ABSTRACT
Mycobacterium chimaera is an opportunistic environmental mycobacterium belonging to the Mycobacterium avium-M. intracellulare complex. Although most commonly associated with pulmonary disease, there has been growing awareness of invasive M. chimaera infections following cardiac surgery. Investigations suggest worldwide spread of a specific M. chimaera clone, associated with contaminated hospital heater-cooler units used during the surgery. Given the global dissemination of this clone, its potential to cause invasive disease, and the laboriousness of current culture-based diagnostic methods, there is a pressing need to develop rapid and accurate diagnostic assays specific for M. chimaera. Here, we assessed 354 mycobacterial genome sequences and confirmed that M. chimaera is a phylogenetically coherent group. In silico comparisons indicated six DNA regions present only in M. chimaera. We targeted one of these regions and developed a TaqMan quantitative PCR (qPCR) assay for M. chimaera with a detection limit of 100 CFU/ml in whole blood spiked with bacteria. In vitro screening against DNA extracted from 40 other mycobacterial species and 22 bacterial species from 21 diverse genera confirmed the in silico-predicted specificity for M. chimaera. Screening 33 water samples from heater-cooler units with this assay highlighted the increased sensitivity of PCR compared to culture, with 15 of 23 culture-negative samples positive by M. chimaera qPCR. We have thus developed a robust molecular assay that can be readily and rapidly deployed to screen clinical and environmental specimens for M. chimaera.
KEYWORDS: mycobacterium, diagnostics, genomics, infectious disease
INTRODUCTION
Mycobacterium chimaera is an environmental mycobacterium and infrequent pathogen, most commonly linked with pulmonary disease (1–8). Interest in M. chimaera has heightened with global reports of invasive infections (including endocarditis and vascular graft infections associated with the use of LivaNova PLC [formerly Sorin Group Deutschland GmbH] Stöckert 3T heater-cooler units [HCUs] during cardiac surgery). The most plausible hypothesis for this widespread contamination is a point source outbreak, although the underlying causative factors are not currently known (9–16). Phylogenetic comparisons of 16S-23S rRNA internal transcribed spacer (ITS) sequences, and/or partial rpoB or hsp65 sequences (2, 5–7, 17, 18), suggest that M. chimaera is a distinct entity within the M. avium-M. intracellulare complex (6), and two recent population genomic analyses have confirmed this relationship (8, 13). The complete 6,593,403-bp genome sequence of M. chimaera ANZ045 revealed a single circular 6,078,672-bp chromosome and five circular plasmids ranging in size from 21,123 to 324,321 bp (8). M. chimaera is slow growing; therefore, current culture-based laboratory methods, followed by Sanger sequencing of amplicons for one or more combinations of conserved sequence regions, or line-probe hybridization assays are not amenable to timely and specific detection of this pathogen. This delay carries significant clinical, health provision, and medicolegal implications, as patients may be exposed to contaminated machines during this turnaround time of up to 6 to 8 weeks. A rapid and reliable diagnostic tool is urgently needed to support clinical management of patients and to establish the efficacy of heater-cooler unit decontamination procedures. Here we addressed this issue by using comparative genomics to identify DNA sequences present in M. chimaera and absent from other mycobacteria. We describe the initial development and validation of a sensitive, specific, and quantitative PCR (qPCR) assay for identification of M. chimaera in both clinical and environmental samples.
RESULTS
Assessment of M. chimaera population structure.
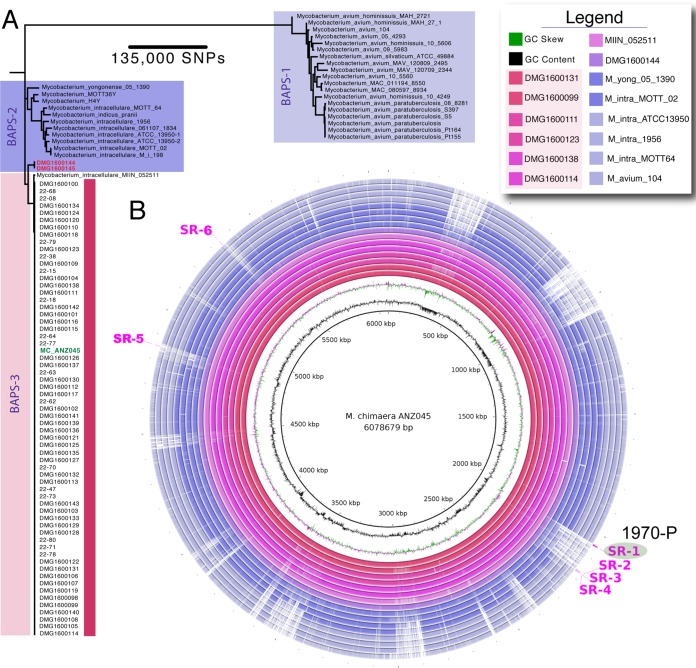
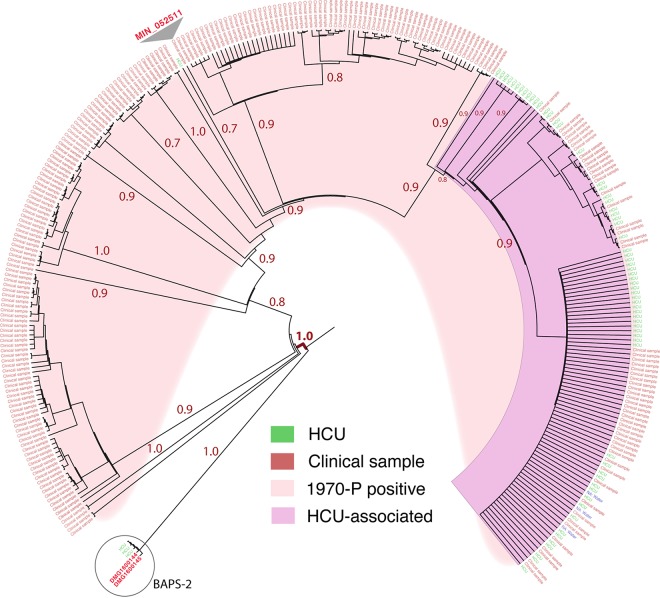
To identify DNA segments present only in M. chimaera genomes, we first assessed the phylogenetic coherence of M. chimaera as a species. Using 96 mycobacterial genome sequences comprising 63 M. chimaera genomes from North America, Australia, and New Zealand and 33 other related, publicly available mycobacterial genomes from the M. avium-M. intracellulare complex, we conducted whole-genome pairwise comparisons of the 96 taxa to the M. chimaera MC_ANZ045 complete reference chromosome. The 63 M. chimaera genomes included 49 HCU-associated and 14 previously described patient isolates, not all of which were associated with Stöckert 3T HCU contamination (8, 12). These comparisons identified 448,878 variable nucleotide positions in a 2,340,885-bp core genome. A robust phylogeny inferred from the alignments strongly suggested that M. chimaera forms a monophyletic lineage within the M. avium-M. intracellulare complex (Fig. 1A) (8, 13). Bayesian analysis of population structure (BAPS) using these same data confirmed this clustering (Fig. 1A). Interestingly, this assessment indicated that a publicly available isolate originally identified as Mycobacterium intracellulare (strain MIN_052511_1280) was in fact M. chimaera. The mean number of single nucleotide polymorphisms (SNPs) between any pair of the 63 M. chimaera isolates and MIN_052511_1280 (BAPS cluster 3 [BAPS-3]), not adjusted for recombination, was 115 (range, 1 to 3,024; interquartile range [IQR], 13 to 31), highlighting restricted core genome variation within this species, particularly given the large 6.5-Mb genome size. In comparison, the mean number of SNPs between 15 M. avium-M. intracellulare complex genomes (BAPS-2) was 24,134 SNPs (range, 13 to 39,109; IQR, 14,780 to 33,138) (Fig. 1A). We then extended this analysis to assess an additional 257 publicly available M. chimaera and related mycobacterial genome sequences (see Table S1 in the supplemental material). Pairwise whole-genome comparisons of the members of this larger data set were performed against the MC_ANZ045 reference genome. Population structure analysis indicated that 303 mycobacterial genomes fell within BAPS-3 (Table S1). Pairwise comparisons were again performed against the MC_ANZ045 using only these 303 genomes, with five other genome sequences from BAPS-2 included for context. The alignment was filtered to remove sites from the alignment affected by recombination, and a phylogeny was inferred from the resulting 10,166 variable nucleotide positions (Fig. 2). The mean number of SNPs between the 303 genomes was 268 (range, 0 to 3,211; IQR, 9 to 62). This analysis confirmed that M. chimaera does indeed form a monophyletic lineage, providing a robust genetic definition for the species. As previously reported, HCU-associated isolates from around the world formed a distinct subclade within this lineage (Fig. 2) (8, 13).
FIG 1.
Phylogenetic and population structure analysis of M. chimaera. (A) Core genome maximum likelihood phylogeny of 63 M. chimaera and 33 other related mycobacterial species based on alignment of 448,878 variable nucleotide positions. The tree was inferred with FastTree using a GTR model of nucleotide substitution. All major branches had FastTree support values of >0.9. Lineages are colored, and BAPS group designations are indicated. The scale bar indicates the number of SNPs represented by the horizontal branches. The location of the MC_ANZ045 M. chimaera reference genome is shown in green typeface. (B) Visualization using a BLAST Ring Image Generator (32) of DNA:DNA whole-genome comparisons among a subset of mycobacterial genomes used to identify M. chimaera-specific regions. The MC_ANZ045 M. chimaera reference chromosome is depicted by the inner black circle. The identity of the subsequent rings is given in the legend. Annotations on the outermost ring show the locations of the six M. chimaera-specific regions, highlighting the region targeted for TaqMan PCR assay development.
FIG 2.
Focused phylogenetic analysis of 303 M. chimaera genomes. Core genome maximum likelihood phylogeny data for 303 M. chimaera genomes (BAPS-3) and genomes of five other, related mycobacterial species (BAPS-2) were determined based on alignment of 10,166 variable nucleotide positions (recombination removed). The tree was inferred with FastTree using a GTR model of nucleotide substitution and rooted using DMG1600144 (BAPS-2, circled) as an outgroup. The FastTree support values for major branches are indicated. Branch lengths have been transformed, and they are proportional but not to scale. The location within the phylogeny of a sequence identified previously as M. intracellulare (MIN_052511) is indicated.
In silico genome comparisons to identify M. chimaera-specific sequences.
A subset of 46 randomly selected M. chimaera genome sequences as defined above became the “training set” to find DNA segments present only in M. chimaera (Fig. 2; see also Table S1). Mapping of DNA sequence reads to the MC_ANZ045 reference genome (refer methods) allowed the identification of 159 genomic segments that were >500 bp in length and covered 510,924 bp that were present across the 46 M. chimaera isolates and absent from 8 M. intracellulare isolates (Fig. 1A). BLAST comparisons of the 159 segments against all entries in the NCBI GenBank nt database and removal of any non-M. chimaera-specific sequence reduced the number to 37 segments (covering a total of 37,890 bp). A larger validation set comprising the 63 M. chimaera genomes described in Fig. 1A and 242 additional, publicly available M. chimaera genomes that satisfied our phylogenomic inclusion criteria described above was then screened (Table S1). Six of the 37 M. chimaera-specific regions (SR), covering a total of 8,292 bp, were present in all 305 M. chimaera genomes (Fig. 1B). The six SRs ranged in length from 531 bp to 4,641 bp, the majority overlapping predicted chromosomal protein-coding sequences (CDS) (Fig. 1B and Table 1). Inferred functions of these CDS are summarized (Table 1). The regions were scanned for sequence polymorphisms, and one of the regions (SR1) that was 100% conserved among all M. chimaera was selected as a template for the design of a TaqMan assay (Fig. 2 and Table 2). SR1 spans two predicted CDS that DNA composition analysis and gene annotation predicted lay within a 35-kb putative prophage or integrative mobile element. A 79-bp TaqMan amplicon (assay identification no. [ID] 1970) was designed within a 2,934-bp CDS (predicted function unknown) (Table 2).
TABLE 1.
Summary of the six putative M. chimaera-specific DNA segments identified by comparative genomics
| Region ID | Position in MC_ANZ045 chromosome | Segment length (bp) | Putative CDS spanning region | TaqMan probe IDa |
|---|---|---|---|---|
| SR1 | 2047981–2052621 | 4,641 | 2× hypothetical proteins | 1970 |
| SR2 | 2168889–2169420 | 531 | 1× hypothetical protein | |
| SR3 | 2171047–2171975 | 928 | 1× hypothetical protein | |
| SR4 | 2174560–2175316 | 756 | No protein-coding region detected | |
| SR5 | 4855157–4855879 | 722 | 1× hypothetical protein | |
| SR6 | 5338898–5339612 | 714 | tRNA-His (GTG) |
The probe ID number is based on the position of the first nucleotide of the TaqMan probe in the DNA segment (refer to Table 2).
TABLE 2.
Sequences of the M. chimaera-specific TaqMan primers and probea
| Probe or primer | ID | Sequence (5′–3′) | Position in ANZ045 chromosome |
|---|---|---|---|
| TaqMan probe | 1970-P | ACTCAAACACCTGACGAGTCA | 2,049,950–2,049,970 |
| Forward primer | 1939-F | ACTTGACGAGGTCTTGCAGG | 2,049,919–2,049,938 |
| Reverse primer | 2017-R | GACGGCATAGAGATTCGCCA | 2,049,978–2,049,997 |
The M. chimaera-specific region for the probe and primers was SR1. The TaqMan amplicon length was 79 bp.
TaqMan assay specificity testing.
The in silico analyses described above predicted that the TaqMan assay would be diagnostic for the presence of M. chimaera. To test this prediction, DNA was prepared from 42 mycobacterial isolates (including 2 M. chimaera isolates) and 22 other bacterial isolates from 21 different genera. A pan-bacterial 16S rRNA PCR was performed first to ensure detectable bacterial DNA was present. All 64 DNA samples were positive by 16S rRNA PCR (data not shown), but only the two M. chimaera isolates were 1970-P TaqMan assay positive, supporting the in silico predictions that these assays are specific for M. chimaera (Table S2).
TaqMan assay efficiency and sensitivity testing.
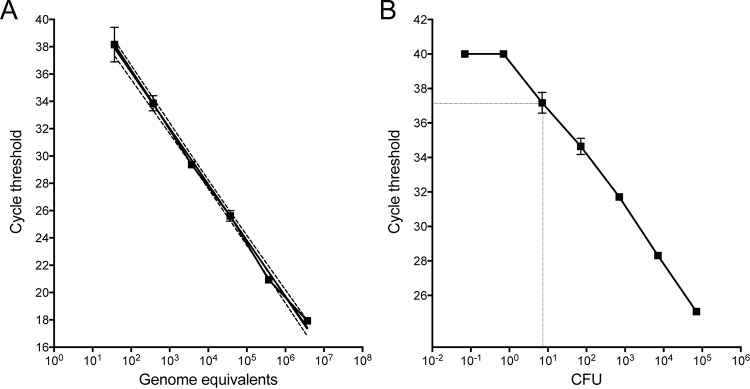
To establish the limit of detection and amplification efficiency for the 1970-P TaqMan assay, 10-fold serial dilutions of purified M. chimaera MC_ANZ045 genomic DNA were tested in triplicate. The 1970-P assay showed excellent performance characteristics, with a very good linear response across 5 orders of magnitude, R2 values of >0.99, amplification efficiency of 94%, and a detection limit of around 20 genome equivalents (GE) (Fig. 3A). The detection limit was also assessed using dilutions of M. chimaera culture spiked into whole blood. Again, the assay showed excellent performance characteristics under this simulated clinical condition, with an absolute detection limit around 10 CFU (equivalent to 100 CFU/ml of blood; Fig. 3B). These experiments indicated that the 1970-P assay is a suitable qPCR assay for sensitive and quantitative detection of M. chimaera DNA.
FIG 3.
Sensitivity testing of 1970-P TaqMan PCR for M. chimaera. (A) Standard curve showing CT values versus M. chimaera 10-fold serial dilutions, expressed as genome equivalents (log scale). Means and standard deviations for triplicate DNA preparations for assay 1970-P are shown. A curve was interpolated using linear regression (R2 = 0.992). Dotted lines indicate 95% confidence intervals. (B) Detection sensitivity of 1970-P for detection of M. chimaera spiked into whole blood, indicating a limit of detection of 10 CFU. Means and standard deviations for triplicate blood samples and DNA extractions for each dilution are shown.
Detection of M. chimaera DNA in environmental samples.
A key requirement for this assay is the ability to screen water and biofilm samples from heater-cooler units for M. chimaera to determine if maintenance procedures had removed the bacteria from contaminated units or had prevented contamination. Having assessed the sensitivity and specificity of the assay with laboratory-prepared samples, we next explored its performance with environmental samples. We screened concentrates from 33 water samples obtained from heater-cooler units at seven hospitals and HCU distributors in our region that had been assessed by culture for M. chimaera. A total of 25 of 33 samples were positive by 1970-P TaqMan PCR, with estimated M. chimaera concentrations ranging from 2 to 102,000 GE/ml of water (Table 3). Using the culture results as a “gold standard,” the estimated sensitivity for the TaqMan assays was high (100%) with all eight of the culture-positive samples that were also positive by the 1970-P TaqMan PCR (Table 4). However, there was poor correspondence between culture-negative samples and qPCR. Fifteen samples that were negative by culture returned 1970-P TaqMan-positive results, with cycle threshold (CT) values for some of these samples being less than 24, indicating high M. chimaera concentrations of above 10,000 GE/ml of original sample (Table 3). Heterotrophic colony counts (HCC) at 37°C are used as a general indicator of water cleanliness and may be used in some settings as a surrogate indicator of decontamination effectiveness (19). However, we observed a poor correlation between HCC and the presence of M. chimaera as measured by qPCR (Spearman's ρ = 0.2813, P = 0.1128) (Table 3), suggesting that HCC is not a suitable surrogate for the presence or absence of M. chimaera.
TABLE 3.
Environmental sampling qPCR, culture, and heterotrophic plate count summary
| Sample ID | 1970-P CT | Total no. of genome equivalents | Vol (ml) of water filtered (qPCR) | No. of genome equivalents/ml water | Culture positive (50 ml) | HCC/ml (log10) | Source |
|---|---|---|---|---|---|---|---|
| 17444 | No CT | NDa | 500 | ND | No | 2.7 | Facility C: HCU |
| 17651 | No CT | ND | 200 | ND | No | 3.2 | Facility F: HCU |
| 17851 | No CT | ND | 216 | ND | No | ND | Facility D: HCU cardioplegia tank |
| 18078 | No CT | ND | 420 | ND | No | ND | Facility G: HCU cardioplegia tank |
| 18298 | No CT | ND | 480 | ND | No | ND | Facility G: HCU cardioplegia tank |
| 18849 | No CT | ND | 220 | ND | No | ND | Facility G: HCU cardioplegia tank |
| 18850 | No CT | ND | 130 | ND | No | ND | Facility G: HCU cardioplegia tank |
| 19130 | No CT | ND | 420 | ND | No | 1 | Facility D: HCU |
| 19330 | 37.76 | 40 | 1,000 | 2 | No | ND | Facility G: HCU cardioplegia tank |
| 17443 | 37.67 | 42 | 34 | 62 | No | 2.8 | Facility C: HCU |
| 14211 | 36.17 | 98 | 270 | 18 | Yes | ND | Facility C: HCU |
| 18079 | 36.04 | 105 | 960 | 5 | No | ND | Facility G: HCU cardioplegia tank |
| 17164 | 35.22 | 167 | 60 | 139 | No | 3.1 | Facility E: HCU |
| 9221 | 34.32 | 277 | 60 | 231 | Yes | ND | Facility A: HCU overflow water bottle |
| 18297 | 34.32 | 277 | 1,000 | 14 | No | ND | Facility G: HCU cardioplegia tank |
| 18080 | 33.79 | 372 | 420 | 44 | No | 1 | Facility G: HCU cardioplegia tank |
| 10895 | 32.96 | 593 | 50 | 593 | Yes | ND | Facility B: HCU-3 patient circuit |
| 10896 | 32.65 | 706 | 70 | 504 | Yes | ND | Facility B: HCU-3 cardioplegia circuit |
| 19128 | 32.53 | 755 | 420 | 90 | No | ND | Facility D: HCU |
| 10892 | 32.26 | 879 | 50 | 879 | Yes | ND | Facility B: HCU-1 cardioplegia circuit |
| 10894 | 31.85 | 1,106 | 30 | 1,843 | Yes | ND | Facility B: HCU-2 cardioplegia circuit |
| 18081 | 30.46 | 2,412 | 960 | 126 | No | 1 | Facility G: HCU cardioplegia tank |
| 17850 | 30.19 | 2,806 | 300 | 468 | No | 1 | Facility D: HCU cardioplegia tank |
| 19129 | 30.13 | 2,902 | 400 | 363 | No | 1 | Facility D: HCU |
| 16569 | 29.19 | 4,917 | 200 | 1,229 | No | 4.3 | Facility D: HCU-1 |
| 18949 | 29.13 | 5,086 | 220 | 1,156 | No | ND | Facility H: HCU cardioplegia circuit |
| 18948 | 28.69 | 6,509 | 275 | 1,183 | No | ND | Facility H: HCU patient circuit |
| 18951 | 28.58 | 6,923 | 280 | 1,236 | No | ND | Facility H: HCU cardioplegia circuit |
| 10897 | 25.98 | 29,765 | 70 | 21,261 | Yes | 1 | Facility B: HCU-1 patient circuit |
| 16571 | 24.82 | 57,055 | 200 | 14,264 | Yesb | 1 | Facility D: HCU-3 |
| 10893 | 23.55 | 116,325 | 80 | 72,703 | Yes | 2.7 | Facility B: HCU-2 patient circuit |
| 18950 | 23.04 | 154,851 | 130 | 59,558 | No | ND | Facility H: HCU patient circuit |
| 16570 | 21.31 | 408,655 | 200 | 102,164 | Yes | 1 | Facility D: HCU-2 |
ND, not done.
Culture contaminated.
TABLE 4.
Correspondence between M. chimaera culture and 1970-P qPCR in HCU samples
| pPCR result | No. of samples |
||
|---|---|---|---|
| Culture positive | Culture negative | Total | |
| + | 10 | 15 | 25 |
| − | 0 | 8 | 8 |
| Total | 10 | 23 | 33 |
DISCUSSION
Since 2012, there have been a small but increasing number of case reports of invasive infection with M. chimaera in individuals who have undergone surgical procedures requiring cardiac bypass (8, 10–16). Almost all cases have involved placement of prosthetic valves or other prosthetic material and are linked to use of a specific type of heater-cooler unit (HCU) in the bypass procedure (8, 13, 14). As contamination of these HCUs may have occurred at or near the time of manufacture and the machines are widely exported, exposure to M. chimaera during cardiac bypass surgery is an emerging issue in infection control that is not restricted by region or country (8). However, while it appears that generation of aerosols when machines are contaminated may be relatively common, so far, the likelihood of infection for any exposed individual has been very low (13). From a clinical perspective, this poses a major diagnostic challenge. Postcardiac surgery M. chimaera infection has a long incubation period and nonspecific symptoms and can be misdiagnosed as a steroid-requiring inflammatory condition with potentially disastrous consequences (10, 15). Moreover, there is a significant case fatality rate even when the infection is correctly identified and current opinion is that early accurate diagnosis is key to achieving the best treatment outcomes (20). Given the nonspecific symptoms and low prior probability of infection in a large exposed population, clinicians urgently need access to a specific and sensitive test for M. chimaera cardiac and extracardiac infection. Infection control practitioners have a different but equally challenging problem with respect to surveying and cleaning contaminated HCUs. In this report, we describe and validate a DNA target that is diagnostic for the presence of M. chimaera. We have shown under simulated conditions that it can accurately detect M. chimaera in human blood samples at low concentrations and outperform culture in detecting M. chimaera in specimens obtained from contaminated HCUs. It may also be possible to improve the detection sensitivity of this assay in human blood to below 100 CFU/ml. Simple changes that may help here include eluting the purified DNA from the Qiagen blood and tissue spin column in a smaller volume or taking a larger volume of the DNA eluate into the TaqMan PCR.
The European Centre for Disease Prevention and Control recommends that M. chimaera identification should be performed by sequencing at least two conserved fragments among 16S-23S rRNA ITS, 16S rRNA, rpoB, and hsp65 (21, 22). Some laboratories have also used the MIN-2 probe in the INNO-LiPA Mycobacteria v2 line probe assay (6, 18). Here we simplify this suite of tests with a M. chimaera-specific PCR assay that has the advantages of providing a rapid yes/no result and an estimate of bacterial concentration. The test could be further enhanced with multiple DNA targets. The other five M. chimaera-specific regions reported here could be used to develop additional diagnostic targets (Fig. 1B). We envisage that this test will be used in conjunction with efforts to culture M. chimaera from specimens, as isolates are required for whole-genome sequencing (WGS) to establish the genetic relatedness of isolates and, potentially, for antimicrobial susceptibility testing (although antimicrobial resistance is not thought to be a major problem). As a previous risk assessment by Public Health England suggested a possible legionellosis risk for staff and patients, we propose that our assay should form part of a “HCU panel” along with Legionella PCR (20, 23). While we have designed an assay to detect all M. chimaera strains, it may also be possible to detect specific M. chimaera lineages by using DNA deletion polymorphisms to discriminate among intraspecies lineages, as in the Mycobacterium tuberculosis complex (24). We are currently exploring this possibility.
The infection risk posed by the water reservoirs within HCUs and the need for regular maintenance have been long recognized (23). Heterotrophic colony counts (HCC) are being considered surrogates to assess the microbiological quality of HCUs (19, 25). We (like others) found a poor correlation between the presence of M. chimaera and HCC in water samples from HCUs (19), with examples of M. chimaera concentrations of 60,000 GE/ml when HCC were below the limit of detection (Table 3). More evaluation of the 1970-P assay is required, but our data suggest that HCC may have an unacceptably high false-negative rate, which significantly reduces its utility for measuring the effectiveness of HCU decontamination procedures.
Screening HCU water samples with our TaqMan assay indicated the widespread presence of M. chimaera and a poor correlation with culture. There are several potential explanations for these observations. Despite the extensive in silico assessments, it is possible that the qPCR assay lacks specificity for M. chimaera or that the culture method lacks sensitivity or that DNA from M. chimaera is perhaps still present but source organisms are no longer viable. Given the extensive in silico validation undertaken here to ensure target specificity and the high prior probability that these HCU water samples contained M. chimaera, the discrepancy between culture and qPCR might best explained by either lower sensitivity of the mycobacterial culture method or PCR detection of intact DNA from nonviable M. chimaera. The later explanation is perhaps the more likely given that some of these PCR-positive and culture-negative samples were obtained from HCUs subjected to extensive decontamination procedures involving extended heating above 70°C.
In summary, we have developed a new diagnostic tool for rapid, sensitive, and specific detection of M. chimaera to help address the urgent need to screen patient and HCU samples.
MATERIALS AND METHODS
Bacterial strains and genome sequences.
Mycobacterium chimaera strain DMG1600125 (a 2016 HCU isolate from New Zealand) was used for spiking experiments (8). The mycobacterial genome sequences used in this study are listed in Table S1 in the supplemental material. M. chimaera was grown on Brown and Buckle whole-egg media, Middlebrook 7H9 broth, or Middlebrook 7H10 agar (Becton Dickinson) supplemented with 10% (vol/vol) oleic acid albumin dextrose complex (OADC; Difco) or Middlebrook 7H10 agar. Cultures were incubated without shaking at 37°C. M. chimaera colony counts were obtained by spotting 3-μl volumes of six 10-fold serial dilutions of M. chimaera culture suspensions in quintuplicate on two Middlebrook 7H10 agar plates. The colonies were counted after incubation for 4 weeks at 37°C.
Genomic DNA extraction methods, M. chimaera culture, and environmental isolation.
Purified M. chimaera genomic DNA for TaqMan assay validation was extracted from 50 mg (wet weight) of cell pellets as described previously (26) and measured by fluorimetry using a Qubit assay kit and a High Sensitivity DNA kit (Thermo Fisher). For spiking experiments in blood, M. chimaera DNA was extracted from 100-μl volumes of whole blood, using a Qiagen Blood & Tissue DNA extraction kit. Purified DNA was eluted from the columns in a 200-μl volume of 10 mM Tris (Qiagen) (pH 8.0). Total bacteria were concentrated from 30- to 1,000-ml volumes of water collected from heater-cooler units by filtration through 47-mm-diameter, 0.22-μM-pore-size mixed cellulose ester Millipore membranes. Immediately after filtration, membranes were aseptically placed in sterile 50-ml plastic tubes and stored at −70°C. DNA was extracted from membrane concentrate using a MoBio PowerWater DNA isolation kit following the instructions of the manufacturer (MoBio) with an additional physical disruption step consisting of 2 centrifugations at 5,000 rpm for 20 s each time in a Precellys 24 tissue homogenizer. To prevent cross-contamination, a sterilized filtration device was used for each sample and sterile, distilled water extraction blanks were filtered and processed (100-ml volumes) at a frequency of one for every 10 test samples. Culture isolation of M. chimaera from 50-ml volumes of water samples was undertaken as described previously (27).
Population structure and phylogenetic analysis.
Snippy v3.1 (https://github.com/tseemann/snippy) was used to align Illumina sequence read data or de novo assembled contigs from M. chimaera and related mycobacterial genomes against the fully assembled, complete MC_ANZ045 reference genome to call core genome single nucleotide polymorphism (SNP) differences and generate pairwise sequence alignments. Hierarchical Bayesian clustering (hierBAPS) was performed using these core whole-genome SNP alignments as input to assess population structure (a prior arrangement of 6 depth levels and a maximum of 20 clusters were specified) (21), with phylogenies inferred using FastTree v2.1.8 and a general time-reversible (GTR) model of nucleotide substitution (22). BAPS clusters were assigned an arbitrary, unique number (e.g., BAPS-1, BAPS-2, or BAPS-3). Pairwise SNP analysis of comparisons between groups of genomes was performed using a custom R script (https://github.com/MDU-PHL/pairwise_snp_differences). Recombination detection was performed using ClonalFrameML v1.7 (28).
In silico subtractive hybridization and target identification.
To identify regions of DNA present in M. chimaera but absent from other mycobacteria, Illumina sequence reads of 46 M. chimaera isolates from Australia and New Zealand and eight publicly available M. intracellulare genomes (Table S1) were aligned using BWA MEM v0.7.15-r1140 (https://arxiv.org/abs/1303.3997) to a complete M. chimaera reference genome (MC_ANZ045) (8). The read depth at each position was examined to identify those positions in the reference genome that were present across all M. chimaera isolates but absent from all M. intracellulare genomes. These genomic regions were extracted from MC_ANZ045 and compared against the NCBI GenBank nonredundant (nt) nucleotide database using NCBI BLAST v2.5.0 with parameters -remote -max_target_seqs 100 -task blastn -outfmt “6 std qcovs staxid ssciname.” Note that GenBank contained 97,300 genus Mycobacterium genomic DNA sequences at the time of analysis. The resulting BLAST hits that were missing a taxon name were retrieved from the NCBI taxonomy database using the taxon ID. Ignoring BLAST hits against bona fide M. chimaera sequences, the query alignment positions for every hit were extracted and were used to obtain all the sequence segments that had no hits against the GenBank nt database and that were greater than 500 bp in length. For this, the bedtools complement and getfasta tools were used (29). The sequence segments thus obtained were considered candidate M. chimaera-specific genomic regions. The presence of these regions across a wider collection of M. chimaera sequences was assessed by downloading all M. chimaera genome sequence reads present in the NCBI sequence read archive (SRA) as of October 2016 (Table S1) and processing through Nullarbor pipeline v1.2 (https://github.com/tseemann/nullarbor). The output information was used to filter out poor-quality or non-M. chimaera read sets on the basis of results showing G+C content significantly below 66%, an average read depth below 30, a total contig length above 8 Mb, more than four predicted rRNA genes, or a total proportion of sequences aligned to the reference genome of below 70% (Table S1). Using Snippy again, all M. chimaera genomes identified as described above were mapped to a version of the MC_ANZ045 reference genome in which the non-M. chimaera-specific sequence regions had been hard masked. The resulting multiple-sequence alignment was parsed using a custom Perl script to identify those M. chimaera-specific regions that were present in all M. chimaera genomes (https://github.com/ezozayav/utility-scripts). These DNA sequences were inspected further for development of M. chimaera TaqMan PCR diagnostic assays. TaqMan primers and probes (Sigma Oligonucleotides) were designed using Primer3 (30), and Primer-BLAST was used against the NCBI nt database to check that the primers and probes that had been designed were specific to M. chimaera. TaqMan probe 1970-P was labeled with the fluorescent dye 6-carboxyfluorescein (FAM) at the 5′ end and a nonfluorescent quencher at the 3′ end (Sigma Oligonucleotides). To assess the context of these M. chimaera-specific regions, AlienHunter v1.4 was used to screen the MC_ANZ045 genome for DNA compositional bias, indicative of horizontally acquired DNA (31).
TaqMan quantitative PCR.
TaqMan PCR mixtures contained 2 μl of template DNA, 0.4 μM concentrations of each primer, a 0.2 μM concentration of the probe, SensiFAST Probe Lo-ROX (1×) mix (Bioline), and TaqMan exogenous internal positive control (IPC) reagents (Applied Biosystems) in a total volume of 20 μl. Amplification and detection were performed with an Mx3005P system (Stratagene) using the following program: 40 cycles of 95°C for 10 s and 60°C for 20 s. DNA extracts were tested in at least duplicate, and negative and positive template controls were included in each run. Standard curves were prepared using eight 10-fold serial dilutions of M. chimaera genomic DNA at an initial concentration of 120 ng/μl, tested in triplicate. The percentage of PCR amplification efficiency (E) for the TaqMan assay was calculated from the slope (C) of the standard curve as follows: E = (10−1/C) × 100. Cycle threshold (CT) values for unknown samples were converted to genome equivalents by interpolation, with reference to the standard curve of CT values versus dilutions of known concentrations of M. chimaera genomic DNA. The mass (in femtograms) of a single M. chimaera genome was estimated to be 6.59 fg, using the formula M = N × 1.096e−21, where M represents the mass of the single double-stranded M. chimaera MC_ANZ045 reference genome and N = 6,593,403, which is the length of the M. chimaera MC_ANZ045 reference genome, assuming that the average molecular weight (MW) of a double-stranded DNA molecule is 660. Analyses were performed using Graphpad Prism v6.0h.
Supplementary Material
ACKNOWLEDGMENTS
We thank the submitting health care facilities and laboratories for providing water samples and mycobacterial isolates. We are grateful to Chris Coulter for provision of materials and critical review of the manuscript.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors. D.A.W., B.P.H., and T.P.S. are supported by National Health and Medical Research Council Fellowships GNT1123854, GNT1105905, and GNT1105525, respectively.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00197-17.
REFERENCES
- 1.Alhanna J, Purucker M, Steppert C, Grigull-Daborn A, Schiffel G, Gruber H, Borgmann S. 2012. Mycobacterium chimaera causes tuberculosis-like infection in a male patient with anorexia nervosa. Int J Eat Disord 45:450–452. doi: 10.1002/eat.20942. [DOI] [PubMed] [Google Scholar]
- 2.Bills ND, Hinrichs SH, Aden TA, Wickert RS, Iwen PC. 2009. Molecular identification of Mycobacterium chimaera as a cause of infection in a patient with chronic obstructive pulmonary disease. Diagn Microbiol Infect Dis 63:292–295. doi: 10.1016/j.diagmicrobio.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Cohen-Bacrie S, David M, Stremler N, Dubus JC, Rolain JM, Drancourt M. 2011. Mycobacterium chimaera pulmonary infection complicating cystic fibrosis: a case report. J Med Case Rep 5:473. doi: 10.1186/1752-1947-5-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mac Aogáin M, Roycroft E, Raftery P, Mok S, Fitzgibbon M, Rogers TR. 2015. Draft genome sequences of three Mycobacterium chimaera respiratory isolates. Genome Announc 3:e01409-15. doi: 10.1128/genomeA.01409-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moon SM, Kim SY, Jhun BW, Lee H, Park HY, Jeon K, Huh HJ, Ki CS, Lee NY, Shin SJ, Koh WJ. 2016. Clinical characteristics and treatment outcomes of pulmonary disease caused by Mycobacterium chimaera. Diagn Microbiol Infect Dis 86:382–384. doi: 10.1016/j.diagmicrobio.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 6.Tortoli E, Rindi L, Garcia MJ, Chiaradonna P, Dei R, Garzelli C, Kroppenstedt RM, Lari N, Mattei R, Mariottini A, Mazzarelli G, Murcia MI, Nanetti A, Piccoli P, Scarparo C. 2004. Proposal to elevate the genetic variant MAC-A, included in the Mycobacterium avium complex, to species rank as Mycobacterium chimaera sp. nov. Int J Syst Evol Microbiol 54:1277–1285. doi: 10.1099/ijs.0.02777-0. [DOI] [PubMed] [Google Scholar]
- 7.Schweickert B, Goldenberg O, Richter E, Gobel UB, Petrich A, Buchholz P, Moter A. 2008. Occurrence and clinical relevance of Mycobacterium chimaera sp. nov., Germany. Emerg Infect Dis 14:1443–1446. doi: 10.3201/eid1409.071032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williamson D, Howden B, Stinear T. 9 February 2017. Spread of Mycobacterium chimaera from units used in cardiac surgery. N Engl J Med doi: 10.1056/NEJMc1612023. [DOI] [PubMed] [Google Scholar]
- 9.Zweifel SA, Mihic-Probst D, Curcio CA, Barthelmes D, Thielken A, Keller PM, Hasse B, Boni C. 18 November 2016. Clinical and histopathologic ocular findings in disseminated Mycobacterium chimaera infection after cardiothoracic surgery. Ophthalmology doi: 10.1016/j.ophtha.2016.09.032. [DOI] [PubMed] [Google Scholar]
- 10.Tan N, Sampath R, Abu Saleh OM, Tweet MS, Jevremovic D, Alniemi S, Wengenack NL, Sampathkumar P, Badley AD. 2016. Disseminated Mycobacterium chimaera infection after cardiothoracic surgery. Open Forum Infect Dis 3:ofw131. doi: 10.1093/ofid/ofw131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sommerstein R, Ruegg C, Kohler P, Bloemberg G, Kuster SP, Sax H. 2016. Transmission of Mycobacterium chimaera from heater-cooler units during cardiac surgery despite an ultraclean air ventilation system. Emerg Infect Dis 22:1008–1013. doi: 10.3201/eid2206.160045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perkins KM, Lawsin A, Hasan NA, Strong M, Halpin AL, Rodger RR, Moulton-Meissner H, Crist MB, Schwartz S, Marders J, Daley CL, Salfinger M, Perz JF. 2016. Notes from the field: Mycobacterium chimaera contamination of heater-cooler devices used in cardiac surgery - United States. MMWR Morb Mortal Wkly Rep 65:1117–1118. doi: 10.15585/mmwr.mm6540a6. [DOI] [PubMed] [Google Scholar]
- 13.Chand M, Lamagni T, Kranzer K, Hedge J, Moore G, Parks S, Collins S, Del Ojo Elias C, Ahmed N, Brown T, Smith EG, Hoffman P, Kirwan P, Mason B, Smith-Palmer A, Veal P, Lalor MK, Bennett A, Walker J, Yeap A, Isidro Carrion Martin A, Dolan G, Bhatt S, Skingsley A, Charlett A, Pearce D, Russell K, Kendall S, Klein AA, Robins S, Schelenz S, Newsholme W, Thomas S, Collyns T, Davies E, McMenamin J, Doherty L, Peto TE, Crook D, Zambon M, Phin N. 7 December 2016. Insidious risk of severe Mycobacterium chimaera infection in cardiac surgery patients. Clin Infect Dis doi: 10.1093/cid/ciw754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sax H, Bloemberg G, Hasse B, Sommerstein R, Kohler P, Achermann Y, Rossle M, Falk V, Kuster SP, Bottger EC, Weber R. 2015. Prolonged outbreak of Mycobacterium chimaera infection after open-chest heart surgery. Clin Infect Dis 61:67–75. doi: 10.1093/cid/civ198. [DOI] [PubMed] [Google Scholar]
- 15.Kohler P, Kuster SP, Bloemberg G, Schulthess B, Frank M, Tanner FC, Rossle M, Boni C, Falk V, Wilhelm MJ, Sommerstein R, Achermann Y, Ten Oever J, Debast SB, Wolfhagen MJ, Brandon Bravo Bruinsma GJ, Vos MC, Bogers A, Serr A, Beyersdorf F, Sax H, Bottger EC, Weber R, van Ingen J, Wagner D, Hasse B. 2015. Healthcare-associated prosthetic heart valve, aortic vascular graft, and disseminated Mycobacterium chimaera infections subsequent to open heart surgery. Eur Heart J 36:2745–2753. doi: 10.1093/eurheartj/ehv342. [DOI] [PubMed] [Google Scholar]
- 16.Achermann Y, Rossle M, Hoffmann M, Deggim V, Kuster S, Zimmermann DR, Bloemberg G, Hombach M, Hasse B. 2013. Prosthetic valve endocarditis and bloodstream infection due to Mycobacterium chimaera. J Clin Microbiol 51:1769–1773. doi: 10.1128/JCM.00435-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Ingen J, Hoefsloot W, Buijtels PC, Tortoli E, Supply P, Dekhuijzen PN, Boeree MJ, van Soolingen D. 2012. Characterization of a novel variant of Mycobacterium chimaera. J Med Microbiol 61:1234–1239. doi: 10.1099/jmm.0.045070-0. [DOI] [PubMed] [Google Scholar]
- 18.Lebrun L, Weill FX, Lafendi L, Houriez F, Casanova F, Gutierrez MC, Ingrand D, Lagrange P, Vincent V, Herrmann JL. 2005. Use of the INNO-LiPA-MYCOBACTERIA assay (version 2) for identification of Mycobacterium avium-Mycobacterium intracellulare-Mycobacterium scrofulaceum complex isolates. J Clin Microbiol 43:2567–2574. doi: 10.1128/JCM.43.6.2567-2574.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.PHLN. 2017. Public Health Laboratory Network: guidance regarding Mycobacterium chimaera and heater-cooler units. Department of Health, Australian Government, Canberra, Australia: http://www.health.gov.au/internet/main/publishing.nsf/Content/83DA1AD46068E894CA258082001C86F0/$File/PHLN-guidance-Mycobacterium-chimaera.pdf. [Google Scholar]
- 20.PHE. 2015. Infections associated with heater cooler units used in cardiopulmonary bypass and ECMO: information for healthcare providers in England. Public Health England, London, United Kingdom: https://www.gov.uk/government/publications/infections-associated-with-heater-cooler-units-used-in-cardiopulmonary-bypass-and-ecmo. [Google Scholar]
- 21.Cheng L, Connor TR, Sirén J, Aanensen DM, Corander J. 2013. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol 30:1224–1228. doi: 10.1093/molbev/mst028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weitkemper HH, Spilker A, Knobl HJ, Korfer R. 2002. The heater-cooler unit–a conceivable source of infection. J Extra Corpor Technol 34:276–280. [PubMed] [Google Scholar]
- 24.Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, Parsons LM, Pym AS, Samper S, van Soolingen D, Cole ST. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A 99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anonymous. 2016. Nontuberculous Mycobacterium (NTM) infections associated with heater-cooler devices (HCD) during cardiothoracic surgery. FDA, Silver Spring, MD, USA: http://www.fda.gov/downloads/AdvisoryCommittees/CommitteesMeetingMaterials/MedicalDevices/MedicalDevicesAdvisoryCommittee/CirculatorySystemDevicesPanel/UCM503716.pdf. [Google Scholar]
- 26.Yip MJ, Porter JL, Fyfe JA, Lavender CJ, Portaels F, Rhodes M, Kator H, Colorni A, Jenkin GA, Stinear T. 2007. Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor. J Bacteriol 189:2021–2029. doi: 10.1128/JB.01442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.PHE. 2016. Protocol for environmental sampling, processing and culturing of water and air samples for the isolation of slow-growing mycobacteria. Department of Health, Public Health England, London, United Kingdom: https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/540325/Air_water_environmental_sampling_SOP_V2.pdf. [Google Scholar]
- 28.Didelot X, Wilson DJ. 2015. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3–new capabilities and interfaces. Nucleic Acids Res 40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vernikos GS, Parkhill J. 2006. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 22:2196–2203. doi: 10.1093/bioinformatics/btl369. [DOI] [PubMed] [Google Scholar]
- 32.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.