ABSTRACT
Identification of species within the Mycobacterium avium complex (MAC) is difficult, and most current diagnostic laboratory tests cannot distinguish between species included in the complex. Differentiation of species within the MAC is important, as Mycobacterium chimaera has recently emerged as a major cause of invasive cardiovascular infections following open heart surgery. A new commercial diagnostic assay, GenoType NTM-DR ver. 1.0, is intended to differentiate between three species within the MAC, namely, Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium chimaera. In this study, we investigated an archival collection of 173 MAC isolates using 16S rRNA and 16S-23S internal transcribed spacer (ITS) gene sequencing, and GenoType NTM-DR was evaluated for identifying M. chimaera and other species belonging to the MAC. Species identification of 157/173 (91%) isolates with the GenoType NTM-DR assay was in agreement with 16S rRNA and 16S-23S ITS gene sequencing results. Misidentification occurred with 16 isolates which belonged to four species included in the MAC that are rarely encountered in clinical specimens. Despite some limitations of this assay, GenoType NTM-DR had 100% specificity for identifying M. chimaera. This novel assay will enable diagnostic laboratories to differentiate species belonging to the Mycobacterium avium complex and to accurately identify M. chimaera. It can produce rapid results and is also more cost efficient than gene sequencing methods.
KEYWORDS: GenoType NTM-DR, Mycobacterium avium complex, Mycobacterium chimaera, genotypic identification
INTRODUCTION
Mycobacterium chimaera is a slow-growing nontuberculous mycobacterium recently recognized as a novel species within the Mycobacterium avium complex (MAC) (1). Similarly to Mycobacterium avium and Mycobacterium intracellulare, M. chimaera is associated with infections primarily in immunosuppressed patients and in those with an underlying lung disease (2–4). However, identifying M. chimaera as the cause of these infections is difficult, as it is often misidentified as M. intracellulare using current phenotypic and molecular tests (5). Many commercial assays such as GenoType Mycobacterium CM (Hain Lifescience, Nehren, Germany), AccuProbe MAC culture identification test (Genprobe, United States), and Inno LiPA Mycobacteria (Immunogenetics, Ghent, Belgium) cannot distinguish between these two closely related species (6).
The importance of accurate identification of M. chimaera is highlighted by its recent emergence as a major cause of life-threatening cardiovascular infections following open heart or vascular surgery (5, 7–11). Recent outbreaks of invasive M. chimaera infections following cardiac surgery have been reported in several countries in Europe and in the United States (10–13). Investigations in Switzerland have demonstrated an airborne transmission route of M. chimaera infection, whereby aerosols from contaminated water in heater-cooler units are transmitted to the patient during open heart surgery (12). Following these reports, many mycobacteriology reference laboratories are assisting national investigations in identification of clinical and environmental isolates of M. chimaera (12, 14).
To date, accurate identification of M. chimaera has required complex molecular techniques based on sequencing the 16S rRNA gene and/or the 16S-23S internal transcribed spacer (ITS) region (1, 5). GenoType NTM-DR ver. 1.0 (Hain Lifescience, Nehren, Germany) is a new commercial assay based on reverse hybridization of a PCR product to a nitrocellulose strip containing immobilized probes which are specific for species belonging to the MAC, as well as to the Mycobacterium abscessus complex. The GenoType NTM-DR ver. 1.0 assay targets the 23S rRNA gene region for the identification of species belonging to the MAC, whereas the erm gene is used to differentiate between members of the M. abscessus complex. The binding of the amplified product to a specific probe results in the development of a colored band on the strip which allows the species to be identified by its banding pattern.
The aim of this study was to identify M. chimaera isolates among MAC isolates in a retrospective collection that were previously identified to the species level with the GenoType Mycobacterium CM assay. In this study, gene sequencing methods were used to reidentify MAC species and the specificity of the GenoType NTM-DR assay for identifying species belonging to the MAC was determined.
RESULTS
Sequencing of both 16S rRNA and ITS genes reidentified 79/144 (55%) of our M. intracellulare isolates as M. chimaera. All nonclinical isolates were also identified as M. chimaera by sequencing the two gene regions. However, only 49/144 (34%) of M. intracellulare identifications by 16S rRNA and ITS gene sequencing were in concordance with the original identifications that were made with the GenoType Mycobacterium CM assay. On the other hand, ITS gene sequencing identified 16/144 (11%) as other species, including Mycobacterium arosiense (n = 2), Mycobacterium timonense (n = 3), Mycobacterium bouchedurhonense (n = 5), and Mycobacterium marseillense (n = 6). All 29 M. avium isolates previously identified by the GenoType Mycobacterium CM assay were also in 100% agreement with 16S rRNA and ITS gene sequencing results. Following this, the 173 MAC isolates were tested with the GenoType NTM-DR assay.
The GenoType NTM-DR assay correctly identified 157/173 (91%) of MAC isolates to the species level, while misidentification occurred in 16 cases (Table 1). Results for all 79 M. chimaera, 49 M. intracellulare, and 29 M. avium isolates were in 100% agreement with 16S rRNA and ITS gene sequencing results. Misidentification of isolates occurred with 2 M. arosiense strains, 3 M. timonense strains, 5 M. bouchedurhonense strains, and 6 M. marseillense strains which were identified only by ITS gene sequencing. Although the GenoType NTM-DR assay does not contain specific probes for these uncommon species, the hybridization patterns identified them as M. intracellulare. The sensitivity and specificity of the GenoType NTM-DR assay for the identification of M. chimaera were 100%.
TABLE 1.
Comparison of gene sequencing methods and GenoType NTM-DR for the identification of 173 Mycobacterium avium complex species
| No. of strains | Gene sequencing result |
GenoType NTM-DR result |
||
|---|---|---|---|---|
| 16S rRNA | 16S-23S ITS | Band patternsa | Identification | |
| 29 | M. avium | M. avium | CC, UC, SP1 | M. avium |
| 49 | M. intracellulare | M. intracellulare | CC, UC, SP2 | M. intracellulare |
| 79 | M. chimaera | M. chimaera | CC, UC, SP2, SP3 | M. chimaera |
| 2 | MACb | M. arosiense | CC, UC, SP2 | M. intracellulare |
| 3 | MAC | M. timonense | CC, UC, SP2 | M. intracellulare |
| 5 | MAC | M. bouchdurhonense | CC, UC, SP2 | M. intracellulare |
| 6 | MAC | M. marseillense | CC, UC, SP2 | M. intracellulare |
A faint band was produced in the universal control for all the isolates tested, and only bands with intensities stronger than the universal control were considered. CC, conjugate control probe; UC, universal control probe; SP1 to SP3, species-specific probes.
MAC, Mycobacterium avium complex.
DISCUSSION
In this study, GenoType NTM-DR was evaluated for the identification of M. chimaera and other Mycobacterium species belonging to the MAC. The novelty of this assay is that it allows differentiation of M. intracellulare from M. chimaera. With the use of 16S rRNA and ITS gene sequencing, 79 M. chimaera isolates were identified from a collection of 144 isolates which were previously identified as M. intracellulare. The ITS sequences obtained showed 100% identity with that of M. chimaera FI-0169T (1). A similar retrospective Belgian study found that 63% of clinical M. chimaera isolates had been incorrectly identified with the GenoType Mycobacterium CM assay (15). Results of recent whole-genome sequencing of respiratory M. chimaera isolates also support the use of 16S rRNA and ITS gene sequencing to distinguish M. intracellulare and M. chimaera, as there is an even greater divergence between these two species at the genomic level (16).
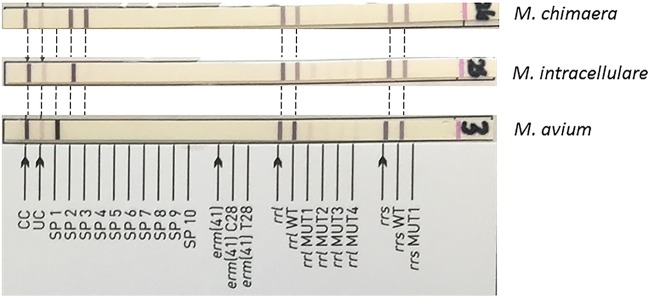
All MAC isolates identified by 16S rRNA and ITS gene sequencing were tested with the GenoType NTM-DR assay. Of 173 isolates, 157 developed the expected banding pattern and 16 were incorrectly identified. A conjugate control band was produced on the strips of all tested isolates, indicating the efficiency of the conjugate binding and substrate reaction for the production of a colored band on the strip. However, faint bands were produced in the universal control (UC) for all the isolates tested (Fig. 1). This may have been due to the inefficient amplification of DNA which was displayed when confirmation of PCR product was performed. All M. chimaera isolates developed strong species-specific (SP) bands at SP probe 2 (SP2) and SP3 only. The interpretation of banding patterns present which represent more than one species must be carefully performed, and only patterns showing strong species-specific bands should be taken into account. This limitation has also been noted with the previous line probe assay GenoType Mycobacterium CM and the AS assay (Hain Lifescience, Nehren, Germany) (17). In our study, some M. chimaera isolates also produced a weak band at SP1, which might have been interpreted as a mixture of M. avium and M. chimaera. However, these were not considered true species-specific bands, as only bands that were much stronger than that seen with the UC were taken into account. Similarly, all M. intracellulare isolates tested produced a strong species-specific band at SP2 only and very few M. intracellulare isolates produced a faint band at SP3. However, these bands were always very faint or fainter than that seen with the UC and were similarly disregarded. Likewise, all M. avium isolates produced a strong SP1 band only, resulting in correct identification. From our study, we would propose that only those bands with intensities that are stronger than that seen with the UC should be considered.
FIG 1.
GenoType NTM-DR test strips of M. chimaera, M. intracellulare, and M. avium. All 79 M. chimaera, 49 M. intracellulare, and 29 M. avium isolates produced the correct species banding pattern. However, the universal control (UC) band were very faint in all the isolates tested and only bands with intensities stronger than those seen with the UC were considered. No antibiotic resistance mutation bands were present in the three test strips shown at the top of the figure.
The MAC also comprises other species which are often misidentified as M. intracellulare with the GenoType Mycobacterium CM assay (18). We observed several other MAC species, including M. arosiense, M. marseillense, M. timonense, and M. bouchedurhonense, which were misidentified as M. intracellulare with the GenoType Mycobacterium CM assay (19). Likewise, GenoType NTM-DR misidentifies these species also, as they cross-react with the M. intracellulare species probes present on the strips. However, those other species are isolated from clinical samples infrequently. Similarly, misidentification of M. arosiense as M. intracellulare with the line probe assays Accuprobe, Inno LiPa Mycobacteria, and GenoType Mycobacterium CM has been previously reported (18, 20). However, there is a lack of published information on these less frequently encountered species and further research is needed to determine their clinical significance and relationship to M. intracellulare. Another limitation to our study is that other MAC species such as Mycobacterium vulneris and Mycobacterium colombiense were not tested with the GenoType NTM-DR assay, and we cannot be sure whether they would react with the probes present on the strip. However, these species are rarely reported and we did not identify these species in our collection of isolates referred between 2007 and 2016.
GenoType NTM-DR also contains probes for detecting genes conferring antibiotic resistance to macrolides and aminoglycosides. Resistance to these classes of antibiotics is encoded by the rrl and rrs genes, respectively, and they have been reported previously in the Mycobacterium abscessus complex (21, 22). Among those studied in our collection of MAC isolates, only two M. intracellulare isolates developed a corresponding mutation band for macrolide resistance (rrl MUT2; mutation A2058G) or aminoglycoside resistance (rrs MUT1; mutation A1408G). Retrospective analysis confirmed that these two isolates were phenotypically resistant to clarithromycin (MIC, >64 μg/ml) or resistant to amikacin (MIC not available). Resistance to macrolides and aminoglycosides was not detected in our collection of M. chimaera and M. avium isolates with this assay (data not shown). Further research is needed to evaluate the performance of this assay for characterizing antibiotic resistance genes.
In conclusion, the GenoType NTM-DR assay allows for rapid and specific detection of M. chimaera isolates and is in good agreement with 16S rRNA and ITS gene sequencing. Despite some of the limitations of this novel assay, it allows M. chimaera to be distinguished from M. intracellulare, with 100% specificity. GenoType NTM-DR is suitable for use in diagnostic laboratories as it is simple, cost-effective, and more accessible than gene sequencing methods. The GenoType NTM-DR kit contains 96 test strips, and the assay can be performed without the need to refer samples to other laboratories for gene sequencing. This assay will be an invaluable test in addition to the GenoType Mycobacterium CM assay for the identification of M. chimaera, as the differentiation of MAC species is becoming more important.
MATERIALS AND METHODS
MAC isolate collection.
All 173 MAC isolates had been referred to the Irish Mycobacteria Reference Laboratory between 2007 and 2016. Of these isolates, 162 were previously recovered from clinical specimens from 119 patients and 11 isolates were recovered from water samples taken from heater-cooler units referred by Irish cardiothoracic surgery centers. The collection of MAC isolates consisted of 144 M. intracellulare and 29 M. avium isolates which were subcultured from frozen pure cultures at −80°C in a Bactec MGIT 960 liquid culture system (Becton Dickinson and Company, USA). Species identification had been performed with the GenoType Mycobacterium CM assay (Hain Lifescience, Nehren, Germany) as part of routine diagnostics. Further identification methods were not in place to identify M. chimaera at the time. A reference strain of M. chimaera DSM 44623 was also cultured in a Bactec MGIT 960 liquid culture system and was included as a positive control for 16S rRNA and ITS gene sequencing and also for the GenoType NTM-DR assay (1).
16S rRNA and ITS gene sequencing.
Identification of MAC isolates to the species level was performed using 16S rRNA and ITS gene sequencing. Mycobacterial DNA was extracted from liquid cultures using a GenoLyse (Hain Lifescience, Nehren, Germany) extraction kit according to the manufacturer's instructions, and the 16S rRNA gene was amplified using Seq1 forward and Seq2 reverse primers as previously described (23). Similarly, the ITS region was amplified using Sp1 forward and Sp2 reverse primers (5, 24). PCR amplification was performed on a Veriti 96-well thermal cycler (Applied Biosystems, USA), and subsequent PCR products were sequenced using 244 and 259 primers for 16S rRNA PCR products as previously described (23, 25). ITS gene products were sequenced with the same primers as those used for DNA amplification. Raw sequence data were analyzed using DNAStar Lasergene software version 12.2, and aligned sequences were compared with sequences available on the National Center for Biotechnology Information (NCBI) website using the Basic Local Alignment Search Tool (BLAST).
GenoType NTM-DR assay.
The GenoType NTM-DR ver. 1.0 assay was performed according to the manufacturer's instructions, and the same DNA extracts as those that had been used for gene sequencing were used for the hybridization assay. For amplification of DNA, a master mix containing 10 μl AM-A and 35 μl AM-B per sample was prepared, and 5 μl of DNA was added to each PCR tube (Hain Lifescience, Nehren, Germany). The amplification profile consisted of 15 min of denaturation at 95°C, followed by 10 cycles consisting of 30 s at 95°C and 2 min at 65°C, and an additional 20 cycles comprising 25 s at 95°C, 40 s at 50°C, and 40 s at 70°C, followed by a final extension for 8 min at 70°C. A positive-control isolate of M. chimaera DSM 44623 was also included for each assay.
Hybridization of amplified products was performed on an automated GT-Blot 48 instrument (Hain Lifescience, Nehren, Germany) after confirmation of amplified products on a 2% agarose gel. Hybridization buffer (1 ml) was added by the instrument to each strip followed by a stop to allow addition of membrane strips. Following the hybridization procedure, the strips were removed from the instrument and were allowed to dry. Each strip was labeled and pasted to an evaluation sheet. Results were then interpreted using a Genotype NTM-DR interpretation chart (Fig. 2).
FIG 2.
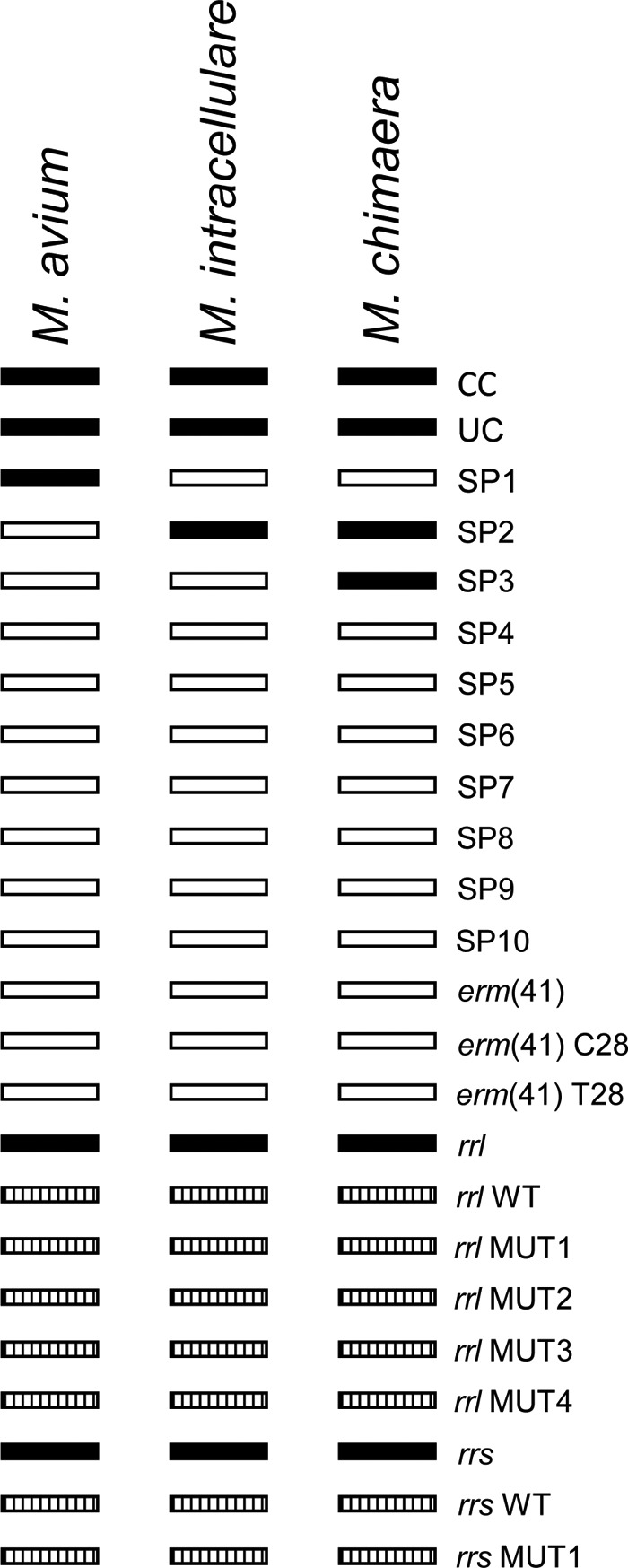
Interpretation chart for the GenoType NTM-DR assay. The specificities of the bands are as follows: CC, conjugate control probe; UC, universal control probe; SP1 to SP10, species-specific probes [specific probes for macrolide resistance mediated by erm(41) which are not applicable to members of the Mycobacterium avium complex, specific probes for the rrl gene and rrs gene, and mutation probes for possible detection of resistance to macrolides and aminoglycosides]. Filled boxes indicate complete staining, striped boxes indicate facultative staining, and blank boxes indicate no staining. WT, wild type.
ACKNOWLEDGMENTS
We acknowledge the many Irish laboratories, and our own hospital's clinicians, for referring isolates and/or samples which generated this archival collection.
We acknowledge support and funding received from the Clinical Microbiology Department, Trinity College Dublin, the Irish Mycobacteria Reference Laboratory and Microbiology Department, and LabMed Directorate, St. James' Hospital, Dublin, Ireland. We also thank Hain Lifescience for reducing the cost of GenoType NTM-DR kits used in this study.
REFERENCES
- 1.Tortoli E, Rindi L, Garcia MJ, Chiaradonna P, Dei R, Garzelli C, Kroppenstedt RM, Lari N, Mattei R, Mariottini A, Mazzarelli G, Murcia MI, Nanetti A, Piccoli P, Scarparo C. 2004. Proposal to elevate the genetic variant MAC-A, included in the Mycobacterium avium complex, to species rank as Mycobacterium chimaera sp. nov. Int J Syst Evol Microbiol 54:1277–1285. doi: 10.1099/ijs.0.02777-0. [DOI] [PubMed] [Google Scholar]
- 2.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Winthrop K, ATS Mycobacterial Diseases Subcommittee, American Thoracic Society Infectious Disease Society of America. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 3.Shah NM, Davidson JA, Anderson LF, Lalor MK, Kim J, Thomas HL, Lipman M, Abubakar I. 2016. Pulmonary Mycobacterium avium-intracellulare is the main driver of the rise in non-tuberculous mycobacteria incidence in England, Wales and Northern Ireland, 2007–2012. BMC Infect Dis 16:195. doi: 10.1186/s12879-016-1521-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle DP, Zembower TR, Reddy S, Qi C. 2015. Comparison of clinical features, virulence, and relapse among Mycobacterium avium complex species. Am J Respir Crit Care Med 191:1310–1317. doi: 10.1164/rccm.201501-0067OC. [DOI] [PubMed] [Google Scholar]
- 5.Schweickert B, Goldenberg O, Richter E, Göbel UB, Petrich A, Buchholz P, Moter A. 2008. Occurrence and clinical relevance of Mycobacterium chimaera sp. nov., Germany. Emerg Infect Dis 14:1443–1446. doi: 10.3201/eid1409.071032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russo C, Tortoli E, Menichella D. 2006. Evaluation of the new GenoType Mycobacterium Assay for identification of mycobacterial species. J Clin Microbiol 44:334–339. doi: 10.1128/JCM.44.2.334-339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sax H, Bloemberg G, Hasse B, Sommerstein R, Kohler P, Achermann Y, Rössle M, Falk V, Kuster SP, Böttger EC, Weber R. 2015. Prolonged outbreak of Mycobacterium chimaera infection after open-chest heart surgery. Clin Infect Dis 61:67–75. doi: 10.1093/cid/civ198. [DOI] [PubMed] [Google Scholar]
- 8.Bills ND, Hinrichs SH, Aden TA, Wickert RS, Iwen PC. 2009. Molecular identification of Mycobacterium chimaera as a cause of infection in a patient with chronic obstructive pulmonary disease. Diagn Microbiol Infect Dis 63:292–295. doi: 10.1016/j.diagmicrobio.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Phadke VK, Hirsh DS, Goswami ND. 2016. Patient report and review of rapidly growing mycobacterial infection after cardiac device implantation. Emerg Infect Dis 22:389. doi: 10.3201/eid2203.150584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Achermann Y, Rössle M, Hoffmann M, Deggim V, Kuster S, Zimmermann DR, Bloemberg G, Hombach M, Hasse B. 2013. Prosthetic valve endocarditis and bloodstream infection due to Mycobacterium chimaera. J Clin Microbiol 51:1769–1773. doi: 10.1128/JCM.00435-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohler P, Kuster SP, Bloemberg G, Schulthess B, Frank M, Tanner FC, Rössle M, Böni C, Falk V, Wilhelm MJ, Sommerstein R, Achermann Y, Ten Oever J, Debast SB, Wolfhagen MJHM, Brandon Bravo Bruinsma GJ, Vos MC, Bogers A, Serr A, Beyersdorf F, Sax H, Böttger EC, Weber R, van Ingen J, Wagner D, Hasse B. 2015. Healthcare-associated prosthetic heart valve, aortic vascular graft, and disseminated Mycobacterium chimaera infections subsequent to open heart surgery. Eur Heart J 36:2745–2753. doi: 10.1093/eurheartj/ehv342. [DOI] [PubMed] [Google Scholar]
- 12.Sommerstein R, Rüegg C, Kohler P, Bloemberg G, Kuster SP, Sax H. 15 June 2016. Transmission of Mycobacterium chimaera from heater-cooler units during cardiac surgery despite an ultraclean air ventilation system. Emerg Infect Dis J doi: 10.3201/eid2206.160045. [DOI] [PMC free article] [PubMed]
- 13.Perkins KM, Lawsin A, Hasan NA, Strong M, Halpin AL, Rodger RR, Moulton-Meissner H, Crist MB, Schwartz S, Marders J, Daley CL, Salfinger M, Perz JF. 2016. Notes from the field: Mycobacterium chimaera contamination of heater-cooler devices used in cardiac surgery - United States. MMWR Morb Mortal Wkly Rep 65:1117–1118. doi: 10.15585/mmwr.mm6540a6. [DOI] [PubMed] [Google Scholar]
- 14.Sommerstein R, Schreiber PW, Diekema DJ, Edmond MB, Hasse B, Marschall J, Sax H. 14 November 2016. Mycobacterium chimaera outbreak associated with heater-cooler devices: piecing the puzzle together. Infect Control Hosp Epidemiol doi: 10.1017/ice.2016.283. [DOI] [PubMed] [Google Scholar]
- 15.Soetaert K, Vluggen C, André E, Vanhoof R, Vanfleteren B, Mathys V. 2016. Frequency of Mycobacterium chimaera among Belgian patients, 2015. J Med Microbiol 65:1307–1310. doi: 10.1099/jmm.0.000359. [DOI] [PubMed] [Google Scholar]
- 16.Mac Aogáin M, Roycroft E, Raftery P, Mok S, Fitzgibbon M, Rogers TR. 2015. Draft genome sequences of three Mycobacterium chimaera respiratory isolates. Genome Announc 3:e01409-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richter E, Rüsch-Gerdes S, Hillemann D. 2006. Evaluation of the GenoType Mycobacterium Assay for identification of mycobacterial species from cultures. J Clin Microbiol 44:1769–1775. doi: 10.1128/JCM.44.5.1769-1775.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tortoli E, Pecorari M, Fabio G, Messinò M, Fabio A. 2010. Commercial DNA probes for mycobacteria incorrectly identify a number of less frequently encountered species. J Clin Microbiol 48:307–310. doi: 10.1128/JCM.01536-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben Salah I, Cayrou C, Raoult D, Drancourt M. 2009. Mycobacterium marseillense sp. nov., Mycobacterium timonense sp. nov and Mycobacterium bouchedurhonense sp nov, members of the Mycobacterium avium complex. Int J Syst Evol Microbiol 59:2803–2808. doi: 10.1099/ijs.0.010637-0. [DOI] [PubMed] [Google Scholar]
- 20.Tortoli E, Adriani B, Baruzzo S, Degl'Innocenti R, Galanti I, Lauria S, Mariottini A, Pascarella M. 2009. Pulmonary disease due to Mycobacterium arosiense, an easily misidentified pathogenic novel mycobacterium. J Clin Microbiol 47:1947–1949. doi: 10.1128/JCM.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nessar R, Reyrat JM, Murray A, Gicquel B. 2011. Genetic analysis of new 16S rRNA mutations conferring aminoglycoside resistance in Mycobacterium abscessus. J Antimicrob Chemother 66:1719–1724. doi: 10.1093/jac/dkr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bastian S, Veziris N, Roux A-L, Brossier F, Gaillard J-L, Jarlier V, Cambau E. 2011. Assessment of clarithromycin susceptibility in strains belonging to the Mycobacterium abscessus group by erm(41) and rrl sequencing. Antimicrob Agents Chemother 55:775–781. doi: 10.1128/AAC.00861-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth A, Fischer M, Hamid ME, Michalke S, Ludwig W, Mauch H. 1998. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J Clin Microbiol 36:139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roth A, Reischl U, Streubel A, Naumann L, Kroppenstedt RM, Habicht M, Fischer M, Mauch H. 2000. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S-23S rRNA gene spacer and restriction endonucleases. J Clin Microbiol 38:1094–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirschner P, Springer B, Vogel U, Meier A, Wrede A, Kiekenbeck M, Bange FC, Böttger EC. 1993. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J Clin Microbiol 31:2882–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]