naegleria sp. in a thermal, acidic stream in Yellowstone National Park. J. Eukaryot. Microbiol. 50:263-265. [DOI] [PubMed] [Google Scholar]
- 33.Sittenfeld, A., M. Mora, J. M. Ortega, F. Albertazzi, A. Cordero, M. Roncel, E. Sánchez, M. Vargas, M. Fernández, J. Weckesser, and A. Serrano. 2002. Characterization of a photosynthetic Euglena strain isolated from an acidic hot mud pool of a volcanic area of Costa Rica. FEMS Microbiol. Ecol. 42:151-161. [DOI] [PubMed] [Google Scholar]
- 34.Steinert, M., U. Hentschel, and J. Hacker. 2002. Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol. Rev. 26:149-162. [DOI] [PubMed] [Google Scholar]
- 35.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 36.Vergin, K. L., M. S. Rappe, and S. J. Giovannoni. 2001. Streamlined method to analyze 16S rRNA gene clone libraries. BioTechniques 30:938-940. [DOI] [PubMed] [Google Scholar]
- 37.White, T. J., T. D. Bruns, S. B. Lee, and J. W. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In N. Innis, D. Gelfand, J. Sninsky, and T. White (ed.), PCR protocols and applications—a laboratory manual. Academic Press, New York, N.Y.
- 38.Winiecka-Krusnell, J., and E. Linder. 1997. Free-living amoebae protecting Legionella in water: the tip of an iceberg? Scand. J. Infect. Dis. 31:383-385. [DOI] [PubMed] [Google Scholar]