Abstract
Eurekanate belongs to the important class of branched-chain carbohydrates present in a wide variety of natural sources. It is a component of avilamycin A, a potent inhibitor of bacterial protein synthesis targeting the 50S ribosomal subunit. The present work provides experimental proof for the function of two genes of the avilamycin biosynthetic gene cluster, aviB1 and aviO2, that are both involved in avilamycin structure modification. The functions of both genes were identified by gene inactivation experiments and nuclear magnetic resonance analyses of extracts produced by the mutants. We suggest that both AviO2 and AviB1 are involved in the biosynthesis of eurekanate within avilamycin biosynthesis. Moreover, two other genes (aviO1 and aviO3) have been inactivated, resulting in a breakdown of avilamycin production in the mutants ITO1 and ITO3, which clearly shows the essential role of both enzymes in avilamycin biosynthesis. The exact functions of both aviO1 and aviO3 remained unknown.
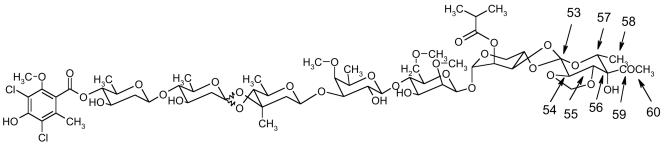
Avilamycin A and avilamycin C (Fig. 1), both belonging to the orthosomycin class of antibiotics, are the major compounds produced by Streptomyces viridochromogenes Tü57 (6). Avilamycin A is currently used as a growth promoter in animal feed, but the European Community decided to prohibit its further use beginning in January of 2006. Like other orthosomycins, avilamycin A inhibits the growth of multidrug-resistant gram-positive bacteria (29). The orthosomycin evernimicin (also called SCH27899 or ziracin) is structurally very similar to avilamycin A. It has been tested in clinical trials as a potential drug for the treatment of serious infectious diseases (11). Both avilamycin A and evernimicin bind exclusively to the 50S ribosomal subunit (19) to inhibit protein synthesis. Recently we reported the characterization of the avilamycin resistance determinants aviRa and aviRb, encoding two rRNA methyltransferases that specifically target 23S rRNA (27). Present evidence indicates that the site of action of the orthosomycins is physically distinct from that of other drugs targeting the 50S ribosomal subunit (18) that makes orthosomycins interesting for the pharmaceutical industry. A complete assignment of the 1H and 13C resonances of avilamycin A, which is an important requirement for biosynthetic studies, has been performed recently. Based on these studies, we were able to elucidate the functions of five methyltransferase genes involved in the biosynthesis of avilamycin A by generating mutants of the wild-type strain. One of these mutants was named S. viridochromogenes GW4 (28). It was producing avilamycin derivatives, named gavibamycin A1 and A3, without a methoxy group at the orsellinic acid moiety. Structural features common to members of the orthosomycin class of antibiotics produced by various actinomycetes are a terminal dichloroisoeverninic acid unit, several deoxysugars, and an eurekanate moiety associated by an unique orthoester linkage (29). The eurekanate moiety belongs to an important class of carbohydrates, the branched-chain sugars, containing a two-carbon side chain at position C4.
FIG. 1.
Structure of avilamycin A and avilamycin C.
The biosynthesis of branched-chain sugars with one carbon side chain is well understood. Several genes encoding S-adenosyl-l-methionine-dependent enzymes have been found in different strains known to catalyze the formation of mostly methylated sugars. Much less is known about the biosynthesis of branched-chain sugars with two carbons. In Yersinia pseudotuberculosis YerE, a thiamine pyrophosphate-dependent flavoprotein catalyzing the conversion of 3,6-dideoxy-4-keto-d-glucose to 3,6-dideoxy-4-acetyl-d-glucose, has been characterized by Liu and coworkers (8). YerE, a bifunctional protein, converts pyruvate to an acetyl carbanion, which then gets attached to the sugar moiety.
To identify genes involved in the biosynthesis and attachment of the two-carbon side chain of eurekanate, we inactivated four genes of the avilamycin biosynthetic gene cluster. As candidates we first chose aviB1, encoding an enzyme resembling the α-chain of several E1 components from different pyruvate dehydrogenase (PDH) multienzyme complexes and aviO1, aviO2, and aviO3, all encoding 2-oxoglutarate-dependent enzymes suggested to be involved in the formation of either C-C or O-C linkages during avilamycin biosynthesis.
Supported by nuclear magnetic resonance (NMR) analysis, the involvement of two gene products (AviO2 and AviB1) in eurekanate biosynthesis was able to be elucidated, while the function of the other two gene products (AviO1 and AviO3) remained unknown.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. S. viridochromogenes Tü57 was grown on 1% malt extract, 0.4% yeast extract, 0.4% glucose, and 1 mM CaCl2, adjusted to pH 7.2 (HA medium) at 37°C. For avilamycin production, S. viridochromogenes Tü57 and all mutants were grown at 28°C in production medium containing 2% glucose, 1% soy-peptone, 0.2% CaCO3, 0.23% l-valine, and CoCl2 (1 mg/liter) adjusted to pH 7.2 (SG medium). DNA manipulation was carried out using Escherichia coli XL1-Blue MRF′ (Stratagene) as a host strain. Before transforming S. viridochromogenes Tü57, plasmids were propagated in E. coli ET12567 (dam− dcm− hsdS Cmr) (10) to obtain unmethylated DNA. E. coli strains were grown on Luria-Bertani agar or liquid medium containing the appropriate antibiotic. pBluescript SK(−) (pBSK−) was from Stratagene. Plasmid pSP1(23) conferring erythromycin resistance was a gift from S. Pelzer, Combinature Biopharm (Berlin, Germany), and pSET152 (3) conferring apramycin resistance was obtained from Eli Lilly and Co. (Indianapolis, Ind.). Plasmid pKC1218 was kindly provided by José A. Salas. Medium components and other chemicals, including antibiotics, were from Roth. Carbenicillin (50 μg ml−1) and apramycin (100 μg ml−1) were used for selective growth of recombinant strains. The 13C-labeled l-methionine and pyruvate for the feeding experiments were from Euriso-Top.
TABLE 1.
List of strains and plasmids used in this study
| Strain or plasmid | Description and/or reference(s) |
|---|---|
| Strains | |
| S. viridochromogenes Tü57 | Producer of avilamycins; 6 |
| S. viridochromogenes GW4 | aviG4-defective mutant, producer of gavibamycins; 27 |
| S. viridochromogenes GW4-ITB1 | aviG4- and aviB1-defective mutant |
| S. viridochromogenes ITO1 | aviO1-defective mutant |
| S. viridochromogenes ITO2 | aviO3-defective mutant |
| S. viridochromogenes ITO3 | aviO3-defective mutant |
| E. coli XL1-Blue MRF′ | Stratagene |
| E. coli ET 12567 | DNA-methylase-negative strains; 9 |
| Plasmids | |
| pBSK− | Stratagene |
| pSP1 | Inactivation plasmid; 22 |
| pSET1cerm | Expression plasmid (integrative); 2, 3 |
| pKC1218 | Expression plasmid (replicative) |
| pSP1-O1/E6S22 | 2.2-kb SacI fragment containing aviO1 ligated into pSP1 |
| AviO2-SK− | aviO2 ligated into the EcoRI and BamHI sites of pBSK− |
| AviO3-SK− | aviO3 ligated into the EcoRI and XbaI sites of pBSK− |
| AviB1-pSP1 | 2.0-kb SacI fragment containing aviB1 ligated into pSP1 |
| IkaviB1 | Defective aviB1 gene ligated into pSP1 |
| IkaviO1 | Defective aviO1 gene ligated into pSP1 |
| IkaviO2 | Defective aviO2 gene ligated into pSP1 |
| IkaviO3 | Defective aviO3 gene ligated into pSP1 |
| O1pSET | aviO1 ligated into pSET1cerm |
| O1pSET | aviO1 ligated into pSET1cerm |
| O1pSET | aviO1 ligated into pSET1cerm |
| B1pKC | aviB1 ligated into pKC1218 |
| H4S112 | 2.0-kb SacI fragment containing aviB1 ligated into pBSK− |
| F4E6 | 6.2-kb EcoRI fragment containing aviO1 ligated into pBSK− |
| P2P13 | 9.7-kb PsI fragment containing aviO2 ligated into pBSK− |
| H4S109 | 8.1-kb SacI fragment containing aviO3 ligated into pBSK− |
General genetic manipulation, PCR, and sequence analysis.
Standard molecular biology procedures were performed as described by Sambrook et al. (24a). Isolation of plasmids was carried out with ion-exchange columns (Nucleobond AX kits; Macherey-Nagel, Düren, Germany) according to the manufacturer's protocol. Southern hybridization was performed on a Hybond-N nylon membrane (Amersham-Pharmacia, Freiburg, Germany) with a digoxigenin-labeled probe using the digoxigenin high-prime DNA labeling and detection starter kit II (Roche Molecular Biochemicals). Isolation of DNA fragments from agarose gel and purification of PCR products were carried out with the NucleoSpin 2 in 1 Extract kit (Macherey-Nagel). Streptomyces protoplast formation, transformation, and protoplast regeneration were performed as described previously (16). PCR was carried out using an Applied Biosystems GeneAmp 9700 thermal cycler and either Taq or Pfu DNA polymerase (Amersham-Pharmacia) according to the supplier's recommendations. DNA sequencing was carried out by 4baselab, Reutlingen, Germany. All oligodeoxynucleotides used for PCR in this study are listed in Table 2.
TABLE 2.
List of primers used in this study
| Primer | Sequence |
|---|---|
| Primers used to generate inactivation plasmids | |
| aviO2F | AAGCACGCCGAATTCTGGCAGGGC |
| aviO2R | TCCGGGGAGGATCCTATCGTTCAC |
| aviO3F | CCACGAATTCCGCGACTACG |
| aviO3R | CAGGTTCTAGACCGCCAGTAC |
| S3A | CTGCTGTGGCGCTACTCG |
| S3B | GAGCTGGCACGGGTAGTC |
| Primers used to screen for mutants | |
| O1Fn | CTCAGCGTCGACGTGCGCTGGCTGG |
| O1Rn | CATCCGAAAGACTCCCTGACCTGCC |
| O2aF | CTACGGGCGAATTCTTCGACATCG |
| O2aR | GGCGCGTCTAGACGGTGAGATTGG |
| O3Fn | CGCCTCCCTCGCGGTCCCCGAATG |
| O3Rn | CACCACCGCCAGTACTCCTCGTGC |
| Primers used to generate complementation plasmids | |
| aviO2FK | GACGAGGTCTAGAGTGCGGCGGTCC |
| aviO2RK | GACGGCCATGGAATTCCCGCTACCG |
| aviO3KF-Eco | GTCGACGAATTCGGCTACACGCTG |
| aviO3KR-Xba | GGAGCGCTCGGTCTAGACCTCCTG |
Construction of gene inactivation plasmids. (i) IKaviO1.
aviO1 was obtained using a 2.2-kb SacI fragment derived from F4E6 (Table 1; Fig. 2). The fragment was ligated into the SacI site of pSP1 to generate pSP1-O1/E6S22. A unique NcoI restriction site inside the gene aviO1 was chosen for targeted inactivation by shifting the reading frame. After NcoI restriction, treatment with T4 DNA polymerase, and religation, the intended alteration (correct fill-in) was confirmed by DNA sequencing.
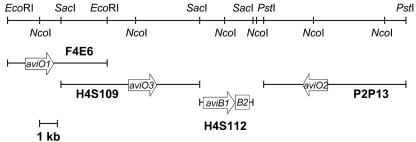
FIG. 2.
Region of the avilamycin biosynthesis gene cluster containing aviO1, aviO2, aviO3, aviB1, and aviB2. Only genes investigated during this study are shown as arrows. Fragments containing each individual gene as well as important restriction sites are indicated.
(ii) IkaviO2.
aviO2 was amplified by PCR using the oligodeoxynucleotides aviO2F and aviO2R (Table 2). The resulting 1.0-kb PCR fragment was ligated into EcoRI and BamHI sites of pBSK−, forming aviO2-SK−. A unique NdeI restriction site inside the gene aviO2 was chosen for targeted inactivation by shifting the reading frame in a way similar to that described for aviO1. The inactivation plasmid was named IkaviO2.
(iii) IkaviO3.
aviO3 was generated by PCR using the oligodeoxynucleotides aviO3F and aviO3R (Table 2). The 1.2-kb PCR fragment was ligated into EcoRI and XbaI sites of pBSK−, forming aviO3SK−. A unique NcoI restriction site inside the gene aviO3 was chosen for targeted inactivation by shifting the reading frame in a way similar to that described above. The inactivation plasmid was named IkaviO3.
(iv) IkaviB1.
aviB1 was obtained using a 2.0-kb SacI fragment of plasmid H4S112 from the avilamycin biosynthesis cluster (Fig. 2). The fragment containing aviB1 was ligated into the SacI site of pSP1 to generate AviB1pSP1. A unique NcoI restriction site inside the gene aviB1 was chosen for targeted inactivation by shifting the reading frame. The inactivation plasmid was named IkaviB1.
Generation of mutants.
The inactivation plasmids were introduced into S. viridochromogenes Tü57 by protoplast transformation using DNA isolated from E. coli ET12567. The integration of the inactivated genes was analyzed by PCR using Taq DNA polymerase. Screening for the integration of the defective aviB1 gene into the chromosome of GW4-ITB1 was carried out using the oligodeoxynucleotides S3A and S3B. For analyzing mutant ITO1, the primer pair O1Fn-O1Rn was used to screen for the integration of the defective aviO1 gene (Table 2). For analyzing mutant ITO2, we used the primer pair O2aF-O2aR. Mutant ITO3 was screened by PCR using the primer pair O3Fn-O3Rn. All oligonucleotides used for generating constructs and for analyzing the mutants are listed in Table 2.
Construction of complementation plasmids.
For the generation of the complementation plasmid O1pSET, plasmid pSP1-O1/E6S22 was digested with EcoRI and XbaI, resulting in a 2.2-kb fragment containing aviO1. This fragment was ligated into pSET-1cerm (2) previously incubated with MunI and XbaI to generate plasmid O1pSET. For complementation of the S. viridochromogenes mutants ITO2 and ITO3, aviO2 and aviO3 were amplified by PCR using Pfu polymerase and chromosomal DNA of the wild type as a template. The two suitable restriction sites, EcoRI and XbaI, were introduced using the primer pairs aviO2FK-aviO2RK and aviO3KF-Eco-aviO3KR-Xba (Table 2), respectively. PCR fragments were ligated into the MunI and XbaI restriction sites of pSET-1cerm to generate the integration plasmids O2pSET and O3pSET. For the complementation of S. viridochromogenes GW4-ITB1, aviB1 was cloned from H4S112 (Fig. 2) of the avilamycin biosynthesis gene cluster using the restriction sites PvuII and BamHI. It was ligated into the EcoRV and BglII sites of the replicative plasmid pKC1218 (Table 1). The resulting ligation product was called B1pKC and transferred into S. viridochromogenes GW4 by protoplast transformation.
Production and analysis of secondary metabolites.
S. viridochromogenes Tü57, S. viridochromogenes GW4, S. viridochromogenes ITO1, S. viridochromogenes ITO2, S. viridochromogenes ITO3, and S. viridochromogenes GW4-ITB1 were cultivated as described above. Cultures were filtered, and the filtrate (200 ml) was extracted with an equal volume of ethylacetate. After evaporation of the solvent, the residue was dissolved in 1 ml of 20% methanol in water and applied to a solid-phase extraction column (SepPak C18; Waters). The column was eluted using a 10 to 100% methanol gradient in water. Avilamycin derivatives could be detected in fractions containing 60 to 80% methanol. After evaporation of the solvent, the avilamycin-containing fractions were redissolved in 0.1 ml of methanol, and 2 μl was analyzed by thin-layer chromatography on silica gel plates (silica gel 60 F254; Merck) with methylene chloride-methanol (9:1, vol/vol) as a solvent. Avilamycin derivatives were detected after treatment with anisaldehyde/H2SO4.
HPLC-ESI-MS analysis.
High-performance liquid chromatography-electrospray ionization-mass spectrometry (HPLC-ESI-MS) was performed on an Agilent 1100 Series System with an electrospray chamber and a quadrupole detector. HPLC analysis was carried out on a Zorbax SB-C18 column (4.6 by 150 mm; 5-μm particle size; Agilent). A 20 to 70% nonlinear gradient of acetonitrile in 0.5% acetic acid over 22 min at a flow rate of 0.5 ml/min was used. The column temperature was 23°C, and the UV detection wavelength was 451 nm. The mass selective detector chamber settings were as follows: drying gas flow, 12 liters/min; nebulizing pressure, 50 lb/in2/g; drying gas temperature, 350°C. The samples were analyzed in a negative scan mode with a mass range of 300 to 1,500 Da.
NMR analysis.
The following experiments were performed with dimethyl sulfoxide-d6 at 295 K on a Bruker DMX 750-MHz spectrometer: 1H-1D, 13C-1D, correlated spectroscopy (COSY) (24), total correlation spectroscopy (1, 5), nuclear Overhauser effect spectroscopy (15), heteronuclear single-quantum coherence (HSQC) (4), heteronuclear multiple-bond correlation (26), heteronuclear multiquantum coherence-COSY (21), HSQC-total correlation spectroscopy (7), and 13C-filtered nuclear Overhauser effect spectroscopy (mixing time, 200 ms) (22). The spectrometer is part of the Bavarian NMR Center in Garching, Germany. All spectra were assigned using the program SPARKY 3 (13).
Bioassay.
Antibacterial activities of the new avilamycin A derivatives were tested using Bacillus subtilis COHN ATCC6051 from the American Type Culture Collection (20a).
Computer-assisted sequence analysis.
The DNASIS software package (Hitachi Software Engineering, San Bruno, Calif.) and the BLAST program were used for sequence analysis and homology searches of the GenBank database, respectively.
RESULTS
Generation of gene replacement mutants.
To inactivate aviB1, aviO1, aviO2, and aviO3, protoplasts of S. viridochromogenes GW4 (aviB1) and S. viridochromogenes Tü57 (aviO1, aviO2, and aviO3) were transformed using the inactivation plasmids IkaviB1, IkaviO1, IkaviO2, and IKaviO3. Several erythromycin-resistant colonies were obtained in each case.
After screening for loss of erythromycin resistance, double-crossover mutants were obtained. PCR fragments obtained from these mutants using primers listed in Table 2 could not be cleaved by NcoI (aviB1, aviO1, and aviO3) and NdeI (aviO2), whereas the PCR fragments obtained from S. viridochromogenes GW4 (aviB1) and S. viridochromogenes Tü57 (aviO1, aviO2, and aviO3) were clearly cut by the enzymes in each case.
aviB1.
The frameshift mutation within aviB1 was confirmed by Southern hybridization. As expected, a 1.4- and a 4.6-kb fragment of NcoI-cleaved chromosomal DNA of S. viridochromogenes GW4 were hybridizing with a 2-kb gene probe, while a 6-kb fragment was hybridizing with chromosomal DNA of mutant S. viridochromogenes GW4-ITB1 (Fig. 2).
aviO1.
The frameshift mutation within aviO1 was also confirmed by Southern blot analysis. When DNA from the wild-type S. viridochromogenes Tü57 cleaved by both NcoI and EcoRI was hybridized with a 2.2-kb probe, two fragments (3.8 and 2.3 kb) were detected. As expected, mutant S. viridochromogenes ITO1 gave only one signal at 6.1 kb (Fig. 2).
aviO2.
The complete digestion of chromosomal DNA by NdeI is not a reliable process. Therefore, the mutant S. viridochromogenes ITO2 was confirmed not by Southern hybridization but by amplifying the disrupted aviO2 gene by use of PCR with the primers O2aF and O2aR. The resulting PCR fragments were cloned into pBluescript SK−. Twenty-five recombinant plasmids were characterized by restriction analysis, and none of these plasmids could be restricted by NdeI. In addition, five plasmids were sequenced, and in all cases the defect within aviO2 was able to be confirmed (Fig. 2)
aviO3.
When NcoI-treated chromosomal DNA of the S. viridochromogenes ITO3 mutant was probed against a 1.2-kb fragment, a 10-kb signal was obtained. In contrast to this, two fragments of 5.4 and 4.6 kb were obtained using DNA of the wild-type strain (Fig. 2).
Complementation of S. viridochromogenes Tü57 mutants ITO1, ITO2, and ITO3 and S. viridochromogenes GW4-ITB1.
To determine clearly that the mutation events in the gene replacement mutants affected only the desired genes, aviO1, aviO2, aviO3, and aviB1 were ligated behind the ermE promoter, cloned into either the integration plasmid pSET152 (aviO1, aviO2, and aviO3) or pKC1218 (aviB1), and introduced by protoplast transformation into the corresponding mutants. Avilamycin A and C production (aviO1, aviO2, and aviO3) and gavibamycin A1 and A3 production (aviB1) were restored as confirmed by HPLC-ESI-MS. Thus, we were able to rule out any upstream or downstream effects.
Identification of new avilamycin derivatives produced by the defective mutants by HPLC-MS.
S. viridochromogenes Tü57, S. viridochromogenes GW4, and the four mutants S. viridochromogenes GW4-ITB1, S. viridochromogenes ITO1, S. viridochromogenes ITO2, and S. viridochromogenes ITO3 were grown under the conditions described in Materials and Methods. Extracts resulting from culture supernatants were analyzed by thin-layer chromatography, HPLC with UV detection, and HPLC-ESI-MS. As main products, all measured in the negative ion modus ([M]−), avilamycin C ([M]− = 1,403) was detected in extracts of S. viridochromogenes Tü57, and gavibamycin A3 ([M]− = 1,389) was detected in extracts of S. viridochromogenes Tü57-GW4. No avilamycin derivative could be detected in the S. viridochromogenes mutants ITO1 and ITO3. Avilamycin derivatives could be detected in extracts of the S. viridochromogenes mutants GW4-ITB1 and ITO2. The mass of the main product of S. viridochromogenes ITO2 was an [M]− value of 1,359, while the mass of the main product produced by S. viridochromogenes GW4-ITB1 was an [M]− value of 1,345. These results are in accordance with avilamycin derivatives lacking the acetyl residue at position C4 of the eurekanate moiety. The mass difference of 14 Da can be explained by the fact that S. viridochromogenes ITO2 was obtained from the wild-type strain and S. viridochromogenes GW4-ITB1 was obtained from S. viridochromogenes GW4 already lacking a methyl group at the dichloroisoeverninic acid moiety.
Identification by NMR of avilamycin derivatives produced by the defective mutants.
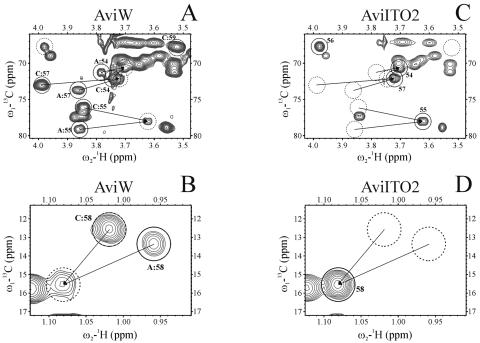
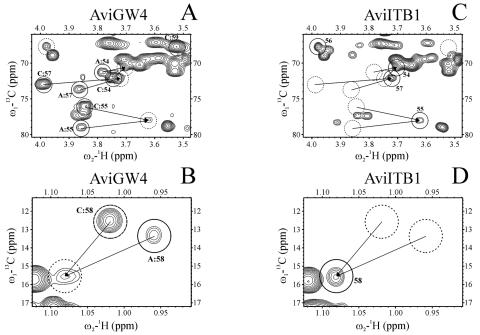
NMR analysis was now performed to demonstrate clearly that both S. viridochromogenes mutants ITO2 and GW4-ITB1 are lacking the acetyl residue at position C4 of the eurekanate moiety (Fig. 3 and 4). The HSQC spectra of the major compounds produced by S. viridochromogenes Tü57 (AviW) and S. viridochromogenes GW4 (AviGW4) show the signals of 54, 55, 57, and 59 (Fig. 3A and Fig. 4A) and 58 and 60 (Fig. 3B and Fig. 4B), corresponding to avilamycin A and C (AviW) and gavibamycin A1 and A3 (AviGW4). These two sets of signals are united in the spectra of AviITO2 (Fig. 3C and D) and AviITB1 (Fig. 4C and D). (The parts of the HSQC spectra with the signals of 56 and 60 have been omitted [for space considerations], but the corresponding results are shown.)
FIG. 3.
Sections of the HSQC spectra of extracts of S. viridochromogenes Tü57 (AviW) (A and B) and S. viridochromogenes ITO2 (AviITO2) (C and D). Interesting peaks are named and encircled. A: and C: indicate signals of avilamycin A and C response. Solid circles: avilamycin A (A and B), AviITO2 (C and D); dashed circles: avilamycin C (A and B); dotted circles: nonacetylated avilamycins (C and D), position of corresponding peaks in Fig. 4 A and B (C and D).
FIG. 4.
Sections of the HSQC spectra of S. viridochromogenes GW4 (AviGW4) (A and B) and S. viridochromogenes ITB1 (AviITB1) (C and D). Interesting peaks are named and encircled. A: and C: indicate signals of GW4 derivative of avilamycin A and C response. Solid circles: GW4 derivative of avilamycin A (A and B), AviITB1 (C and D); dashed circles: GW4 derivative of avilamycin C (A and B); dotted circles: nonacetylated GW4 derivative of avilamycin (C and D), position of corresponding peaks in Fig. 5 top and bottom (C and D).
DISCUSSION
AviB1 and AviB2, components of an incomplete PDH complex.
AviB1 shows convincing sequence similarities to thiamine PPi (TPP)-dependent enzymes. Enzymes that use TPP as a cofactor are found in all organisms, where they catalyze reactions involving the formation or cleavage of a carbon-carbon bond adjacent to an oxo group. The biologically active form of vitamin B1 is an essential cofactor in biocatalysis and is involved in numerous metabolic pathways, such as the oxidative and nonoxidative decarboxylation of α-keto acids (pyruvate decarboxylase, PDH), the formation of amino acid precursors (acetohydroxyacid synthase), electron transfer reactions (pyruvate oxidase), and ketol transfer between sugars (transketolase) (25). In all thiamine-dependent enzymes, there is a common sequence motif of about 30 residues that generates a common structural motif involved in the binding of the cofactor (14). Within the amino acid sequence of AviB1, we found the highly conserved TPP fingerprint binding region GDG and the highly conserved sequence EXXXXAXXXXPXXXNNKY. This motif is found in alpha chains of E1 components from PDH multienzyme complexes of several bacteria. In the avilamycin biosynthetic gene cluster, we also found aviB2. The deduced amino acid sequence of aviB2 resembles beta chains of E1 components of PDH complexes. Depending on the organism, the E1 component is an E1α2 homodimer typical for gram-negative bacteria or an E1α2β2 heterotetramer common in gram-positive bacteria. It was recently reported that the E1β subunit of the heterotetrameric type of E1 components is not functional in the absence of its α-subunit counterpart since it cannot bind cofactors or assemble into the appropriate quaternary structure (9, 17). Multiple sequence alignments of heterotetrameric E1 components indicated a common site that appeared to be conserved in E1α subunits (12). This region lies about 50 amino acids downstream of the TPP binding motif. When analyzing the amino acid sequence of AviB1, we found this consensus sequence, indicating that AviB1 and AviB2 form a heterotetrameric E1 (α2β2) complex.
AviO1, AviO2, and AviO3 are similar to α-ketoglutaric acid-dependent enzymes.
AviO1, AviO2, and AviO3 show some degree of similarity to α-ketoglutaric acid-dependent, nonheme iron-requiring enzymes. These enzymes contain two conserved histidine-signature sequences determined as His-1 and His-2 motifs (20). These motifs, named the 2-His-1-carboxylate facial triad, are thought to play a crucial role in accommodating a variety of catalytic reactions as desaturative cyclization or oxidative ring expansion.
Characterization of compounds produced by mutants ITO2 and GW4-ITB1.
The inactivation of the avilamycin biosynthetic genes aviO2 and aviB1 resulted in the production of novel avilamycin derivatives. Based on MS data, we could conclude that the acetyl group at the eurekanate moiety (ring H) was missing in extracts of both mutants. Based on our complete assignment of wild-type avilamycin A, the signals of the interesting atoms in ring H of the samples AviW and GW4 could be assigned. Starting from the characteristic signals of methylene group 61 in the spectra of AviITO2 and AviITB1, unambiguous nuclear Overhauser effect contacts to adjacent hydrogens 54 and 55 were found. Based on these assignments, the COSY spectra showed a yet-unassigned five-spin system that could be identified as C54H-C55H-C56H-C57H-C58H3. This spin system was already visible in the spectra of AviW and GW4 as an impurity not yet assigned. Therefore, the two reference samples contained both the normal and nonacetylated compounds. As indicated by our NMR results, S. viridochromogenes mutants ITO2 and ITB1 produce only nonacetylated avilamycin derivatives. The presence of nonacetylated derivatives in the reference samples leads to the assumption that in these cases the biosynthesis was aborted, and the molecules therefore were not completed. Additionally, sample AviW contains both avilamycin A and C, whereas sample AviGW4 contains the corresponding derivatives lacking the methyl group at the orsellinic acid moiety. As the difference between avilamycin A and C (or between the corresponding derivatives) is located in the interesting residue connected to C56, the signals of the atoms of ring H show two sets of different chemical shifts, one with the normal resonance frequencies for avilamycin A and another with slightly perturbed frequencies and an additional proton signal. These belong to avilamycin C, and the new signal is that of the hydrogen at C59. The two sets of signals collapse into one for AviITO2 and AviITB1, since they both lack the residue attached to C56.
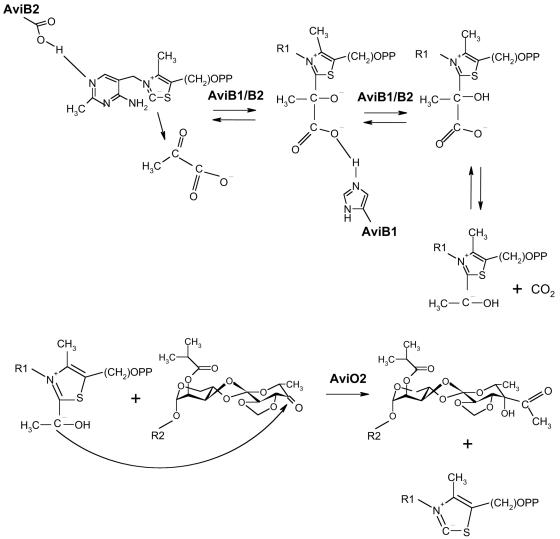
Function of AviB1/AbiB2 and AviO2.
The aviB1 mutant accumulates avilamycin derivatives lacking the acetyl group at position C4 of the eurekanate. In view of this result as well as sequence homology, the aviB1- and aviB2-encoded proteins must catalyze the conversion of pyruvate to an acetyl carbanion. The disruption of aviO2 also led to a mutant producing nonacetylated avilamycin derivatives. Based on this result, we speculate that AviO2 attaches the acetyl carbanion to the sugar moiety (Fig. 5). However, at present, we cannot exclude the possibility that AviO2 is involved in the oxidation of the 4-OH group to initiate the condensation step catalyzed by AviB1/AviB2. Surprisingly, derivatives produced by both the aviB1 and the aviO2 mutants lack the keto group at position C4 and contain a hydroxyl group instead. From the mechanistic point of view, the 4-keto group is required for the attachment of the carbanion. We believe that the 4-keto derivatives become reduced after the condensation step by a so-far-unrecognized, possibly unspecific ketoreductase. The functions of neither AviO1 nor AviO3 could be determined within this study. Inactivation of aviO1 or aviO3 resulted in a complete breakdown of avilamycin biosynthesis, and extracts of both mutants did not show any antibiotic activity, indicating an essential function of both genes for avilamycin A biosynthesis or its regulation.
FIG. 5.
Hypothetical mechanism of the enzymatic activation and attachment of a two-carbon branch to the eurekanate moiety. (Top) AviB1 and AviB2 are involved in the formation of an acetyl carbanion which is bound to thiamine diphosphate. (Bottom) AviO2 is involved in the attachment of the acetyl carbanion to form the branched-chain sugar moiety of avilamycin A.
Acknowledgments
Gisella Grabellus is thanked for her excellent technical assistance.
This work was supported by a grant of the BMBF, called “Genomforschung an Bakterien für den Umweltschutz, die Landwirtschaft und die Biotechnologie,” to A.B.
REFERENCES
- 1.Bax, A., and D. G. Davis. 1985. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 65:355-360. [Google Scholar]
- 2.Bibb, M. J., G. R. Janssen, and J. M. Ward. 1985. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene 38:215-226. [DOI] [PubMed] [Google Scholar]
- 3.Biermann, M., R. Logan, K. O'Brien, E. T. Seno, R. Nagaraja, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 4.Bodenhausen, G., and D. J. Ruben. 1980. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 69:185-189. [Google Scholar]
- 5.Braunschweiler, L., and R. R. Ernst. 1983. Coherence transfer by isotropic mixing: application to proton correlation spectroscopy. J. Magn. Reson. 53:521-528. [Google Scholar]
- 6.Buzzetti, F., F. Eisenberg, H. N. Grant, W. Keller-Schierlein, W. Voser, and H. Zähner. 1968. Avilamycin. Experientia 24:320-324. (In German.) [DOI] [PubMed] [Google Scholar]
- 7.Cavanagh, J., A. G. Palmer, P. E. Wright, and M. Rance. 1991. Sensitivity improvement in proton-detected 2-dimensional heteronuclear relay spectroscopy. J. Magn. Reson. 91:429-436. [Google Scholar]
- 8.Chen, H., Z. Guo, and H. Liu. 1998. Biosynthesis of yersiniose: attachment of the two-carbon branched-chain is catalyzed by a thiamine pyrophosphate-dependent flavoprotein. J. Am. Chem. Soc. 120:11796-11797. [Google Scholar]
- 9.Danson, M. J., A. R. Fersht, and R. N. Perham. 1978. Rapid intramolecular coupling of active sites in the pyruvate dehydrogenase complex of Escherichia coli: mechanism for rate enhancement in a multimeric structure. Proc. Natl. Acad. Sci. USA 75:5386-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flett, F., V. Mersinias, and C. P. Smith. 1997. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155:223-229. [DOI] [PubMed] [Google Scholar]
- 11.Foster, D. R., and M. J. Rybak. 1999. Pharmacologic and bacteriologic properties of SCH-27899 (Ziracin), an investigational antibiotic from the everninomicin family. Pharmacotherapy 19:1111-1117. [DOI] [PubMed] [Google Scholar]
- 12.Fries, M., H. J. Chauhan, G. J. Domingo, H. I. Jung, and R. N. Perham. 2003. Site-directed mutagenesis of a loop at the active site of E1 (α2β2) of the pyruvate dehydrogenase complex. A possible common sequence motif. Eur. J. Biochem. 270:861-870. [DOI] [PubMed] [Google Scholar]
- 13.Goddard, T. D., and D. G. Kneller. 2001. Sparky 3. University of California, San Francisco, Calif.
- 14.Hawkins, C. F., A. Borges, and R. N. Perham. 1989. A common structural motif in thiamin pyrophosphate-binding enzymes. FEBS Lett. 255:77-82. [DOI] [PubMed] [Google Scholar]
- 15.Jeener, J., B. H. Meier, P. Bachmann, and R. R. Ernst. 1979. Investigation of exchange processes by two-dimensional NMR-spectroscopy. J. Chem. Phys. 71:4546-4553. [Google Scholar]
- 16.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. and Hoopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 17.Kleiger, G., J. Perry, and D. Eisenberg. 2001. 3D structure and significance of the GPhiXXG helix packing motif in tetramers of the E1β subunit of pyruvate dehydrogenase from the archeon Pyrobaculum aerophilum. Biochemistry 40:14484-14492. [DOI] [PubMed] [Google Scholar]
- 18.Mankin, A. S. 2001. Ribosomal antibiotics. Mol. Biol. 35:509-520. [Google Scholar]
- 19.McNicholas, P. M., D. J. Najarian, P. A. Mann, D. Hesk, R. S. Hare, K. J. Shaw, and T. A. Black. 2000. Evernimicin binds exclusively to the 50S ribosomal subunit and inhibits translation in cell-free systems derived from both gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 44:1121-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myllyla, R., V. Gunzler, K. I. Kivirikko, and D. D. Kaska. 1992. Modification of vertebrate and algal prolyl 4-hydroxylases and vertebrate lysyl hydroxylase by diethyl pyrocarbonate. Evidence for histidine residues in the catalytic site of 2-oxoglutarate-coupled dioxygenases. Biochem. J. 286(Pt. 3):923-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 21.Norwood, T. J., J. Boyd, J. E. Heritage, N. Soffe, and I. D. Campbell. 1990. Comparison of techniques for H-1-detected heteronuclear H-1-N-15 spectroscopy. J. Magn. Reson. 87:488-501. [Google Scholar]
- 22.Otting, G., H. Senn, G. Wagner, and K. Wüthrich. 1986. Editing of 2D 1H NMR spectra using X half-filters: combined use with residue-selective 15N-labeling of proteins. J. Magn. Reson. 70:500-505. [Google Scholar]
- 23.Pelzer, S., W. Reichert, M. Huppert, D. Heckmann, and W. Wohlleben. 1997. Cloning and analysis of a peptide synthetase gene of the balhimycin producer Amycolatopsis mediterranei DSM5908 and development of a gene disruption/replacement system. J. Biotechnol. 56:115-128. [DOI] [PubMed] [Google Scholar]
- 24.Rance, M., O. W. Sorensen, G. Bodenhausen, G. Wagner, R. R. Ernst, and K. Wüthrich. 1983. Improved spectral resolution in COSY 1H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun. 117:479-485. [DOI] [PubMed] [Google Scholar]
- 24a.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Schellenberger, A. 1998. Sixty years of thiamin diphosphate biochemistry. Biochim. Biophys. Acta 1385:177-186. [DOI] [PubMed] [Google Scholar]
- 26.Summers, M. F., L. G. Marzilli, and A. Bax. 1986. Complete 1H and 13C assignments of coenzyme B12 through the use of new two-dimensional NMR experiments. J. Am. Chem. Soc. 108:4285-4294. [Google Scholar]
- 27.Treede, I., L. Jakobsen, F. Kirpekar, B. Vester, G. Weitnauer, A. Bechthold, and S. Douthwaite. 2003. The avilamycin resistance determinants AviRa and AviRb methylate 23S rRNA at the guanosine 2535 base and the uridine 2479 ribose. Mol. Microbiol. 49:309-318. [DOI] [PubMed] [Google Scholar]
- 28.Weitnauer, G., A. Muhlenweg, A. Trefzer, D. Hoffmeister, R. D. Sussmuth, G. Jung, K. Welzel, A. Vente, U. Girreser, and A. Bechthold. 2001. Biosynthesis of the orthosomycin antibiotic avilamycin A: deductions from the molecular analysis of the avi biosynthetic gene cluster of Streptomyces viridochromogenes Tu57 and production of new antibiotics. Chem. Biol. 8:569-581. [DOI] [PubMed] [Google Scholar]
- 29.Wright, D. E. 1979. The orthosomycins, a new family of antibiotics. Tetrahedron 35:1207-1237. [Google Scholar]