Abstract
Streptococcus mutans has been recognized as an important etiological agent in human dental caries. Some strains of S. mutans also produce bacteriocins. In this study, we sought to demonstrate that bacteriocin production by S. mutans strains GS5 and BM71 was mediated by quorum sensing, which is dependent on a competence-stimulating peptide (CSP) signaling system encoded by the com genes. We also demonstrated that interactions with some other oral streptococci interfered with S. mutans bacteriocin production both in broth and in biofilms. The inhibition of S. mutans bacteriocin production by oral bacteria was stronger in biofilms than in broth. Using transposon Tn916 mutagenesis, we identified a gene (sgc; named for Streptococcus gordonii challisin) responsible for the inhibition of S. mutans bacteriocin production by S. gordonii Challis. Interruption of the sgc gene in S. gordonii Challis resulted in attenuated inhibition of S. mutans bacteriocin production. The supernatant fluids from the sgc mutant did not inactivate the exogenous S. mutans CSP as did those from the parent strain Challis. S. gordonii Challis did not inactivate bacteriocin produced by S. mutans GS5. Because S. mutans uses quorum sensing to regulate virulence, strategies designed to interfere with these signaling systems may have broad applicability for biological control of this caries-causing organism.
Cell-cell communication is a fundamental activity performed by most types of cells. Previously, bacteria were considered to exist as individual cells that scavenge for nutrients and multiply independently. However, recent studies have shown that bacteria are capable of coordinated activities, such as quorum sensing (13). Quorum sensing is the control of gene expression in response to cell density. The mediators for this intercellular communication among bacterial cells are self-secreted signaling molecules called autoinducers. These signaling molecules include N-acylhomoserine lactones, used by gram-negative bacteria (8), and small peptides, effectors of gram-positive bacteria (6). Gram-positive bacteria use two-component signal transduction systems for detection of autoinducers (13). Quorum sensing also regulates virulence in many human pathogens (2). Examples of quorum-sensing-dependent systems in gram-positive bacteria include the formation of biofilms (18, 33), the development of genetic competence (10, 16), the acid tolerance response (18), and the production of antimicrobial peptides (12), which may all provide a competitive advantage over other organisms. Recent studies have shown that quorum sensing modulates both intra- and interspecies cell-cell communications. For example, there is cross-inhibition by quorum-sensing pheromones between Staphylococcus aureus and Staphylococcus epidermidis (21). S. epidermidis pheromones inhibited the S. aureus agr response, which may explain the predominance of S. epidermidis on the skin. Interaction between Bacillus subtilis and Erwinia carotovora provides another example (5), since the former produces an enzyme (AiiA) that inactivates the E. carotovora autoinducers. Inactivation of the autoinducers by AiiA attenuated the virulence of E. carotovora.
In the oral cavity, bacteria interact with each other as they attempt to establish themselves in this environment. Oral streptococci are gram-positive, facultative anaerobes that initiate the formation of biofilms (known as dental plaque) on tooth surfaces. Streptococcus mutans, one of the colonizers of dental plaque, exhibits an acid tolerance response (16). Some S. mutans strains are naturally transformable (14, 15) and produce bacteriocins (3, 22-24). Recent studies have demonstrated that biofilm formation, acid tolerance, and the transformation of S. mutans are mediated by quorum sensing (4). This signaling system depends on a competence-stimulating peptide (CSP) and a two-component signal transduction system, which are encoded by the comCDE genes, corresponding to the CSP precursor, a histidine kinase (receptor for CSP), and a response regulator, respectively (4, 13, 20). By interfering with this cell-cell signaling mechanism, caries-causing S. mutans bacteria that use quorum sensing to control virulence could potentially be rendered avirulent.
Many gram-positive bacteria produce antimicrobial peptides called bacteriocins (11, 28). Although these peptide molecules are not required for growth, they may help the microorganisms that produce them to compete for the limited nutrients in their environment (31). For example, a mixed culture of bacteriocin-producing S. mutans SW31 with sensitive Streptococcus sanguis Ny101 resulted in a plaque composed primarily of S. mutans SW31. A nonbacteriocinogenic mutant of S. mutans SW31 allowed substantial growth of S. sanguis Ny101 (32). It has been shown that production of these antimicrobial peptides is frequently regulated by a two-component regulatory system through a quorum-sensing system in some gram-positive bacteria (12). However, this has not yet been demonstrated for S. mutans bacteriocin production.
Fabio et al. (7) studied a sample of human streptococci (mainly S. mutans species) from dental plaques to evaluate the production frequency and activity spectrum of bacteriocins. Most of the 55 S. mutans strains (89%) produced substances with a wide spectrum of antibacterial activity. The bacteriocins produced showed a marked inhibitory activity against gram-positive bacteria. How interspecies interactions with other oral bacteria affects S. mutans bacteriocin production is not yet understood. The objectives of the present study were to demonstrate that S. mutans bacteriocin production is a quorum-sensing-controlled phenomenon and to establish the role of interactions with other oral bacteria on bacteriocin production by S. mutans.
MATERIALS AND METHODS
Bacterial strains and media.
S. mutans GS5 and its comC, comD, and comE mutants (33), S. mutans BM71 and its com mutants (obtained from D. G. Cvitkovitch, University of Toronto, Toronto, Ontario, Canada), S. mutans NG8, Streptococcus gordonii Challis, S. gordonii 10558, S. sanguis 10556, Streptococcus mitis 10712, and Streptococcus oralis KS32AR were maintained on tryptic soy agar (TSA) (Difco, Detroit, Mich.) plates supplemented with erythromycin (10 μg/ml) where indicated. Bacteria were routinely cultured in Todd-Hewitt broth (THB) (Difco). A group C streptococcal strain RP66 was used as an indicator for assays of S. mutans bacteriocin activity (22). Escherichia coli DH5α competent cells (Life Technologies, Gaithersburg, Md.) were used in construction of an S. gordonii Challis inactivation vector.
Bacteriocin production and assays. (i) Agar plate assays.
S. mutans GS5 or BM71 cells in the presence or absence of other oral streptococcal cells were inoculated into THB agar plates with or without the addition of exogenous synthetic CSP (3 μg in 3 μl) of S. mutans (the amino acid sequence of CSP was SGSLSTFFRLFNRSFTQALGK [17]) (synthesized by Sigma-Genosys, The Woodlands, Tex.) for stab cultures. After 24 h of incubation at 37°C under anaerobic conditions, the plates were overlaid with indicator RP66 cells (106 CFU) in 3 ml of THB with 1% low-melting-point agarose (BioProducts, Rockland, Maine). After 24 more hours of incubation at 37°C under anaerobic conditions, the diameters of the clearance zones surrounding the inoculated bacteria (which indicated the presence of bacteriocin produced by S. mutans) were measured.
(ii) THB broth assays.
S. mutans GS5 or BM71 cells (107 CFU) or mixtures with other oral streptococcal cells at equal densities were cultured in 1.0-ml portions of THB supplemented with 3% yeast extract (THBY) with or without the addition of exogenous synthetic CSP (final concentration, 1.0 μg/ml) for 24 h at 37°C. Supernatant fluids containing bacteriocin from the cultures were filtered through 0.22-μm-pore-size filters and either assayed immediately or frozen at −20°C for subsequent assays. RP66 cells (2 × 105 CFU) in 0.7 ml of THB were grown in the presence of the supernatant fluids mentioned above and incubated at 37°C for 5 to 6 h. The optical density at 600 nm (OD600) was measured and indicated the presence or absence of bacteriocin. The presence or absence of bacteriocin using arbitrary units (microliters of supernatant used to obtain complete inhibition of RP66 growth) was scored as follows: 10 μl, +++; 100 μl, ++; 300 μl, +; and no inhibition, −.
(iii) Agar well assays.
The supernatant fluids containing bacteriocin from S. mutans GS5 cultures were added into precut wells in THB agar plates and incubated at 37°C for 24 h. The wells were then filled using THB with 1% low-melting-point agarose, and the plates were overlaid with the indicator strain RP66 (106 CFU) in 3 ml of THB with 1% low-melting-point agarose. After 24 more hours of incubation at 37°C under anaerobic conditions, the diameters of the inhibition zones surrounding the wells were measured.
(iv) Killing assays.
A 100-μl aliquot from S. mutans GS5 or BM71 or mixtures with S. gordonii Challis from broth or biofilm cultures was mixed with 5 × 104 CFU of RP66 indicator cells (in 100 μl of THB) and incubated at 37°C for 1 h (modified version of the method of Paul and Slade [22]). Two-microliter aliquots of the cultures were plated on the surfaces of TSA plates and incubated for another 24 h at 37°C under anaerobic conditions. The percent killing of RP66 cells was calculated as follows: [100 − (number of colonies in the presence of supernatants/number of colonies in THB)] × 100.
Effects of oral streptococci on the CSP of S. mutans.
Supernatant fluids from various oral streptococci grown to the stationary phase in THB were filtered through 0.22-μm-pore-size filters to eliminate cellular components. The supernatant fluids were then neutralized with NaOH to pH 7.0 to 7.5. Exogenous synthetic S. mutans CSP (final concentration, 2.5 μg/ml) was incubated with supernatant fluid at 37°C for 0.5 to 3 h. For some experiments, part of the supernatant fluid was boiled for 10 min before the incubation with CSP. CSP thus treated was added to S. mutans GS5 comC mutant cells (107 CFU) in 1.0 ml of THBY to a final concentration of 0.25 μg/ml. After a 24-h incubation at 37°C, the supernatant fluids from the cultures were passed through 0.22-μm-pore-size filters, and the bacteriocin level produced by the S. mutans GS5 comC mutant was determined by the agar well assays.
Transposon mutagenesis of S. gordonii Challis.
S. gordonii Challis cells were randomly mutagenized by transformation with pAM620, a plasmid containing transposon Tn916 (9). The individual clones of the transformants on TSA plates supplemented with tetracycline (10 μg/ml) were cultured in THB. Mixtures with S. mutans GS5 in THB (0.7:1 by volume at the stationary phase) were inoculated into THB agar plates for stab cultures, and the clones with decreased ability to inhibit S. mutans GS5 bacteriocin production were selected for further investigation.
Sequencing of chromosomal DNA flanking Tn916 from insertional mutants of S. gordonii Challis.
Chromosomal DNA was extracted from a Tn916 insertion mutant of S. gordonii Challis (designated 41A9) using a Puregene DNA isolation kit (Gentra System, Inc., Minneapolis, Minn.). The DNA was digested with BclI (with a cutting site 2.2 kb from the N terminus of Tn916) and a combination of BclI with SalI, HindIII, PstI, KpnI, SacI, or EcoRI. Southern blot analysis was performed according to the protocol for the ECL detection system (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom) using PCR products of the N-terminal end of Tn916 as the probe. The DNA fragments digested with PstI and BclI were ligated with pUC19 (PstI and BamHI digested) using T4 ligase. The ligation mixtures were then used as PCR templates. PCR was performed using one primer of the Tn916 sequence (5′-CGTCGTATCAAAGCTC-3′) and another primer from the pUC19 vector sequence (5′-AGCTCACTCATTAGGCAC-3′). Both primers were designed in the 3′ direction toward the chromosomal DNA flanking Tn916. The resulting PCR fragments were sequenced by the sequencing facility at the Roswell Park Cancer Institute (Buffalo, N.Y.).
Construction of the S. gordonii Challis protease knockout mutant BYW1.
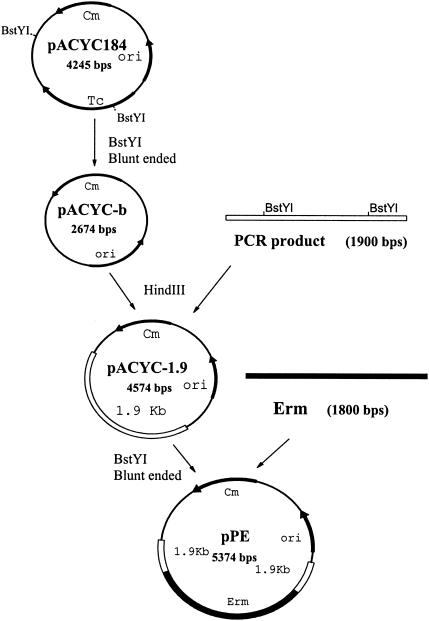
pPE (Challis protease knockout inactivation vector) was constructed as described in Results (see Fig. 3). A plasmid, pACYC184, that replicates only in E. coli, was digested with BstYI. The resulting 2.67-kb fragment was blunt ended with T4 DNA polymerase and self-ligated with T4 ligase to obtain pACYC-b. A 1.9-kb PCR product, obtained using primers at the N-terminal end of the S. gordonii Challis sgc gene (forward primer, 5′-ACGAAGCTTTACGAGGTGTCAG-3′; reverse primer, 5′-CAAGTCGCAAGCTTCTGAAG-3′), was digested with HindIII (HindIII sites were designed into the primers) and ligated with pACYC-b (digested with HindIII), using T4 ligase. The resulting plasmid, pACYC-1.9, was then digested with BstYI to delete a 1.0-kb fragment of the sgc gene fragment. The remaining 3.57-kb pACYC-1.9 fragment was blunt ended using T4 DNA polymerase and ligated to the SmaI-digested Erm cassette (released from pEMnP [B. Wang, personal communication]), using T4 ligase. The final plasmid, pPE, contained two 400-bp fragments of the sgc gene flanking the Erm cassette. S. gordonii Challis strains with knockout mutations of the sgc gene were constructed via a double crossover event into the Challis chromosome at the predicted sgc site by transforming Challis using XbaI-linearized pPE. The mutants were selected on TSA plates supplemented with erythromycin (10 μg/ml).
FIG. 3.
Construction of the S. gordonii Challis vector pPE. As described in the text, pPE was constructed containing an interrupted sgc gene of S. gordonii Challis.
Zymography of supernatant fluids from S. gordonii Challis and its protease mutants.
The supernatant fluids from broth cultures of S. gordonii Challis or sgc mutants grown overnight were filtered through 0.22-μm-pore-size filters and precipitated with 40% (wt/vol) ammonium sulfate at room temperature for 1 h, with constant shaking. After 30 min of centrifugation at 15,000 × g, the pellets were resuspended in phosphate-buffered saline (PBS) and 30-K Centricon ultrafilters (Millipore Co., Bedford, Mass.) were utilized to further concentrate the supernatant fluids. The supernatant fluids (concentrated 500-fold) were applied to ready-made casein gels (Bio-Rad, Hercules, Calif.), and zymography was performed following the supplier's protocol.
Azocasein assays for the protease activity of S. gordonii Challis.
Azocasein assays were performed by a modified version of the method of Sojar et al. (27). Briefly, 50-ml cultures of S. gordonii Challis or its sgc mutants grown overnight were centrifuged at 4,000 × g for 10 min. The supernatant fluids from these 50-ml cultures were then concentrated as described above for the zymography experiments. The cell pellets and the concentrated supernatant fluids from 50-ml cultures were utilized in the azocasein assays. The OD420 measured in these assays indicated the amount of protease present.
Biofilm formation.
S. mutans BM71 cells (107 CFU) or mixtures with other oral streptococcal cells were inoculated into 2-ml portions of one-quarter strength THB supplemented with 0.01% mucin (Sigma Chemical Co., St. Louis, Mo.) and 0.1% glucose (biofilm medium [BM]) (17) in six-well polystyrene microtiter plates to form biofilms. After 24 h of incubation at 37°C under aerobic conditions, the planktonic cells were discarded, and 1.5 ml of THBY was added to each well containing biofilm cells and incubated at 37°C for another 24 h for bacteriocin production by S. mutans BM71 cells.
Reverse transcription-PCR.
Total RNA from S. mutans BM71 or GS5 alone and in mixtures with other streptococci was isolated from 15-ml samples of cultures (OD600 readings of 1.0 to 1.2). The cells were centrifuged at 4,000 × g for 10 min at 4°C. The cell pellets were resuspended in diethyl pyrocarbonate-treated water, and the suspensions were mixed vigorously with Lysing Matrix B (Q. Biogene, Carlsbad, Calif.) and Trizol (Invitrogen, Carlsbad, Calif.). After two washes with chloroform, RNA was precipitated with isopropanol. The RNA pellets were washed with 70% ethanol and resuspended in diethyl pyrocarbonate-treated water. After DNase (Promega, Madison, Wis.) treatment, 1 μg of RNA from each sample was utilized for reverse transcription, using SuperScript III RNase H− reverse transcriptase (Invitrogen) and a forward primer for the S. mutans GS5 bacteriocin gene (5′-CTGCAGTACTCCGGCATGTGCAATT-3′) (H. Yonezawa and H. K. Kuramitsu, submitted for publication), a forward primer for the S. mutans GS5 regulatory region of the bacteriocin operon (5′-TCCGAGCTCTGTGTGGAAACAAGGACTGATAA-3′), or a primer for S. mutans fructosyltransferase (ftf) gene (5′-GATCAAGCAGCTACCGTAAC-3′) (26). PCR was then performed using the cDNAs indicated as templates and primers for the S. mutans GS5 bacteriocin gene (forward, 5′-CTGCAGTACTCCGGCATGTGCAATT-3′; reverse, 5′-CCCTTTATTTCCCAATACAATG-3′), the S. mutans GS5 bacteriocin operon regulatory region (forward, 5′-TCCGAGCTCTGTGTGGAAACAAGGACTGATAA-3′; reverse, 5′-GGGGTACCTCATCTCTGCTATCCAGA-3′), or the S. mutans ftf gene (forward, 5′-TTGGGATTCTTGGCCAGTTC-3′; reverse, 5′-CAATGACATGTGGATCCGC-3′).
RESULTS
Bacteriocin production by S. mutans GS5 and BM71 is mediated by the CSP.
S. mutans GS5, BM71, or NG8 and their comC, comD, and comE knockout mutants were assayed for the production of bacteriocin using the agar plate assays. Of three strains surveyed, the wild-type S. mutans GS5 and BM71 produced significant amounts of bacteriocin as indicated by the diameter of the inhibition zone of the indicator strain RP66. However, their com mutants produced minimum amounts of bacteriocin (Table 1). The addition of exogenous CSP of S. mutans in the bacterial stab cultures restored the bacteriocinogenic ability of the comC mutant, but not that of the comD or comE mutant. These results indicated that bacteriocin production by S. mutans GS5 or BM71 is a CSP-dependent phenomenon. Recent results have demonstrated that BM71 produces the same bacteriocin as GS5 (Yonezawa and Kuramitsu, submitted). However, S. mutans NG8 did not produce detectable bacteriocin against RP66, even with the addition of exogenous CSP (data not shown). Therefore, the dependence (on CSP) of the production of the antimicrobial substances active against RP66 is strain specific.
TABLE 1.
Production of bacteriocin by S. mutans GS5 or BM71 in agar plate assaysa
| Bacterium | Diameter of inhibition zone (mm)b
|
|||
|---|---|---|---|---|
| Without CSP
|
With CSP
|
|||
| GS5 | BM71 | GS5 | BM71 | |
| Wild-type | 38.3 ± 1.7 | 39.0 ± 0.8 | 39.0 ± 2.4 | 39.3 ± 1.2 |
| comC mutant | 3.7 ± 2.9 | 1.7 ± 1.2 | 38.7 ± 2.1 | 39.0 ± 0.8 |
| comD mutant | 2.7 ± 2.1 | 1.3 ± 0.5 | 2.3 ± 1.7 | 1.7 ± 0.5 |
| comE mutant | 2.3 ± 1.7 | 1.7 ± 0.5 | 3.0 ± 2.2 | 2.3 ± 0.5 |
S. mutans GS5 or BM71 or their com mutant cells were inoculated into THB agar plates with or without the addition of exogenous synthetic CSP (3 μg in 3 μl) for stab cultures. After 24 h of incubation at 37°C under anaerobic conditions, the plates were overlaid with indicator RP66 cells (106 CFU) in 3 ml of THB with 1% low-melting-point agarose. After 24 more hours of incubation at 37°C under anaerobic conditions, the diameters of the clearance zones surrounding the inoculated bacteria were measured.
Values are means ± standard deviations from three experiments.
To confirm the results of the agar plate assays, another assay system, using THB, was used to investigate the production of bacteriocin in broth by S. mutans GS5 or BM71. Similar to the results in agar plate assays, S. mutans GS5 or BM71 produced bacteriocin in broth, but not its com mutants (data not shown). The addition of exogenous synthetic CSP also restored bacteriocin production by S. mutans GS5 comC mutant or BM71 comC mutant but did not restore bacteriocin production by the comD or comE mutant.
Inhibition of S. mutans bacteriocin production by other oral streptococci.
To investigate interactions among oral streptococci relative to bacteriocin production by S. mutans, other oral streptococcal cells were mixed with S. mutans GS5 cells (1:1 by volume at the stationary phase) and inoculated into THB agar plates with or without the addition of exogenous synthetic CSP. The inhibition zones of overlaid indicator strain RP66 were measured and are shown in Table 2. All of the other oral streptococci, except S. mutans NG8, inhibited bacteriocin production by S. mutans GS5 to different degrees. The addition of exogenous CSP fully restored bacteriocin production by S. mutans GS5 in the indicated mixed stab cultures. All of these other species of oral streptococci did not produce detectable bacteriocin against RP66 or GS5 in agar plate assays (data not shown).
TABLE 2.
Inhibition of S. mutans GS5 bacteriocin production by other oral streptococci in agar plate assaysa
| Bacterial mixturesb | Diameter of inhibition zone (mm)c
|
|
|---|---|---|
| Without CSP | With CSP | |
| GS5 + NG8 | 38.5 ± 2.5 | 39.0 ± 3.0 |
| GS5 + Challis | 5.5 ± 2.5 | 38.5 ± 2.5 |
| GS5 + 10558 | 20.5 ± 3.5 | 38.5 ± 2.5 |
| GS5 + 10556 | 22.5 ± 1.0 | 38.5 ± 2.5 |
| GS5 + 10712 | 14.0 ± 5.0 | 39.0 ± 3.0 |
| GS5 + KS32AR | 22.0 ± 6.0 | 38.5 ± 2.5 |
S. mutans GS5 cells in the presence or absence of other oral streptococcal cells were inoculated into THB agar plates with or without the addition of exogenous synthetic CSP (3 μg in 3 μl) for stab cultures. After 24 h of incubation at 37°C under anaerobic conditions, the plates were overlaid with indicator RP66 cells (106 CFU) in 3 ml of THB with 1% low-melting-point agarose. After 24 more hours of incubation at 37°C under anaerobic conditions, the diameters of the clearance zones surrounding the inoculated bacteria were measured.
Mixtures of S. mutans GS5 and NG8, S. gordonii Challis and 10558, S. sanguis 10556, S. mitis 10712, and S. oralis KS32AR were used.
Values are means ± standard deviations from two experiments.
THB broth assays were also performed to confirm the results from the agar plate assays. All of the oral streptococci, except S. mutans NG8, completely inhibited bacteriocin production by S. mutans GS5 or BM71 in broth (data not shown). S. mutans NG8 did not inhibit bacteriocin production by S. mutans GS5 or BM71 at all. The addition of exogenous CSP fully restored bacteriocin production in the mixed cultures to the level of wild-type S. mutans GS5 or BM71. We also examined the potential effects of the streptococci on the viability of S. mutans in mixed broth cultures. The cell numbers of S. mutans GS5 or BM71 in the mixtures with other species were comparable to those in the mixtures with NG8 (data not shown). These results indicated that inhibition of bacteriocin production by other species in the mixed cultures was not due to a reduction in cell numbers of S. mutans GS5 or BM71, since the amounts of bacteriocin produced by the mixtures of S. mutans GS5 or BM71 with NG8 were similar to those produced by S. mutans GS5 or BM71 single strain cultures.
Effects of supernatant fluids from oral streptococci on the CSP activity of S. mutans.
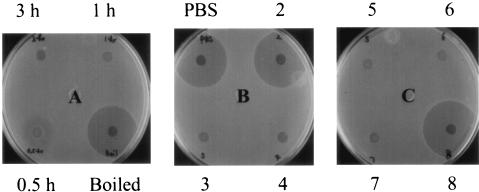
Next, experiments were performed to determine whether supernatant fluids from other species of oral streptococci could inactivate exogenous synthetic S. mutans CSP. S. mutans GS5 comC mutant cells were used in this experiment as an indicator of the levels of active CSP, since the mutant requires exogenous CSP to restore bacteriocin production (Table 1). As shown in Fig. 1A, incubating CSP with the supernatant fluid from S. gordonii Challis for more than 1 h completely inactivated CSP, while a 0.5-h incubation only partially inactivated CSP. Heating the supernatant fluid from S. gordonii Challis at 100°C for 10 min destroyed the inhibitory activity on S. mutans CSP, indicating that the substance(s) responsible for S. mutans CSP inactivation is heat sensitive. Figure 1B demonstrated that the supernatant fluid from S. gordonii Challis mutant 41A9 (described below) lost its ability to inactivate S. mutans CSP, since S. mutans GS5 comC cells produced a similar amount of bacteriocin in the presence of CSP treated with either 41A9 supernatant fluid or PBS. Supernatant fluids from other oral streptococci, namely, S. gordonii 10558, S. sanguis 10556, S. mitis 10712, and S. oralis KS32AR, also inactivated S. mutans CSP as did the supernatant fluid from S. gordonii Challis (Fig. 1B and C). However, the supernatant fluid from S. mutans GS5 did not inactivate its own CSP (Fig. 1C).
FIG. 1.
Determination of the effect of supernatant fluids from oral streptococci on S. mutans CSP in agar well assays. Exogenous synthetic S. mutans CSP (final concentration, 2.5 μg/ml) was incubated with the supernatant fluids from oral streptococci at 37°C for 0.5 to 3 h. CSP thus treated was added to S. mutans GS5 comC mutant cells (107 CFU) in 1.0 ml of THBY to a final concentration of 0.25 μg/ml. After a 24-h incubation at 37°C, the supernatant fluids from the cultures were passed through 0.22-μm-pore-size filters, and the bacteriocin level produced by the S. mutans GS5 comC mutant was determined by the agar well assays. (A) Supernatant fluid from S. gordonii Challis. Part of the supernatant fluid was boiled for 10 min before the incubation with CSP. (B and C) PBS, a CSP positive control treated with PBS instead of supernatant fluid; zone 2, supernatant fluid from S. gordonii Challis mutant 41A9; zones 3 to 7, supernatant fluid from S. gordonii Challis, S. gordonii 10558, S. sanguis 10556, S. mitis 10712, and S. oralis KS32AR, respectively; and zone 8, supernatant fluid from S. mutans GS5. CSP was incubated with the supernatant fluid for 3 h in panels B and C.
S. gordonii Challis does not inactivate the S. mutans GS5 bacteriocin.
To determine whether S. gordonii Challis inactivated the bacteriocin activity of S. mutans GS5 as it inactivated the CSP of S. mutans, supernatant fluids (containing bacteriocin) from GS5 grown in broth cultures in THBY supplemented with 1% glucose, 1% sucrose, or 0.1 μg of CSP per ml were mixed with S. gordonii Challis (107 CFU) and incubated in precut wells in THB agar plates for 24 h at 37°C. If S. gordonii Challis inactivates S. mutans GS5 bacteriocin present in the supernatant fluids, there would be a reduction in the inhibition zone diameters in the presence of S. gordonii Challis cells compared to the zones in the absence of S. gordonii Challis cells. The presence of S. gordonii Challis cells grown in the S. mutans supernatant fluids did not reduce the diameter of the inhibition zones (Table 3), indicating that S. gordonii Challis has limited ability to inactivate the bacteriocin produced by S. mutans GS5.
TABLE 3.
Effect of S. gordonii Challis on bacteriocin produced by S. mutans GS5a
| Supplement in THBY | Diameter of inhibition (mm)b
|
|
|---|---|---|
| Without Challis | With Challis | |
| None | 0 ± 0 | 0 ± 0 |
| Glucose | 0 ± 0 | 0 ± 0 |
| Sucrose | 24.5 ± 0.5 | 24.0 ± 1.0 |
| CSP | 39.5 ± 0.5 | 39.5 ± 0.5 |
Supernatant fluids from S. mutans GS5 grown in broth cultures in THBY supplemented with 1% glucose, 1% sucrose, or 0.1 μg of CSP per ml were mixed with S. gordonii Challis (107 CFU) and incubated in precut wells in THB agar plates for 24 h at 37°C. The wells were then filled using THB with 1% low-melting-point agarose, and the plates were overlaid with the indicator strain RP66 (106 CFU) in 3 ml of THB with 1% low-melting-point agarose. After 24 more hours of incubation at 37°C under anaerobic conditions, the diameters of the inhibition zones surrounding the wells were measured.
Values are means ± standard deviations from two experiments.
Isolation of an S. gordonii Challis mutant defective in attenuating bacteriocin production by S. mutans GS5.
To determine the mechanism of the interspecies inhibition of S. mutans GS5 bacteriocin production, S. gordonii Challis cells were randomly mutagenized using Tn916. The resulting insertion mutants were screened for mutants that displayed decreased ability to inhibit S. mutans GS5 bacteriocin production. Of 1,500 clones screened, one mutant, named 41A9, exhibited this property in agar plate bacteriocin assays (Fig. 2, zone 3). The transformants of wild-type S. gordonii Challis with chromosomal DNA extracted from 41A9 all showed the same phenotype as the original 41A9 transformant in the plate bacteriocin assays, with 19- to 24-mm inhibition zone diameters. These results confirmed that the phenotype of 41A9 was determined by the insertional mutation of Tn916 into the host chromosome.
FIG. 2.
Screening Tn916 insertional mutants of S. gordonii Challis in agar plate bacteriocin assays. S. mutans GS5 alone or in mixtures with S. gordonii Challis or with S. gordonii Challis mutant 41A9 were inoculated into THB agar plates. After 24 h of incubation at 37°C under anaerobic conditions, the plates were overlaid with indicator RP66 cells (106 CFU) in 3 ml of THB with 1% low-melting-point agarose. After 24 more hours of incubation at 37°C under anaerobic conditions, the diameters of the clearance zones surrounding the inoculated bacteria were measured. Zones: 1, S. mutans GS5 alone; 2, mixture of wild-type S. gordonii Challis and S. mutans GS5; and 3, mixture of S. gordonii Challis mutant 41A9 and S. mutans GS5.
Sequencing the chromosomal DNA flanking Tn916 from mutant 41A9.
As described in Materials and Methods, restriction enzyme digestions and Southern blot analysis revealed a 1.4-kb fragment of 41A9 chromosomal DNA that flanked the transposase proximal end of Tn916. Using appropriate primers, PCR of the ligation mixture of digested 41A9 DNA fragments with pUC19 resulted in the predicted 1.4-kb PCR product. Subsequent sequencing of the PCR product and analysis in The Institute for Genomic Research (TIGR) S. gordonii Challis unfinished sequence database (http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=genome&gi=5078) revealed that Tn916 interrupted a putative subtilisin-like serine protease gene in S. gordonii Challis. The complete sequence of this protease gene (4,541 bp) was identified in the unpublished TIGR database (M. M. Vickerman, personal communication). This protease has 53% identity and 66% similarity to PstS, a cell envelope protease in Streptococcus thermophilus and 49% identity and 63% similarity to CspA, a cell-surface-associated protease in a group B streptococcus. We have named this gene sgc (for S. gordonii challisin) gene, as it codes for the challisin protease.
Construction of the S. gordonii Challis sgc mutant.
As depicted in Fig. 3, pPE, a vector containing the disrupted sgc gene was constructed so that it could be used to inactivate the sgc gene in S. gordonii Challis after selection with erythromycin. To confirm sgc inactivation in Challis transformants, chromosomal DNA was extracted from wild-type Challis and its sgc mutant (BWY1). The chromosomal DNA was digested with three different restriction enzymes with known digestion sites that flank the integration site of the Erm cassette but with no digestion sites in the Erm cassette. AflIII, EcoRI, and PvuII digestions of the chromosomal DNA produced the predicted fragments after Southern blot analysis (2.3-, 2.1-, and 1.3-kb fragments for Challis and 4.1-, 3.9-, and 3.1-kb fragments for BYW1, respectively).
The S. gordonii Challis sgc mutant BYW1 exhibited the same phenotype as the insertional mutant, 41A9.
Agar plate and agar well assays were performed to determine whether the BYW1 mutant had the same phenotype as 41A9, the Tn916-insertion-inactivated mutant of the S. gordonii Challis sgc gene. Strain BYW1, compared to its parent, Challis, indeed exhibited attenuation in its ability to inhibit S. mutans GS5 bacteriocin production in the agar plate assay to the same extent as 41A9. Both the BYW1 and 41A9 mutants yielded 19- to 24-mm inhibition zone diameters compared to the parent strain Challis (3- to 8-mm inhibition zone diameters) in the mixed stab culture with S. mutans GS5 in agar plate assays.
The effects of S. gordonii Challis and its sgc mutants on S. mutans CSP were compared using the agar well assays. The synthetic S. mutans CSP was incubated with supernatant fluids from S. gordonii Challis, 41A9, or BYW1 or with PBS (as a positive control) for 2 h, since more than 1-h incubation was sufficient to inactivate CSP by Challis supernatant fluid (Fig. 1). The treated CSP was then used to restore bacteriocin production by the S. mutans comC mutant cells as described earlier in this study. The Challis supernatant fluid completely inactivated S. mutans CSP, while those from 41A9 or BYW1 did not inactivate S. mutans CSP, since the S. mutans comC cells responded to the CSP treated with the mutant supernatant fluids to a degree similar to that seen with the CSP positive control (data not shown).
Protease activity of S. gordonii Challis resides in its supernatant fluids.
To determine whether S. gordonii Challis primarily secretes the challisin protease or expresses it on the cell surface, azocasein assays were performed as described above. The supernatant fluids exhibited higher protease activity than the cell pellets from equivalent numbers of cells (Table 4), indicating that there was a higher level of protease activity in the supernatant fluid than in the cell surface. These experiments also demonstrated that the levels of protease activity of protease mutants, 41A9 and BYW1, were lower than that of the parental strain Challis.
TABLE 4.
Protease activity of S. gordonii Challis by azocasein assaysa
| S. gordonii strain | OD420b
|
|
|---|---|---|
| Cell pellet | Supernatant | |
| Challis | 0.174 ± 0.055 | 0.452 ± 0.044 |
| 41A9 | 0.011 ± 0.034 | 0.255 ± 0.016 |
| BYW1 | 0.103 ± 0.031 | 0.233 ± 0.013 |
The cell pellets and supernatant fluids were from cultures of S. gordonii Challis grown overnight in THB. The supernatant fluids were concentrated as described in Materials and Methods. All of the cell pellets and the concentrated supernatant fluids from 50-ml cultures were utilized in the assays.
Values are means ± standard deviations from three experiments.
A 168-kDa protease was missing in sgc mutants of S. gordonii Challis.
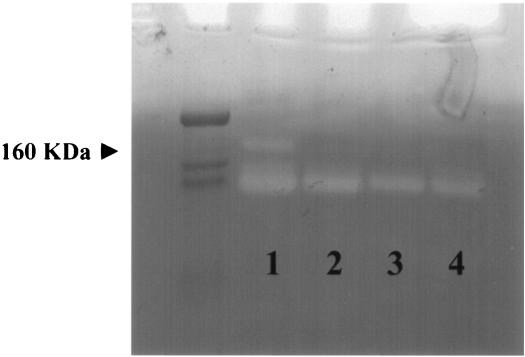
To further characterize the proteases secreted by S. gordonii Challis, concentrated supernatant fluids from Challis and its sgc mutants were assayed by zymography. As shown in Fig. 4, a protease present in the supernatant fluid of parental strain Challis at approximately 160 kDa could not be detected in the supernatant fluids of the protease mutants. Since the size of the S. gordonii Challis challisin gene is 4,541 bp (M. M. Vickerman, personal communication) and therefore a predicted protein size for challisin protease is 168 kDa, the missing 160-kDa activity in the sgc mutants detected by zymography is consistent with the suggestion that sgc gene expression had been interrupted.
FIG. 4.
Casein zymography of S. gordonii protease activity. The supernatant fluids from broth cultures of S. gordonii Challis or sgc mutants grown overnight were concentrated 500-fold and applied to ready-made casein gels. Concentrated supernatant fluids from parental strain Challis (lane 1), insertional mutant 41A9 (lane 2), and two clones of knockout mutant BYW1 (lanes 3 and 4) were used. The approximate position of the 160-kDa band is indicated by the arrowhead.
Comparison of bacteriocin production in broth or biofilms by S. mutans in mixed cultures with either wild-type S. gordonii Challis or sgc mutant BYW1.
To compare bacteriocin production in broth or biofilms, S. mutans BM71 was used instead of S. mutans GS5, since BM71, but not GS5, consistently formed stable biofilms in six-well polystyrene plates in BM. S. mutans BM71 or S. gordonii Challis or BYW1 were cultured in THB to a density of 0.5 at OD600. For broth bacteriocin production, strain BM71 alone (40 μl) or in mixtures with either Challis or BYW1 (40 μl of BM71 and 20 μl of Challis or BYW1) were cultured in 1.0 ml of THBY at 37°C for 24 h. For biofilm bacteriocin production, the same amounts of cells were inoculated in 2 ml of BM in six-well plates to form biofilm. THBY was used subsequently to obtain biofilm bacteriocin supernatant fluids as described earlier in this study. We used different media for biofilm bacteriocin production, since S. mutans does not form stable biofilms in rich medium (THBY) and does not produce as much bacteriocin in BM medium. The numbers of cells counted on mitis salivarius agar plates for broth cultures and biofilm cultures were comparable (Table 5). Killing assays were performed to compare the amounts of bacteriocin produced by S. mutans BM71. There was no difference in bacteriocin production by S. mutans BM71 in biofilms or broth cultures when it was the single strain (Table 5). Supernatant fluids from mixtures of S. mutans BM71 with wild-type S. gordonii Challis in biofilms or broth cultures exhibited no killing of RP66 indicator cells. However, the killing percentage of RP66 cells by supernatant fluid from a mixture of S. mutans BM71 and mutant BWY1 cells in broth culture was similar to that from S. mutans BM71 single cultures, while the supernatant fluids from mixtures of S. mutans BM71 and mutant BWY1 in biofilms exhibited 52.9% killing (Table 5).
TABLE 5.
Comparison of bacteriocin production by mixed cultures of S. mutans BM71 S. gordonii Challis in broth or biofilmsa
| Supernatant from strain(s) | % Killing of indicator RP66b
|
|
|---|---|---|
| Broth | Biofilms | |
| BM71 alone | 98.3 ± 0.3 | 99.6 ± 0.4 |
| BM71 + Challis | 0 ± 0 (537/406) | 0 ± 0 (517/353) |
| BM71 + BYW1 | 97.1 ± 1.0 (447/356) | 52.9 ± 7.0 (479/146) |
A 100 μl aliquot from S. mutans BM71 alone or in a mixture with S. gordonii Challis from a broth or biofilm culture was mixed with 5 × 104 CFU of RP666 indicator cells (in 100 μl of THB) and incubated at 37°C for 1 h. Two micro liters of the culture was plated on the surface of a TSA plate and incubated for another 24 h at 37°C under anaerobic conditions.
Values are means ± standard deviations from two experiments. The values in parentheses are the total number of S. mutans cells (106)/total number of S. gordonii cells (106) counted on mitis salivarius plates.
S. gordonii Challis down-regulated bacteriocin gene expression in S. mutans.
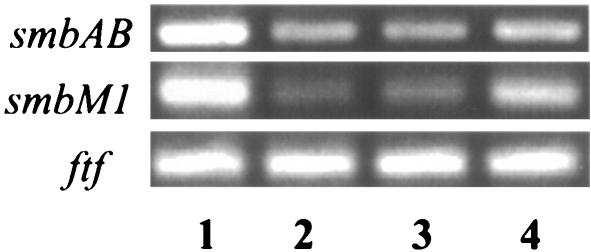
S. mutans BM71 alone and in mixtures with S. gordonii Challis, the sgc mutant BYW1, and the S. mutans BM71 comC mutant were cultured in THBY. Total RNA was extracted, and RT-PCR was performed to examine the expression of the S. mutans bacteriocin gene smbAB, its modification gene smbM1 (Yonezawa and Kuramitsu, submitted), or the ftf gene (the internal control). These three genes are expressed only in S. mutans and not in S. gordonii (data not shown). The expression of S. mutans smbAB and smbM1 genes was significantly down-regulated in the S. mutans BM71 comC mutant compared to its wild type (Fig. 5), indicating that CSP also regulates the expression of S. mutans bacteriocin genes. S. gordonii Challis down-regulated the expression of smbAB and smbM1 genes of S. mutans BM71 in the mixed cultures to an extent similar to that of the S. mutans BM71 comC mutant, while the sgc mutant BYW1 only slightly down-regulated the expression of these genes (Fig. 5). This is another indication that the expression of S. mutans bacteriocin genes is regulated by CSP, since Challis completely inactivated S. mutans CSP, while the sgc mutants did not (Fig. 1). DNase-treated RNA without reverse transcription (as negative controls) did not produce the 274- or 357-bp PCR products for S. mutans smbAB and smbM1 genes, respectively (data not shown). The ftf gene of S. mutans was used as an internal control for RT-PCR, and the expression of the ftf gene did not exhibit regulation in the mixed cultures as the S. mutans bacteriocin gene and its modification gene did.
FIG. 5.
S. mutans bacteriocin gene expression in mixed cultures. Total RNA from S. mutans BM71 alone or in a mixture with S. gordonii or from S. mutans BM71 comC mutant cells was isolated from broth cultures in THBY (OD600 readings of 1.0 to 1.2). Reverse transcription-PCR products were electrophoresed in 2% agarose gels and demonstrated bands of 274, 357, and 482 bp for the smbAB, smbM1, and ftf genes, respectively. Lanes: 1, S. mutans BM71; 2, S. mutans BM71 comC mutant, 3, S. mutans BM71 mixed with S. gordonii Challis; 4, S. mutans BM71 mixed with S. gordonii mutant BYW1.
DISCUSSION
Previous studies have shown that many clinical isolates and laboratory strains of S. mutans produce antimicrobial substances termed bacteriocin (mutacins) that exhibit various degrees of inhibition of other bacteria. The operons coding for four mutacins (mutacins I, II, III, and IV) have been identified (3, 23, 24). On the other hand, bacteriocins produced by many S. mutans strains have been identified only by the bacterial strains that they inhibited and have not yet been further characterized. S. mutans GS5 produces a mutacin that inhibits a group C streptococcal strain RP66, among other sensitive indicator bacteria (22). Among the three S. mutans strains tested in the present study, GS5 and BM71 inhibited the growth of RP66, while their corresponding com mutants did not. These results indicated that bacteriocin production by S. mutans, at least in the two strains tested, is a quorum-sensing-dependent phenomenon (4). The addition of exogenous CSP to the com mutants restored the ability of bacteriocin production only in the comC mutant, not in the comD or comE mutant. These results further confirmed that S. mutans bacteriocin production is a CSP-dependent phenomenon, since the comC mutant is defective in CSP itself, while the comD and comE mutants are defective in the CSP receptor and response regulator, respectively.
The inhibition zone sizes measured in plate assays have been previously used by many investigators to evaluate production of bacteriocin by S. mutans (3, 24). Since the bacteriocin from strain GS5 has not been purified, it is not clear to what extent zone size is directly related to bacteriocin concentration. Nevertheless, it is a reliable method to indicate relative levels of bacteriocin produced by S. mutans. In our study, the stab cultures of the com mutants of S. mutans resulted in much smaller zones than those of the wild type (0 to 6 mm compared to 36 to 40 mm), indicating that the com mutants of S. mutans produce minimal amounts of bacteriocin. This was further confirmed by another semiquantitative method (broth assays) using supernatant fluids from broth cultures. However, the plate assay does have limitations, since bacteriocin produced by S. mutans has to diffuse through agar. In broth assays, the addition of exogenous CSP resulted in enhanced bacteriocin production by wild-type S. mutans, which was not reflected in the plate assays.
Mutacin production is affected by the composition of the culture medium. Paul and Slade (22) reported that S. mutans GS5 consistently produced bacteriocin in Trypticase soy broth (TSB) supplemented with 5% horse serum, but not in THB plus horse serum. However, Rogers (25) reported that the addition of yeast extract enhanced bacteriocin production of S. mutans. We utilized THB supplemented with 3% yeast extract in our experiments of broth bacteriocin production to compare the results with those of agar stab culture assays. THBY, compared with TSB plus serum, produced an even larger amount of bacteriocin when assayed in broth assays (data not shown). THBY that had been filtered through 0.22-μm-pore-size filters instead of being autoclaved produced a slightly increased amount of bacteriocin (data not shown).
The addition of 1% sucrose in THBY significantly increased bacteriocin production by S. mutans GS5 in broth (Table 3). This seems to be correlated with aggregations of S. mutans GS5, since gtfB (glucosyltransferase) (1) and gtfC (30) mutants of GS5 did not produce as much bacteriocin as their parent strain did in the presence of sucrose, while SP2 (a spontaneous mutant of GS5 with gtfB and gtfC genes that had undergone recombination [29]) did not exhibit enhanced bacteriocin production in the THBY supplemented with sucrose (data not shown). Agar well assays used in Table 3 allow bacteriocins in the supernatant fluids to diffuse into agar surrounding the wells. The assays produced reproducible results without much variation. However, the agar well assays were not as sensitive as other assays. The supernatant fluids of S. mutans GS5 from either THBY or THBY plus 1% glucose did not produce detectable inhibition zones in agar well assays, while they inhibited the growth of RP66 in broth or viable count assays. Agar well assays were valuable for the detection of bacteriocin activities beyond a certain threshold (e.g., the supernatant fluids from S. mutans GS5 cultures in THBY plus 1% sucrose or in the presence of exogenous CSP), since the agar well assay is quantitative and exhibits little variation.
Mixtures of S. mutans GS5 with other oral streptococci resulted in attenuated bacteriocin production by S. mutans GS5. Bacteriocin production by S. mutans GS5 was completely abolished in mixed broth cultures for all species but was inhibited by different degrees in mixed agar stab culture assays. This may be explained by the difference in the differential inhibition of the growth of other species by S. mutans GS5. S. mutans GS5 did not inhibit S. gordonii Challis growth in agar stab culture assays, while it weakly inhibited the growth of other oral bacteria (data not shown). This difference might be detected only in stab culture assays, since the inoculated bacteria were in much smaller numbers in stab culture assays than in broth assays.
Supernatant fluids from other oral streptococci inactivated CSP, but not the bacteriocin of S. mutans (Fig. 1 and Table 3). The concentration of CSP (2.5 μg/ml) that was inactivated by the supernatants from other oral streptococci was 25 times higher than that sufficient to restore the ability of bacteriocin production in the S. mutans GS5 or BM71 comC mutant (0.1 μg/ml). This indicated that a substance(s) in the supernatant fluids from other oral streptococci was responsible for this interspecies interaction. The subsequent identification of the sgc gene and the construction of sgc mutants confirmed that this substance was produced by S. gordonii Challis.
A relatively high concentration (3 μl of 1 mg/ml) of CSP was added to the mixed stab cultures in plate assays. We have titrated the CSP and discovered that as little as 3 μl of 10-μg/ml CSP (100-fold diluted) could induce bacteriocin production by the comC mutant of S. mutans GS5 or BM71 in plate assays (data not shown). Therefore, even though some CSP was degraded by protease produced by other species of oral streptococci, the remaining CSP would still be sufficient to induce maximal amounts of bacteriocin production by S. mutans in the mixed stab cultures.
There are proteases present in S. gordonii Challis other than the challisin protease (Fig. 4 and Table 4). Therefore, knocking out the sgc gene in S. gordonii Challis would not completely abolish its interference with the CSP-dependent phenomenon of S. mutans. This was demonstrated in Fig. 2 where the mixed stab culture of S. mutans GS5 and the insertional mutant 41A9 produced a smaller zone of inhibition than did S. mutans GS5 itself. This was also true for the knockout mutant BYW1 and 41A9.
Inhibition of S. mutans bacteriocin production by other oral streptococci was more significant in biofilms than in broth. This was demonstrated in Table 5, where BYW1 exhibited no inhibition of S. mutans bacteriocin production in broth but around 50% inhibition of that in mixed biofilms.
Recently, the bacteriocin genes of S. mutans GS5 have been identified and sequenced and appear to code for a dipeptide mutacin (Yonezawa and Kuramitsu, submitted). Using primers for the newly identified S. mutans GS5 bacteriocin genes, we demonstrated that S. gordonii Challis, but not its sgc mutant, significantly down-regulated bacteriocin gene expression in strain GS5 (data not shown). Strain BM71 appears to contain the same bacteriocin genes as GS5, as was demonstrated in the present study.
Quorum-sensing systems in S. mutans govern the competence of genetic transformation (17), biofilm formation (19, 33), and acid tolerance (18). Therefore, the CSP-dependent quorum-sensing system and the virulence response it mediates offer new targets for strategies to reduce the cariogenicity of organisms, such as S. mutans. We demonstrated here that S. mutans bacteriocin production is another quorum-sensing-dependent phenomenon. We also demonstrated that the interactions with other oral streptococci interfered with S. mutans bacteriocin production. At this time, we are also examining the inhibitory effects of other oral streptococci on S. mutans genetic transformation. An understanding of interactions among oral streptococci may provide valuable insight into the prevention of oral infections, such as dental caries.
Acknowledgments
We thank F. Scannapieco for helpful discussions. We also thank M. M. Vickerman for assistance in identifying the complete sgc gene sequence of S. gordonii Challis in the unpublished TIGR database. B.-Y.W. is grateful to the members of the Kuramitsu laboratory, especially B. Wang, W. Chen, and M. S. Qi, for their support and advice.
These studies were supported in part by grants DE03258 and T32-DE07034 from the National Institute of Dental and Craniofacial Research.
REFERENCES
- 1.Aoki, H., T. Shiroza, M. Hayakawa, S. Sato, and H. K. Kuramitsu. 1986. Cloning of a Streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesis. Infect. Immun. 53:587-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 3.Chen, P., F. Qi, J. Novak, and P. W. Caufield. 1999. The specific genes for lantibiotic mutacin II biosynthesis in Streptococcus mutans T8 are clustered and can be transferred en bloc. Appl. Environ. Microbiol. 65:1356-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cvitkovitch, D. G., Y. H. Li, and R. P. Ellen. 2003. Quorum sensing and biofilm formation in streptococcal infections. J. Clin. Investig. 112:1626-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong, Y. H., J. L. Xu, X. Z. Li, and L. H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunny, G. M., and B. A. Leonard. 1997. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 51:527-564. [DOI] [PubMed] [Google Scholar]
- 7.Fabio, U., M. Bondi, G. Manicardi, P. Messi, and R. Neglia. 1987. Production of bacteriocin-like substances by human oral streptococci. Microbiologica 10:363-370. [PubMed] [Google Scholar]
- 8.Fuqua, C., and E. P. Greenberg. 1998. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr. Opin. Microbiol. 1:183-189. [DOI] [PubMed] [Google Scholar]
- 9.Harris, G. S., S. M. Michalek, and R. Curtiss III. 1992. Cloning of a locus involved in Streptococcus mutans intracellular polysaccharide accumulation and virulence testing of an intracellular polysaccharide-deficient mutant. Infect. Immun. 60:3175-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Havarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jack, R. W., J. R. Tagg, and B. Ray. 1995. Bacteriocins of gram-positive bacteria. Microbiol. Rev. 59:171-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleerebezem, M., and L. E. Quadri. 2001. Peptide pheromone-dependent regulation of antimicrobial peptide production in Gram-positive bacteria: a case of multicellular behavior. Peptides 22:1579-1596. [DOI] [PubMed] [Google Scholar]
- 13.Kleerebezem, M., L. E. Quadri, O. P. Kuipers, and W. M. de Vos. 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24:895-904. [DOI] [PubMed] [Google Scholar]
- 14.Kuramitsu, H. K., and C. M. Long. 1982. Plasmid-mediated transformation of Streptococcus mutans. Infect. Immun. 36:435-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuramitsu, H. K., and V. Trapa. 1984. Genetic exchange between oral streptococci during mixed growth. J. Gen. Microbiol. 130:2497-2500. [DOI] [PubMed] [Google Scholar]
- 16.Li, Y.-H., M. N. Hanna, G. Svensäter, R. P. Ellen, and D. G. Cvitkovitch. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 183:6875-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, Y.-H., P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Y.-H., P. C. Y. Lau, N. Tang, G. Svensäter, R. P. Ellen, and D. G. Cvitkovitch. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 184:6333-6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, Y.-H., N. Tang, M. B. Aspiras, P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrison, D. A. 1997. Streptococcal competence for genetic transformation: regulation by peptide pheromones. Microb. Drug Resist. 3:27-37. [DOI] [PubMed] [Google Scholar]
- 21.Otto, M., H. Echner, W. Voelter, and F. Götz. 2001. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69:1957-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul, D., and H. D. Slade. 1975. Production and properties of an extracellular bacteriocin from Streptococcus mutans bacteriocidal for group A and other streptococci. Infect. Immun. 12:1375-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi, F., P. Chen, and P. W. Caufield. 2001. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl. Environ. Microbiol. 67:15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi, F., P. Chen, and P. W. Caufield. 1999. Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthesis genes. Appl. Environ. Microbiol. 65:3880-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers, A. H. 1972. Effect of the medium on bacteriocin production among strains of Streptococcus mutans. Appl. Microbiol. 24:294-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiroza, T., and H. K. Kuramitsu. 1988. Sequence analysis of the Streptococcus mutans fructosyltransferase gene and flanking regions. J. Bacteriol. 170:810-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sojar, H. T., J.-Y. Lee, G. S. Bedi, and R. J. Genco. 1993. Purification and characterization of a protease from Porphyromonas gingivalis capable of degrading salt-solubilized collagen. Infect. Immun. 61:2369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tagg, J. R., A. S. Dajani, and L. W. Wannamaker. 1976. Bacteriocins of gram-positive bacteria. Bacteriol. Rev. 40:722-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueda, S., and H. K. Kuramitsu. 1988. Molecular basis for the spontaneous generation of colonization-defective mutants of Streptococcus mutans. Mol. Microbiol. 2:135-140. [DOI] [PubMed] [Google Scholar]
- 30.Ueda, S., T. Shiroza, and H. K. Kuramitsu. 1988. Sequence analysis of the gtfC gene from Streptococcus mutans GS-5. Gene 69:101-109. [DOI] [PubMed] [Google Scholar]
- 31.Vining, L. C. 1990. Functions of secondary metabolites. Annu. Rev. Microbiol. 44:395-427. [DOI] [PubMed] [Google Scholar]
- 32.Weerkamp, A., L. Bongaerts-Larik, and G. D. Vogels. 1977. Bacteriocins as factors in the in vitro interaction between oral streptococci in plaque. Infect. Immun. 16:773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]