Abstract
The ichthyocidal activity of Pfiesteria piscicida dinospores was examined in an aquarium bioassay format by exposing fish to either Pfiesteria-containing environmental sediments or clonal P. piscicida. The presence of Pfiesteria spp. and the complexity of the microbial assemblage in the bioassay were assessed by molecular approaches. Cell-free water from bioassays that yielded significant fish mortality failed to show ichthyocidal activity. Histopathological examination of moribund and dead fish failed to reveal the skin lesions reported elsewhere. Fish larvae within “cages” of variable mesh sizes were killed in those where the pore size exceeded that of Pfiesteria dinospores. In vitro exposure of fish larvae to clonal P. piscicida indicated that fish mortality was directly proportional to the dinospore cell density. Dinospores clustered around the mouth, eyes, and operculi, suggesting that fish health may be affected by their direct interaction with skin, gill epithelia, or mucous surfaces. Molecular fingerprinting revealed the presence of a very diverse microbial community of bacteria, protists, and fungi within bioassay aquaria containing environmental sediments. Some components of the microbial community were identified as potential fish pathogens, preventing the rigorous identification of Pfiesteria spp. as the only cause of fish death. In summary, our results strongly suggest (i) that this aquarium bioassay format, which has been extensively reported in the literature, is unsuitable to accurately assess the ichthyocidal activity of Pfiesteria spp. and (ii) that the ichthyocidal activity of Pfiesteria spp. is mostly due to direct interactions of the zoospores with fish skin and gill epithelia rather than to soluble factors.
Pfiesteria piscicida is a heterotrophic dinoflagellate that has been associated with fish mortality and human health problems in estuaries along the Atlantic shore of the United States (6, 11, 19; http://www.epa.gov/OWOW/estuaries/pfiesteria/, http://www.pfiesteria.seagrant.org/). Evidence of this association has relied on the detection of motile P. piscicida cells in water samples collected from sites where fish mortality has occurred (11, 13, 29) or close to areas where unusual dermal, respiratory, and neurological symptoms were reported by individuals upon exposure to environmental water (2, 17, 38). The ichthyocidal activity of P. piscicida, routinely assessed in an aquarium bioassay format (12), was reported to become apparent only when the vegetative forms (dinospores) emerge from resting cysts present in environmental sediments in the presence of fish (7). By analogy to other dinoflagellates, it was proposed that P. piscicida produces one or more toxins that affect fish and other organisms, including mammals (5, 8, 27, 34, 36). It has been hypothesized that the action of these toxins on fish would result in skin lesions, loss of neural function, and eventually death (16, 22, 28). Despite significant efforts by several laboratories directed towards the isolation and identification of the proposed toxin(s), no success has been reported in the peer-reviewed literature to date. The situation has been complicated further by the description of nontoxic or temporarily nontoxic strains that revert to toxicity under certain environmental cues (8, 9) and by an unusually complex life cycle with 24 stages of variable toxicity (8, 10). However, rigorous experimental data in support of the aforementioned claims have been lacking, and the existence of the amoeba stage and Pfiesteria toxin(s) has recently been questioned in detailed studies (1, 24, 32, 42). A recent in vitro study of the ichthyocidal activity of clonal P. piscicida in a culture flask bioassay format indicated that active proliferation of the dinospores can be associated with fish death, although the cause(s) remains unknown (33).
The goal of this study was to examine the ichthyocidal activity of P. piscicida dinospores in a controlled laboratory setting, using the standard aquarium bioassay format, to gain insight into possible icthyocidal mechanisms. The report that the presence of live fish during the excystment of P. piscicida zoospores from the sediment is a requirement for its toxicity or ichthyocidal activity (8, 9) was taken into account as a key factor in the experimental design. Accordingly, live fish were exposed either to sediments that contained Pfiesteria spp. or to P. piscicida clonal cultures, their health was monitored throughout the experiments, and mortalities were recorded. The presence of Pfiesteria spp. and the complexity of the microbial assemblage in the experimental system were examined by molecular approaches, and water quality parameters were monitored to assess their potential modulatory role in the effects of P. piscicida on fish. In vitro experiments were conducted to study in further detail the possible mechanism(s) of the observed ichthyocidal activity of P. piscicida.
MATERIALS AND METHODS
Clonal dinoflagellate cultures.
Clonal P. piscicida, a generous gift from K. A. Steidinger (Florida Department of Environmental Protection, St. Petersburg), was isolated in 1997 by K. A. Steidinger and J. M. Burkholder (Center for Applied Aquatic Ecology, North Carolina State University) from the Chicamacomico River, Maryland. It was propagated in f/2 medium at a salinity of 7 or 15 ppt with artificial seawater (ASW) (Instant Ocean; Aquarium Systems Inc., Mentor, Ohio) (18) at 23°C under a 14-h light, 10-h dark cycle (white fluorescent; 150 mol of photon m−2 s−3) and was fed with live cryptomonads (Rhodomonas sp. strain CCMP 768, obtained from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton, West Boothbay Harbor, Maine). P. piscicida was harvested or used for experiments after it reached cell density of above 105 cells ml−1. Karlodinium micrum was provided by D. K. Stoecker (Center of Environmental Studies, University of Maryland, Cambridge) and D. W. Coats (Smithsonian Environmental Research Center, Edgewater, Md.). COMB4872, a presumptive Oxyrrhis strain, was isolated from a water sample (collection site unknown) provided by the Maryland Department of Natural Resources as a part of the “Pfiesteria and Fish Health” field monitoring program (http://www.dnr.state.md.us/bay/cblife/algae/dino/pfiesteria/monitoring_updates.html) by the Core Facility for Culture of Toxic Dinoflagellates at the Center of Marine Biotechnology (COMB), University of Maryland Biotechnology Institute, Baltimore.
Environmental samples.
Sediment and water samples were collected from selected fish ponds at Hyrock Fish Farm, an aquaculture facility along the Manokin River, Princess Anne, Md., and at several sites along the Neuse River in North Carolina, in which fish kills attributed to harmful algal blooms associated with P. piscicida have been reported (7; http://www.pfiesteria.org/). Sediment samples from all ponds tested positive for P. piscicida by a species-specific PCR assay (see below). Sediment samples from ponds 1, 4, 8, and 10 were selected for this study based on the frequency of recent fish kill events.
Experimental fish.
Adult (2- to 3-cm-long) and larval (1- to 2-day-old) sheepshead minnows (Cyprinodon variegatus) were purchased from Aquatic Biosystems Inc. (Fort Collins, Colo.) and gradually acclimated to a salinity of 7 ppt (pH 8.0) at 23°C for at least 2 weeks prior to use.
Bioassay for ichthyocidal activity in aquarium format. (i) Biosafety procedures.
Bioassays were carried out in a biosafety level 3 laboratory equipped with a glove box (model 818-GB; PLAS-LABS Inc., Lansing, Mich.) and a class II type B2 biological safety cabinet (Purifier Class II Total Exhaust; LABCONCO Co., Kansas City, Mo.). Exhaust air exiting the cabinet was processed through pleated bag dust filters, separate HEPA filters, and nuclear-grade charcoal filters before being exhausted from the building (http://www.cdc.gov/od/ohs/biosfty/bmbl/section3.htm). All experiments were conducted according to established standard operating procedures for Pfiesteria cultivation and experimental methodologies, in accordance with Center for Disease Control and Prevention (Atlanta, Ga.) guidelines (http://www.cdc.gov/mmwr/preview/mmwrhtml/00049554.htm) as administered by the University of Maryland Department of Environmental Health and Safety.
(ii) Experimental aquarium design.
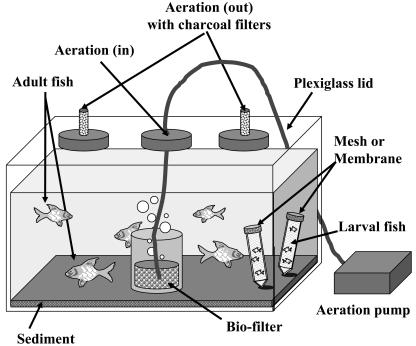
Aquaria (8 or 40 liters) were filled with 6 or 30 liters of ASW at a salinity of 7 ppt and fitted with either “naíve” or “activated” crushed-coral filters (200 g of coral in a filter) for aeration with a standard air pump (500 ml min−1). Activated biofilters were prepared by placing naíve filters in a 40-liter aquarium with 15 fish for at least 2 weeks, to allow for bacterial colonization of the substrate surface. Aquaria were sealed with tightly fitting Plexiglas lids specifically designed to avoid any release of potentially toxic aerosols generated by the aeration devices to the environment (Fig. 1). For this purpose, lids were fitted with inlets for air and feeding, and air outlets were fitted with built-in activated-charcoal filters. Aquaria were placed in a glove box, and all experiments were performed within the biosafety level 3 facility.
FIG. 1.
Experimental aquarium design. Aquaria were fitted with Plexiglas lids with ports for charcoal-filtered vents, feeding, and dead fish retrieval. Activated or naíve crushed coral filters and aeration pumps were used to maintain water quality and oxygen concentration in the bioassay at desired levels. For the cage experiment, standard 50-ml Falcon tubes fitted with membranes of selected pore sizes (60 μm, 10 μm, and 30 kDa) were filled with sterile ASW, fish larvae were introduced, and the tubes were placed in the experimental aquarium.
(iii) Bioassay for ichthyocidal activity.
Sediment samples (20 or 100 ml) were resuspended with ASW (salinity, 7 ppt; pH 8.0; aerated overnight and filter sterilized [0.2-μm-pore-size filter]; sediment/ASW ratio, 1:5) into a slurry and sequentially sieved through a coarse metal mesh and a 60-μm-pore-size nylon filter (Spectrum Laboratories, Inc., Savannah, Ga.). The sieved sediment slurry was placed in the bioassay aquaria (100 ml in 8-liter aquaria or 500 ml in 40-liter aquaria), and the volume was completed with ASW. Fish were introduced into the bioassay aquaria to a final density of 2 fish liter−1, and their health and behavior were monitored throughout the experimental period. Dead fish were immediately removed from the aquarium and replaced by the same number that died in order to maintain the fish density at 2 fish liter−1. Sediment sample controls were prepared by autoclaving (steam sterilization at 121°C and 15 lb/in2 gauge for 20 min) the sediment slurry (sediment/ASW ratio, 1:5) and processed as described above. Experimental control aquaria were inoculated with either clonal culture of P. piscicida to a final density of 103 cells ml−1, autoclaved sediment, or ASW without a dinoflagellate or sediment inoculum.
(iv) Emergence of P. piscicida dinospores from sediment.
A slurry (50 ml) of pond 10 sediment processed as described above, with an additional 450 ml of ASW, was incubated in a 750-ml vented cap culture flask with or without one fish for a period of 30 days. The appearance of P. piscicida dinospores in the water column was monitored by PCR, and Pfiesteria-like dinoflagellates were identified by direct observation under an optical microscope. Water samples for DNA extraction (15 ml) and for cell counts (1 ml) were collected from each experimental flask at 18 time points during the experimental time period. P. piscicida-specific PCR was carried out under conditions reported elsewhere (37). A quantitative assessment of P. piscicida dinospores at selected time points was carried out by PCR on serial dilutions of the DNA samples spiked with an internal standard (2 × 103 copies in a 20-μl reaction mixture) as described by Saito et al. (37).
(v) Water quality analyses.
Concentrations of ammonia, nitrate, and nitrite in experimental aquaria were measured with colorimetric assay kits (HACH Co., Loveland, Colo.) as reported earlier (33), but with the assays adapted to a 96-well plate format.
(vi) Cell counts.
Dinoflagellate cell densities were assessed by direct counting of formalin-fixed cells in a hemocytometer. Water samples (15 ml) were collected from the bioassay aquaria (every 7 days) and concentrated by centrifugation at 2,000 × g followed by removal of 14 ml of supernatant. The cell pellet was resuspended in the remaining supernatant (1 ml) and fixed with 4% formalin.
(vii) Histopathological examination of fish.
Histopathological examinations were carried out on euthanatized moribund fish or immediately after death in the experimental tanks. After the gross necropsy examination was performed, specimens were fixed in 10% buffered formalin, embedded into paraffin, and sectioned (5-μm sections). The physiological and pathological examinations were carried out on sections stained with hematoxylin and eosin.
(viii) Cell-free ichthyocidal activity bioassay.
Water samples (500 ml) from experimental and control aquaria were filtered through a 0.2-μm-pore-size filter (Nalge Nunc International Co., Rochester, N.Y.) and transferred into a 750-ml culture flask fitted with aeration. One small fish (approximately 1 cm in length) was introduced into each flask, and its health and behavior were monitored throughout the exposure period (10 days).
(ix) Size-selective exposure bioassay.
“Cages” for exposure of fish larvae in the bioassay aquaria were constructed by cutting a 3-cm-diameter hole into the lids of 50-ml plastic tubes (BD Biosciences, Bedford, Mass.). The tubes were filled with 7-ppt-salinity ASW, and fish larvae (1 to 2 days old; 5 or 30 larvae cage−1) were introduced. The tube opening was covered with a nylon mesh or cellophane membrane of the selected pore size, and the lids were screwed onto the tubes over the mesh or membrane covers. Tubes were placed into the bioassay aquaria of control fish, fish with clonal P. piscicida (aquarium CPP), and fish with sediment pond 10 (aquarium P10), and survival of larvae was examined after 16 h of exposure.
PCR-based detection assay for P. piscicida.
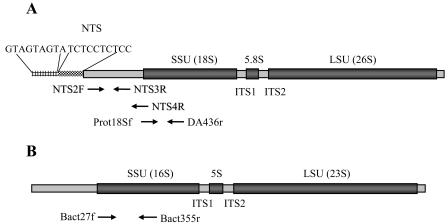
The presence of P. piscicida in the bioassay aquaria was examined by a species-specific PCR-based detection assay reported elsewhere (37). DNA was extracted from cell pellets obtained from water samples (15 ml) with the FastDNA spin kit for soil (Q · BIOgene, Inc., Carlsbad, Calif.). PCR primers (NTS2F and NTS4R in Fig. 2) that amplify a 523-bp target sequence in the nontranscribed spacer (NTS) within the intergenic spacer of the P. piscicida rRNA locus were used for the detection. Integrity of the DNA templates was confirmed by PCR amplification with universal actin primers designed to amplify actin genes from lower eukaryotes to vertebrates (amplicons, ≅730 bp; primers, G-480 and G-482 [kindly provided by G. W. Warr, Medical University of South Carolina, Charleston]).
FIG. 2.
Organization of P. piscicida and prokaryotic rRNA genes and locations of the molecular probes used in this study. The primers for P. piscicida species-specific PCR (NTS2F, NTS3R, and NTS4R) and for ALH fingerprinting and cloning (protists, Pro18Sf and DA436r; eubacteria, Bact27f and Bact355r) are in panels A and B, respectively.
Characterization of microbial assemblages by ALH fingerprinting. (i) DNA extraction and PCR conditions for ALH fingerprinting.
Whole community genomic DNA was extracted as described above for the species-specific PCR-based detection assay (37). PCR amplification of the first two variable regions of the small-subunit (SSU) rRNA was performed with a fluorescently (6-carboxyfluorescein) labeled forward primer and a nonfluorescent reverse primer (Fig. 2). These primers were a conserved eukaryotic primer, Prot18Sf (5′-6-carboxyfluorescein-GGTTGATCCTGCCAGTAGTCATATGCTTG-3′) and a primer that was conserved in both amoebae and dinoflagellates, DA436r (5′-TTRCGCGCCTGCTGCYTTCCTT-3′). For positive controls, DNAs from pure laboratory cultures were used, while in negative controls, the DNA was replaced with diethyl pyrocarbonate-treated water. Final concentrations or amounts in PCR mixtures were 1× PCR buffer, 0.25 mM MgSO4, 0.25 mM deoxynucleoside triphosphates (Boehringer Gmbh, Mannheim, Germany), 0.5 μM forward and reverse primers, 0.25 U of Tf1 DNA polymerase (Promega Corp., Madison, Wis.), 0.1% (wt/vol) bovine serum albumin, (fraction V; ICN Biomedicals Inc., Aurora, Ohio), 10 to 40 ng of DNA, and DEPC-treated water to make up the final volume. Amplicon length heterogeneity (ALH) PCR products were diluted, mixed with 1.5 μl of internal standard (GeneScan-1000 ROX; Applied Biosystems, Foster City, Calif.), denatured in deionized formamide (98%; Sigma, St. Louis, Mo.), and loaded directly onto 4.25% denaturing polyacrylamide gels (bisacrylamide/acrylamide ratio, 19:1; Bio-Rad, Richmond, Calif.) on an ABI 377 instrument (Applied Biosystems) for 7 h.
(ii) Data analyses.
ALH fingerprint profiles were analyzed by using the ABI Prism GeneScan and ABI Prism Genotyper software (Applied Biosystems) and Microsoft Excel (Microsoft Corp.). In GeneScan, the ALH-PCR profiles were analyzed by using analysis parameters set to the local Southern size calling, no peak correction for the shorter products, and leftmost peak correction for the longer products. The minimum noise threshold was set at 50 fluorescence units. A Visual Basic routine written in Microsoft Excel was used to calculate the relative abundance of each peak in the fingerprint by dividing each individual peak area by the total peak area of each electropherogram.
(iii) Cloning and sequencing of the microbial assemblages.
Samples from experimental bioassay aquaria were pooled, and whole-community genomic DNA was extracted as described above for ALH fingerprinting. PCR amplification of the first two variable regions of the SSU rRNA for both the eubacterial community and the protist community was performed with either the eukaryotic primer set Prot18Sf and DA436r (see “DNA extraction and PCR conditions for ALH fingerprinting” above) or the standard eubacterial primer set Bact27f and Bact355r (25, 41).
Prior to cloning, PCR amplicons from each pooled tank community were cleaned with a Wizard Prep kit (Promega) according to the manufacturer's protocol. The concentrated PCR amplicons were sized via electrophoresis on a low-melting-temperature agarose gel. Amplicons within a selected size range were excised from the gel and isolated with a Wizard purification kit as described in the Promega Notes bulletin 118, section IV, for cloning. Ligation, transformation, incubation, and screening of the inserted fragments were done with either pCR 2.1 or TOPO II vector (Invitrogen Corp., Carlsbad, Calif.) according to the manufacturer's protocol.
Sequence data were obtained for all isolates with ABI BigDye Terminator version 2 according to the protocol of the manufacturer (Applied Biosystems). Contigs obtained from all clones were assembled with Sequencher 4.1 software (Gene Codes Co., Ann Arbor, Mich.). The sequence data for each community in this study were then used for BLAST searches in GenBank (National Center for Biotechnology Information, Rockville, Md.). The results of the BLAST search were parsed with a PERL script to enumerate the multiple hits for each community.
Exposure of fish to high cell densities of P. piscicida dinospores.
P. piscicida dinospore densities of above 6 × 105 cells ml−1 were obtained from a standard culture (1 × 105 to 2 × 105 cells ml−1) by selectively isolating a broad layer, approximately 1 cm from the bottom of the flask, where the P. piscicida dinospores concentrate during stationary culture. The isolated culture layer (approximately 30 ml) was adjusted to the experimental cell densities (0.001 × 105 to 1 × 105, 1 × 105 to 4 × 105, or 4 × 105 to 7 × 105 cells ml−1) by dilution in f/2 medium (salinity, 15 ppt) and transferred into a 25-ml culture flask, and a small fish (approximately 1 cm in length) was introduced. Control fish were maintained in f/2 medium (salinity, 15 ppt) in the absence of P. piscicida dinospores. Experiments were carried out at room temperature (around 21 to 23°C), and fish health, behavior, and time of death monitored for 16 h. Interactions of P. piscicida dinospores with fish were examined in six-well plates containing one fish larva (1 to 2 days old) and P. piscicida dinospores (2 × 105 cells ml−1) in 3 ml of f/2 medium (salinity, 15 ppt) in each well. Wells were examined under an inverted light microscope (magnification, ×100 to ×200), and interactions were documented by digital photography.
RESULTS
Characterization of the aquarium bioassay format. (i) Optimization of the biofiltration system.
Aquaria fitted with naíive crushed-coral filtration systems and inoculated with sediments from ponds 1, 4, 8, and 10 (Hyrock Fish Farm) revealed that dinoflagellates appeared in the water column within 24 to 48 h, with dinospore densities reaching maximal levels in 5 to 10 days (data not shown). The increase in dinospore density was concurrent with fish death, which ranged from approximately 20% (pond 1) to 55% (ponds 4 and 10). Pond 10 showed the highest fish mortality relative to dinospore density. The water quality assessment revealed that ammonia concentrations gradually increased, reaching the highest levels (about 6 to 12 mg of N liter−1) within 5 to 10 days, and later declined as the nitrite concentrations increased, presumably by microbial conversion of the former to the latter. Although the control aquarium yielded 20% fish mortality, this was the only one in which ammonia continued to increase throughout the course of the experiment, reaching levels toxic to fish (>6 to 10 mg of N liter−1 within a pH range of 7.8 to 8.0).
Aquaria fitted with activated crushed-coral filters, containing either fresh or autoclaved sediments, showed little or no increase in ammonia and nitrite concentrations, likely due to the activity of the microbial biofilm present on the surface of the coral. Aquaria inoculated with fresh pond 10 sediments maintained constant low levels of ammonia and nitrites and remained positive for P. piscicida throughout the 35-day experiment. Aquaria inoculated with autoclaved pond 10 sediments exhibited a water quality profile similar to those from aquaria with naive filters described above: ammonia levels increased at about day 5 and declined at day 20, the time at which nitrite levels increased (data not shown). This suggests that the microbial flora present in the sediments is a major contributor to water quality. As expected, no P. piscicida was detected in the water of the latter aquaria.
(ii) Effects of the presence of fish on emergence of P. piscicida dinospores from sediments.
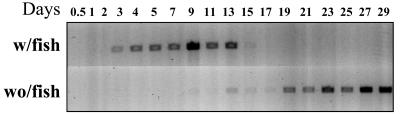
The effect of fish or fish products on the emergence of dinospores from sediments was examined in the flask format bioassay (33) by incubating pond 10 sediments in ASW either with or without fish and assessing the presence of P. piscicida in the water column by use of a species-specific PCR. The earlier detection of P. piscicida in the former samples suggests that the presence of fish may promote cyst germination from the sediment, followed by either dinospore proliferation or grazing pressure reduction (Fig. 3).
FIG. 3.
Detection of P. piscicida dinospores emerged from sediment with (w/) and without (wo/) fish by species-specific PCR. The emergence of P. piscicida from Hyrock Farm pond 10 sediment with fish (top panel) or without fish (bottom panel) was examined in a flask bioassay format. Water samples (1 and 15 ml) were collected for detection of P. piscicida and counting of Pfiesteria-like dinoflagellates, respectively, following the time course (days 0.5, 1, 2, 3, 4, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, and 29).
(iii) Ichthyocidal activity in bioassay aquaria inoculated with sediments.
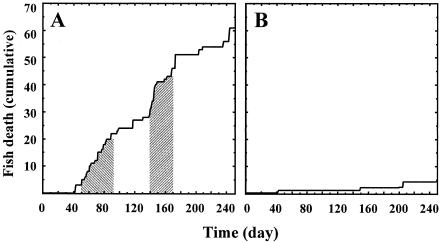
The ichthyocidal activity of microorganisms present in sediments from the Maryland fish farm and the Neuse River, North Carolina, was examined in the aquarium bioassay format characterized above, with the goal of identifying and isolating soluble factors potentially toxic to fish that may be secreted or excreted by P. piscicida dinospores or another component(s) of the microbial assemblage. Aquaria (40 liters) fitted with activated crushed-coral filters were inoculated with sediment from either pond 10 from the Maryland fish farm (aquarium P10), sediment from Neuse River (aquarium NR), or clonal P. piscicida (aquarium CPP) and monitored for the presence of P. piscicida and fish death. Within 24 to 48 h postinoculation, Pfiesteria-like dinoflagellates were observed in the sediment-inoculated aquaria, and the water tested positive for P. piscicida by PCR. All aquaria remained positive for P. piscicida throughout the experiment (240 days), although dinospore densities were lower (≤102 cells ml−1) than those observed in the preliminary experiments described above. At day 41 from the start of the experiment, the first fish died in aquarium P10. During the course of the experiment, fish died in aquarium P10 at intervals varying from two to three fish per day to one fish in 2 weeks, with two periods in which fish deaths increased in frequency: phase I (days 50 to 90) and phase II (days 140 to 170) (Fig. 4A). In contrast, only occasional fish deaths occurred in aquarium CPP (Fig. 4B). No fish deaths were observed in aquarium NR (data not shown). Histopathological analysis of tissues from moribund or dead fish from this experiment (aquaria P10 and CPP) failed to reveal skin lesions or any common signs that would indicate a unique cause for death, such as the action of a soluble toxin. In general, the liver, adipose, intestine, and swim bladder tissues were histologically normal. In some specimens, granulomatous branchitis (an inflammatory process in the gills), presence of protozoan cysts in skin and mesentery, necrosis in melanomacrophages in kidney, and focal granulomatous encephalitis were observed. Water quality (ammonia, nitrate, and nitrite levels) remained within limits (nitrite, <0.005 mg of N liter−1; ammonia, <0.02 mg of N liter−1) compatible with fish health in the three aquaria.
FIG. 4.
Bioassay in aquarium format with an activated biofilter. Hyrock Farm (Maryland) pond 10 sediments (A) or clonal P. piscicida culture (B) and Neuse River (North Carolina) sediments (not shown) were inoculated into bioassay aquaria. Fish mortality was recorded for 240 days. Shaded areas indicate phases I and II of ichthyocidal activity. Water samples from all three aquaria tested positive for P. piscicida, and water quality remained compatible with fish health throughout the experimental period. Samples for analysis of microbial assemblage by ALH and denaturing gradient gel electrophoresis were taken from assay aquaria P10 and CPP.
(iv) Cell-free ichthyocidal activity bioassay.
In order to elucidate the cause(s) of the ichthyocidal activity observed in aquarium P10, the presence of a putative soluble factor(s) in the water column was examined by exposing fish to “cell-free” P10 and CPP water collected during the period at which high fish mortality was observed in aquarium P10 (150 days) (Fig. 4). Cell-free exposure experiments were carried out in culture flasks filled with filtered (0.2-μm-pore-size filter) aquarium water, with one adult fish per flask. In all experiments fish remained healthy during a 10-day exposure, with no particular signs of stress, while fish deaths continued to occur in aquarium P10.
(v) Size-selective exposure bioassay.
This assay enabled the selective exposure of the fish larvae to soluble components (30-kDa-cutoff cellophane membrane), bacteria or small dinoflagellates (10-μm-pore-size nylon mesh), or other larger protozoa or dinoflagellates (60-μm-pore-size nylon mesh) that may be present in the bioassay system. Greater fish mortality occurred in the P10 aquarium (60% total mortality; n = 105 fish larvae) than in CPP aquarium (3% total mortality; n = 105 fish larvae) and the control aquarium (0% total mortality; n = 105 fish larvae) within a 16-h exposure period. In aquarium P10, fish mortality was higher in those cages with larger-pore-size mesh: 20% mortality in the cage with a ≤30-kDa cutoff, 60 to 80% in the cage with a ≤10-μm cutoff, and 90 to 100% in the cage with a ≤60-μm cutoff. Microscopic examination of the cage contents revealed the presence of Pfiesteria-like dinoflagellates within the 10 and 60-μm-cutoff mesh cages.
(vi) Characterization of the microbial assemblage by ALH fingerprinting.
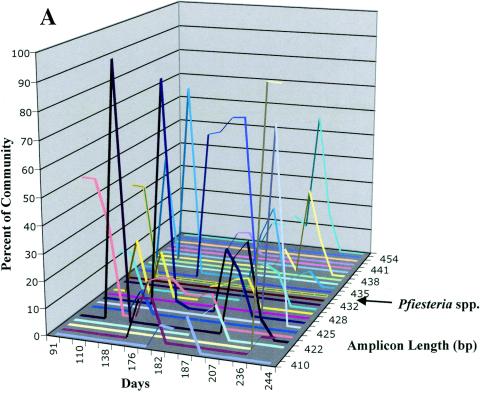
In order to examine which organism(s) may be associated with the fish deaths observed in the bioassay and cage experiments described above, we attempted to characterize the composition and dynamics of the microbial assemblage in aquarium P10. To assess the biodiversity present in the water column, selected samples (days 91 through 244) were analyzed by ALH fingerprinting. The samples selected represent a time period in which we observed two bursts of ichthyocidal activity in the P10 tank bioassay (phase I, days 50 to 90; phase II, days 140 to 170). ALH fingerprinting characterizes the organisms in a community by amplifying variable regions of the rRNA and separating the natural variation of amplicon length on a denaturing polyacrylamide gel. Each amplicon peak in the profile represents the presence of very few distinct species, and the peak area is proportional to the abundance of those species in the community. The ALH fingerprints depict the protist community at days 91, 187, and 222.
The predicted amplicon size for both P. piscicida and P. shumwayae was 434 bp. Pfiesteria spp. were a major component of the community at day 91 and were associated with the phase I ichthyocidal activity. In contrast, Pfiesteria spp. were not a major component of the community during the rest of the P10 tank assay, although both P. piscicida and P. shumwayae were still detectable with our standard PCR assay (Fig. 5) throughout the entire tank bioassay. The phase I ichthyocidal activity was also correlated with the presence of other protist taxa, as there were two other major amplicon peaks present at this time period. In fact, over 50% of the community fingerprint was represented by an amplicon of 424 bp, thus precluding any correlation of ichthyocidal activity to the abundance of Pfiesteria spp. The ALH profile at day 187 reveals that the Pfiesteria amplicon had been reduced to less than 1% of the ALH community profile (see shoulder on the peak at 436 bp), and seven other amplicon peaks had emerged at this time period, with only one (at 431 bp) shared with the day 91 profile. Finally, the ALH profile from day 222 shows the emergence of three new amplicons (peaks at 423, 431, and 439 bp) and the loss of four amplicons (peaks at 410, 422, 424, and 428 bp). Figure 5A illustrates the overall diversity of the protist community cycling through the selected sampling period. A total of 26 different amplicon lengths (each potentially revealing the presence and abundance of at least one distinct taxon) were observed to cycle throughout the test period, with some taxa persisting for extended time periods (i.e., amplicons at peaks of 424 and 477 bp), while others appeared for only short time periods (i.e., amplicons at peaks of 427 and 444 bp). Some taxa appeared only once (i.e., amplicons at peaks of 426 bp), while others (i.e., amplicons at peaks of 430 bp) appeared at several time points during the course of the experiment.
FIG. 5.
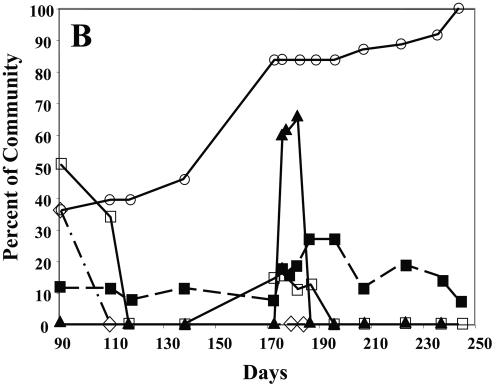
Representative ALH fingerprints of P10 aquarium bioassay. A: Total community fingerprints of the P10 aquarium bioassay. Samples of the P10 bioassay aquarium were collected from day 91 to 244, and ALH fingerprinting was performed. A percentage of the community for each amplicon length was calculated and plotted. The amplicon length (434 bp) representing Pfiesteria spp. is indicated on the plot. B: Plot of the accumulated fish death in the P10 bioassay aquarium versus the total community diversity. Samples of the P10 bioassay aquarium were analyzed by ALH fingerprinting as described for panel A. The accumulated fish death (○) was plotted as a percentage of the total community abundance. The number of amplicons at each time point (▪) was plotted as a percentage of the total community diversity. The relative abundances of peaks at 424 (□), 434 (Pfiesteria spp.) (⋄), and 435 (▴) bp was also plotted. The pooled PCR amplicons obtained with universal primer sets for eubacterium and protist SSU rRNAs were cloned and sequenced. The results of BLAST searches with sequences are listed in Table 1 (bacteria) and Table 2 (protists).
The correlation of the abundance (blooms) of specific microbial taxa with phase I and II ichthyocidal activity in aquarium P10 is shown in Fig. 5B. The number of killed fish per day was plotted as a percentage of the total accumulated dead fish, and the number of taxa (amplicon lengths) present at each day was plotted as a percentage of the total community diversity (26 taxa), along with the community dynamics of three selected peaks. A fairly complex community was observed during the first phase of ichthyocidal activity, and two small protist blooms occurred at days 190 and 220, neither of which correlated with phase II of ichthyocidal activity at day 180. The bloom of the amplicon at the peak of 434 bp, which corresponded to the predicted size for P. piscicida and P. shumwayae, was associated with phase I but not with phase II of ichthyocidal activity. In fact, the putative Pfiesteria peak at 434 bp had virtually disappeared from community by 110 days, although Pfiesteria spp. were still present in small numbers as determined by PCR. The bloom of the amplicon at peak 435 bp correlated with phase II of ichthyocidal activity but was not present during the first phase. Interestingly, the amplicon at the peak of 424 bp correlated with both phase I and II of ichthyocidal activity. Although it was by far the most abundant component of the protist community during phase I of ichthyocidal activity (>50%), it was only a minor community component during phase II of ichthyocidal activity (∼16%). The identity of either the amplicon at 424 bp or the amplicon at 435 bp has yet to be determined.
To identify the components of the diverse microbial assemblage present in P10 and CPP aquaria, DNAs extracted from pooled water samples were amplified, cloned, and sequenced. Based on sequencing of randomly selected clones, at least 95 organisms were identified by BLAST search (Tables 1 and 2). Three bacteria were found in both aquaria and were identified as Flexibacter aggregans and a Cytophaga sp. (both Flexibacteracae) and a Pseudomonas sp. (γ-Proteobacteria). Brevibacillus borstelensis and Pseudoaltermonas spp., which are categorized by the Advisory Committee on Dangerous Pathogens as group 1 pathogens; Vibrio vulnificus, which is a widely distributed pathogen that causes disease in fish, shellfish, and humans; and cyanobacteria, which can also be toxic or pathogenic, were detected only in the CPP aquarium. It is noteworthy that P. piscicida was the only protist common to both the P10 and CPP aquaria.
TABLE 1.
Bacterial communities in experimental aquaria
| Aquarium and accession no. | No. of hits | Closest relative
|
|
|---|---|---|---|
| Species | Taxonomic description | ||
| P10 | |||
| AB078038 | 3 | Flexibacter aggregans | Flexibacteraceae |
| AJ224416 | 2 | Cytophaga sp. | Sphingobacteria; Flexibacteraceae |
| AJ400946 | 1 | Methylobacterium sp. strain NP19 | α-Proteobacteria |
| AF414882 | 1 | α-Proteobacterium P708 | α-Proteobacteria |
| AJ419674 | 1 | Pseudomonas sp. strain 2N1-1 | γ-Proteobacteria |
| AJ430587 | 1 | Caldithrix abyssi | Anaerobic bacterium |
| AJ244689 | 1 | Cyclobacterium sp. strain V4.MS.32 | Sphingobacteria; Flexibacteraceae |
| AY241567 | 1 | Marine bacterium HP34 | Sphingobacteria; Flexibacteraceae |
| AF029039 | 1 | Benzene-mineralizing consortium clone SB-1 | Environmental |
| X72770 | 1 | Methylococcus capsulatus | γ-Proteobacteria |
| CPP | |||
| AY183115 | 43 | Cyanobium sp. strain LB03 | Cyanobacteria |
| AY162047 | 15 | α-Proteobacterium PI_GH2.1.D5 | α-Proteobacteria |
| X76084 | 7 | Nanochlorum eucaryotum | Green microalgae; Chlorophyta |
| AY154889 | 6 | Flavobacterium sp. strain SRI | Flavobacteria |
| AJ224414 | 4 | Cytophaga sp. | Sphingobacteria; Flexibacteraceae |
| AB078038 | 3 | Flexibacter aggregans | Sphingobacteria; Flexibacteraceae |
| AB000478 | 3 | Aquaspirillum itersonii | α-Proteobacteria |
| AF182021 | 2 | Cytophaga sp. strain BAL50 | Sphingobacteria; Flexibacteraceae |
| AE016801 | 2 | Vibrio vulnificus CMCP6 | γ-Proteobacteria |
| AE016795 | |||
| AF330253 | 2 | Synechococcus sp. strain BS 5 | Cyanobacteria |
| AB069650 | 1 | Rhizobium sp. JEYF16 | α-Proteobacteria; Rhizobium-Agrobacterium group |
| AY217769 | 1 | Agrobacterium sp. strain SP25 | α-Proteobacteria; Rhizobium-Agrobacterium group |
| AY162083 | 1 | α-Proteobacterium GMD37F4 | α-Proteobacteria |
| AY162115 | 1 | α-Proteobacterium GMD13F07 | α-Proteobacteria |
| AF041446 | 1 | Bradyrhizobium sp. strain SH 283012 | α-Proteobacteria |
| AY190148 | 1 | Sphingobium sp. strain S14 | α-Proteobacteria |
| AF408866 | 1 | Pseudomonas sp. strain NZWM5 | γ-Proteobacteria |
| AY187028 | 1 | Pseudoalteromonas sp. strain MMM18 | γ-Proteobacteria |
| AB064358 | 1 | Aeromonas sp. strain AER 102 | γ-Proteobacteria |
| AF430120 | 1 | Aeromonas sp. strain VKM B-2261 | γ-Proteobacteria |
| AF539686 | 1 | Aeromonas sp. strain Ni46 | γ-Proteobacteria |
| X74677 | 1 | Aeromonas hydrophila | γ-Proteobacteria |
| AJ431219 | 1 | Proteobacterium BH160-11 | Proteobacteria |
| M62797 | 1 | Flavobacterium aquatile | Flavobacteria |
| M28236 | |||
| D89036 | 1 | Microcystis holsatica | Cyanobacteria |
| AF025552 | 1 | Cytophagales strain MED9 | Bacteroidetes-Chlorobi group |
| AY241563 | 1 | Marine bacterium HP28 | Sphingobacteria; Flexibacteraceae |
| AB015264 | 1 | Cytophaga sp. | Sphingobacteria; Flexibacteraceae |
| AY275498 | 1 | Pedobacter sp. strain MSB3023 | Sphingobacteria |
| D78456 | 1 | Brevibacillus borstelensis | Paenibacillaceae |
| AF538743 | 1 | Bacterium CAGY1 | Eubacteria |
| AF001655 | 1 | Environmental clone OCS31 | Environmental |
TABLE 2.
Protist communities in experimental aquaria
| Aquarium and accession no. | No. of hits | Closest relative
|
|
|---|---|---|---|
| Species | Taxonomic description | ||
| P10 | |||
| X80341 | 11 | Neocallimastix frontalis | Fungi |
| AF194410 | 8 | Halteria grandinella | Ciliates |
| AF164241 | 7 | Powellomyces variabilis | Fungi |
| U97112 | 5 | Strombidium purpureum | Ciliates |
| AF192386 | 4 | Pseudoperkinsus tapetis | Ichthyosporea |
| AF368505 | 3 | Basidiobolus microsporus | Fungi |
| AF374481 | 3 | Thalassiosira pseudonana | Diatoms |
| Y17504 | 2 | Hyaloraphidium curvatum | Fungi |
| AF290539 | 2 | Cryothecomonas aestivalis | Heterotrophic flagellates |
| AF448162 | 2 | Proscoloplos cygnochaetus | Polychaetes |
| AY212807 | 2 | Parauronema longum | Ciliates |
| Y16938 | 2 | Ankistrodesmus bibraianus | Green algae |
| AY112746 | 1 | Pfiesteria piscicida | Dinophyceae |
| AF335515 | 1 | Epistylis wenrichi | Ciliates |
| AY103190 | 1 | Uronema elegans | Ciliates |
| AF508769 | 1 | Oxytricha sp. strain Steamboat Hot Springs | Ciliates |
| AJ310493 | 1 | Gonostomum strenuum | Ciliates |
| AJ488910 | 1 | Novistrombidium testaceum subsp. ligusticum | Ciliates |
| AB041250 | 1 | Phyllosticta pyrolae | Fungi |
| AF164237 | 1 | Spizellomyces kniepii | Fungi |
| AF322420 | 1 | Penicillidia sp. strain VH-2001 | Fungus-metazoan group |
| L10824 | 1 | Diaphanoeca grandis | Choanoflagellates |
| AF421220 | 1 | Mastigamomeba sp. strain ATCC50617 | Mastigamoebidae |
| AF462059 | 1 | Thalassiosira rotula | Thalassiosiraceae |
| AF457128 | 1 | Lecudina sp. strain BSL-2002 | Apicomplexans |
| AY039208 | 1 | Aurelia aurita | Jellyfishes; Scyphozoa |
| AF534709 | 1 | Chlamydaster sterni | Centrohelids |
| X74324 | 1 | Phytophthora capsici | Heterokonts (plant disease) |
| X79096 | |||
| U20320 | 1 | Minchinia teredinis | Alveolates |
| CPP | |||
| AY212807 | 23 | Parauronema longum | Ciliates |
| L27633 | 17 | Cafeteria roenbergensis | Bicosoecids |
| M32704 | 12 | Ochromonas danica | Golden algae |
| J02950 | |||
| X06425 | 6 | Nanochlorum eucaryotum | Green algae |
| AB022864 | 5 | Paraphysomonas foraminifera | Golden algae |
| AY112746 | 4 | Pfiesteria piscicida | Dinophyceae |
| X56104 | 4 | Scenedesmus vacuolatus | Green algae |
| AF290540 | 2 | Cryothecomonas longipes | Cryothecomonas |
| AF174364 | 2 | Cafeteria roenbergensis | Bicosoecids |
| AF174365 | 2 | Cafeteria sp. strain EWM2 | Bicosoecids |
| U52357 | 1 | Scrippsiella nutricula | Symbiotic dinoflagellates; Dinophyceae |
| AF289081 | 1 | Nuclearia-like filose amoeba N-Por | Cercozoa |
| AF513373 | 1 | Scenedesmus sp. strain SEV3VF49 | Green algae |
| AF388379 | 1 | Dimorphococcus lunatus | Green algae |
| AB037097 | 1 | Tetradesmus wisconsinensis | Green algae |
| AY220081 | 1 | Nannochloris sp. strain ANR-9 | Green algae |
| AY197621 | 1 | Scenedesmid sp. strain Mary 9/21 BT-16w | Green algae |
| AF164261 | 1 | Chytridiales sp. strain JEL207 | Fungi |
| X54864 | 1 | Podospora anserina | Fungi |
| AF047888 | 1 | Paracanthonchus caecus | Nematoda |
| Y15814 | 1 | Ostreococcus tauri | Prasinophytes |
| L27633 | 1 | Cafeteria roenbergensis | Bicosoecids |
Exposure of fish to high cell densities of P. piscicida dinospores.
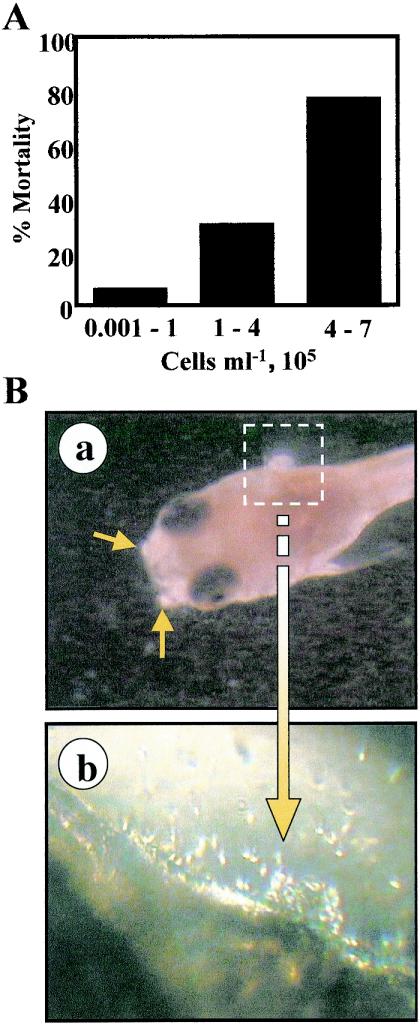
In the course of these experiments and previous work (33), we observed that fish mortality could be associated with increased densities of Pfiesteria-like dinoflagellates and/or Pfiesteria spp. To investigate a possible correlation between P. piscicida dinospore density and fish mortality, we exposed fish to various cell densities of clonal P. piscicida and monitored fish health and mortality within a 16-h experimental period. We obtained high densities of clonal P. piscicida by selectively collecting the lower layers of the culture flask, where dinospores accumulate during stationary culture. Results of the high-density exposure experiments are summarized in Fig. 6A. About 80% of the fish exposed to P. piscicida dinospore densities of 4 × 105 to 7 × 105 cells ml−1 died within 16 h. A significant portion of these died within 5 h of exposure, during which they showed increasing signs of anoxia, such as gulping for air at the surface. Although dissolved oxygen was relatively low in the experimental flasks (1.5 mg of O2 liter−1), they were comparable to the control flasks in that the unexposed fish lived for extended periods of time without showing any signs of anoxia.
FIG. 6.
Fish mortality after exposure to a high density of P. piscicida. A: Fish mortality within 16 h upon exposure to increasing densities of P. piscicida dinospores (n = 39). B: Distribution of P. piscicida dinospores around fish larvae. Fish larvae were exposed to clonal P. piscicida in a six-well plate bioassay format. P. piscicida dinospores clustered around the mouth (a) as well as the eyes and operculi (b).
Observation of fish larvae exposed to P. piscicida dinospores in six-well plates revealed that within a few minutes high numbers of P. piscicida dinospores concentrated around the larvae, in particular the operculi, eyes, mouth, and fins, clearly attaching to the skin (Fig. 6B). The fish appeared to be irritated by the swarming and attachment of the dinospores and would swim erratically, becoming obviously stressed as the exposure time increased. This behavior was not observed in fish maintained in the absence of P. piscicida.
DISCUSSION
In this study we examined the potential ichthyocidal activity of P. piscicida in an aquarium format bioassay and investigated the possibility that soluble factors secreted or excreted by this dinoflagellate species may be responsible for such activity. Previous work from our laboratory (33), aimed at assessing the ichthyocidal activity of P. piscicida in a small-scale and high-throughput flask format assay, revealed the association of fish death with proliferation of P. piscicida within the first 2 weeks of the experiment course. However, the deteriorated water quality and proliferation of multiple microorganisms after that initial period hindered the unambiguous establishment of cause-effect relationships linking P. piscicida and fish death. Furthermore, the relatively small volumes of crude samples produced in those experiments precluded the development of rigorous and systematic fractionation and purification procedures that may lead to the identification and isolation of the toxins or bioactive compounds that may have caused fish deaths. The aquarium bioassay format characterized in the present study was developed with the goal of addressing the abovementioned problems. Although the presence of Pfiesteria spp. and the observation of fish death in the bioassay revealed similarities with bioassays reported elsewhere (6, 10, 13), the detailed analysis and interpretation of the results questioned the unambiguous association of Pfiesteria spp. with fish deaths and suggested that a more complex analysis is required to explain the observations.
The presence of fish in the bioassay induced a rapid appearance of Pfiesteria spp. and Pfiesteria-like dinoflagellates in the water column, most likely by excystment from pond sediments and proliferation. Because some of the sediments tested were from dry ponds, it is unlikely that dinospores observed in the water column were already present in interstitial spaces between the sediment particles but rather is likely that they emerged from resting cysts. This is consistent with earlier reports suggesting that P. piscicida dinospores rapidly emerge from cysts present in sediments and enhance proliferation of dinospores in the presence of fish (13), although the induction mechanism(s) remains unknown.
The absence of skin lesions in the moribund and dead fish in the bioassay was inconsistent with previous reports describing skin ulcers as the result of toxic activity by Pfiesteria spp. (5). A recent study suggests that these are probably due to the fungal pathogen Aphanomyces invadens, the causative agent of epizootic ulcerative syndrome (3, 4, 23). In the present study, histological analysis indicated pathologies commonly associated with bacterial and protozoan infections.
The lack of even noticeable changes in fish health upon exposure to dinospore-free water collected from aquaria where fish deaths were observed suggested that either the putative ichthyocidal soluble factor was extremely labile, as proposed elsewhere (6, 27), or fish mortalities were caused by biotic components of the bioassay system. Furthermore, experiments designed to examine possible correlations between particle size and fish death revealed that fish were killed at the highest rates in those cages where the pore size exceeded that of medium-sized protists, including Pfiesteria dinospores, suggesting that the largest contribution to the fish deaths was from direct interaction with components of the microbial assemblage. However, a 10% increase in fish death relative to the controls was observed in cages fitted with a membrane through which only soluble factors could diffuse, suggesting that a soluble factor(s) present in the bioassay may have a limited yet significant contribution to the overall fish mortality observed. However, no evidence that would support attributing its source to Pfiesteria spp. or to any particular component of the bioassay microbial flora was obtained.
ALH fingerprinting, a robust and well-established method for analyzing biocomplexity (14, 35, 41), was used to examine the composition and dynamics of the microbial flora in experimental and control bioassay systems and the possible association of Pfiesteria spp., other protists, or bacteria with the fish deaths. Although a major peak of Pfiesteria spp. obtained by ALH coincided with phase I of ichthyocidal activity, it was not present in significant amounts during phase II. Furthermore, Pfiesteria was not the only taxon correlated with phase I, suggesting that although it may play a part in some events under specific conditions (i.e., when present at high density), its mere presence should not immediately imply causality. In addition to P. piscicida, at least 95 species of prokaryotic and eukaryotic organisms, including other dinoflagellate species, ciliates, algae, fungi, nematodes, and diatoms, were tentatively identified by BLAST analysis on the ALH amplicon library. Among the diverse components of the assemblage, the presence of potentially pathogenic bacteria and toxin producers was noteworthy because of their possible deleterious effects on fish health. ALH analysis also confirmed the fluctuation of dinospore densities in the water column of the experimental and control bioassays detected by direct cell counts. The natural cycling of heterotrophic dinoflagellates from dinospore to either division, digestion, or conversion resting cysts (24, 30, 31) may be modulated by the availability of prey and physical environmental conditions, such as temperature, light, salinity, etc. (13, 20), and could be responsible for this fluctuation. A second possible cause is the potential effect of grazing of Pfiesteria spp. by other components of the microbial assemblage (21). Pfiesteria spp. are vulnerable to predation by protozoan ciliates, rotifers, and the microcrustacean copepod zooplankton (39, 40). Our preliminary studies suggest that Oxyrrhis sp., a large dinoflagellate species identified in the bioassay system, is an active grazer on P. piscicida dinospores (data not shown).
The development and optimization of the flask bioassay format revealed the association of P. piscicida dinospore densities in the water column and fish deaths (33). Because the populations of Pfiesteria dinospores fluctuated in a similar fashion within the aquarium bioassay format, it became critical to experimentally test this possible association. Our results not only clearly confirm the proposed quantitative relationship between Pfiesteria dinospore density and fish mortality but also suggest that relatively large numbers of dinospores (>105 cells ml−1) are required to kill fish larvae. Microscopic observation of the interactions of the dinospores with the fish indicated that high numbers of P. piscicida dinospores concentrated around the larvae, in particular the operculi, eyes, mouth, and fins, clearly attaching to the skin. This behavior is similar to that reported by Vogelbein et al. (42) for P. shumwayae but is in sharp contrast with their findings that P. piscicida cultured in the laboratory did not attach to, feed on, or exhibit pathogenicity for fish. Studies by others have confirmed our observations concerning the aggressive behavior of P. piscicida towards fish (W. Litaker, personal communication), suggesting that genetic and/or environmental factors may affect the behavior of Pfiesteria spp. towards prey.
In conclusion, this study documents for the first time that in addition to Pfiesteria spp., a very diverse assemblage of bacteria, protists, and fungi, including potential pathogens of fish, are present in the aquarium bioassay. In the absence of histopathological signs that could be specifically assigned to Pfiesteria spp., this observation makes it impossible to unambiguously attribute the cause of fish death to Pfiesteria spp. Based on field observations and experimental evidence from various bioassay formats (5, 8-11, 13, 26), it has been proposed that Pfiesteria spp. kill fish by the release of one or more toxins. However, results have been inconsistent between laboratories, and the existence of a Pfiesteria toxin has yet to be demonstrated. Association does not necessarily imply causality, and bioassay systems need to be examined rigorously and alternative hypotheses need to be tested before conclusions are drawn. Some of the problems in bioassay interpretation reside in the lack of experimental evidence demonstrating that a clonal axenic culture of P. piscicida is ichthyocidal and that a cell-free supernatant from this culture is ichthyotoxic. Because in this study only a minor portion of the fish death could be attributed to soluble components, it is unlikely that under the experimental conditions established, the fish deaths in an aquarium bioassay are caused by a toxin, as proposed by others for similar bioassay formats (6, 8, 27). Moreover, considering the complexity of the microbial assemblage, there is no valid reason to attribute to the Pfiesteria spp. present the synthesis and secretion or excretion of such a soluble component(s). The evidence presented here also supports reexamination of the rationale for the interpretation of results obtained in cell receptor assays using bioassay supernatants, as well as environmental water (15). Our results suggest that, as described for P. shumwayae (43), any contribution of Pfiesteria spp. to fish death in the bioassay system would be mostly mediated by direct interactions of the dinospore with fish external surfaces, as a result of their feeding behavior. The relevance of this behavior to ichthyocidal activity in the natural environment remains to be examined.
Acknowledgments
We acknowledge the expertise provided by K. A. Steidinger (Florida Department of Environmental Protection, St. Petersburg) and D. W. Coats (Smithsonian Environmental Research Center, Edgewater, Md.) and helpful suggestions from D. E. Terlizzi (COMB, University of Maryland Sea Grant Extension, Baltimore), M. Alavi (COMB, University of Maryland Biotechnology Institute, Baltimore), and A. Mazzaccaro (Hyrock Farm, Princess Anne, Md.).
This study was partially supported by grants NIEHS 5-P01-ES09563, ECOHAB NA860P0192, NSF DEB-9972093, and CTSG LIS LR/LR-5.
REFERENCES
- 1.Berry, J. P., K. S. Reece, K. S. Rein, D. G. Baden, L. W. Haas, W. L. Ribeiro, J. D. Shields, R. V. Snyder, W. K. Vogelbein, and R. E. Gawley. 2002. Are Pfiesteria species toxicogenic? Evidence against production of ichthyotoxins by Pfiesteria shumwayae. Proc. Natl. Acad. Sci. USA 99:10970-10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bever, C. T., Jr., L. Grattan, and J. G. Morris. 1998. Neurologic symptoms following Pfiesteria exposure: case report and literature review. Maryland Med. J. 47:120-123. [PubMed] [Google Scholar]
- 3.Blazer, V. S., J. H. Lilley, W. B. Schill, Y. Kiryu, C. L. Densmore, V. Panyawachira, and S. Chinabut. Aphanomyces invadans in Atlantic menhaden along the East Coast of the United States. J. Aquat. Anim. Health 14:1-10.
- 4.Blazer, V. S., W. K. Vogelbein, C. L. Densmore, E. B. May, J. H. Lilley, and D. E. Zwerner. 1999. Aphanomyces as a cause of ulcerative skin lesions of menhaden from Chesapeake Bay tributaries. J. Aquat. Anim. Health 11:340-349. [Google Scholar]
- 5.Burkholder, J. M. 2002. Pfiesteria: the toxic Pfiesteria complex, p. 2431-2447. In G. Bitton (ed.), The encyclopedia of environmental microbiology. Wiley & Sons, Inc., New York, N.Y.
- 6.Burkholder, J. M., and H. B. Glasgow. 2001. History of toxic Pfiesteria in North Carolina estuaries from 1991 to the present. BioScience 51:827-841. [Google Scholar]
- 7.Burkholder, J. M., and H. B. Glasgow. 1997. Trophic controls on stage transformations of a toxic ambush-predator dinoflagellate. J. Eukaryot. Microbiol. 44:200-205. [DOI] [PubMed] [Google Scholar]
- 8.Burkholder, J. M., H. B. Glasgow, and N. J. Deamer-Melia. 2001. Overview and present status of the toxic Pfiesteria complex (Dinophyceae). Phycologia 40:186-214. [Google Scholar]
- 9.Burkholder, J. M., H. B. Glasgow, N. J. Deamer-Melia, J. Springer, M. W. Parrow, C. Zhang, and P. J. Cancellieri. 2001. Species of the toxic Pfiesteria complex, and the importance of functional type in data interpretation. Environ. Health Perspect. 109:667-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burkholder, J. M., and H. B. Glasgow, Jr. 1997. Pfiesteria piscicida and other Pfiesteria-like dinoflagellates: behavior, impacts, and environmental controls Limnol. Oceanogr. 42:1052-1075. [Google Scholar]
- 11.Burkholder, J. M., H. B. Glasgow, Jr., and C. W. Hobbs. 1995. Fish kills linked to a toxic ambush-predator dinoflagellate: distribution and environmental conditions. Mar. Ecol. Prog. Ser. 124:43-61. [Google Scholar]
- 12.Burkholder, J. M., H. G. Marshall, H. B. Glasgow, D. W. Seaborn, and N. J. Deamer-Melia. 2001. The standardized fish bioassay procedure for detecting and culturing actively toxic Pfiesteria, used by two reference laboratories for Atlantic and Gulf Coast states. Environ. Health Perspect. 109:745-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burkholder, J. M., E. J. Noga, C. W. Hobbs, and H. B. Glasgow, Jr. 1992. New ‘phantom’ dinoflagellate is the causative agent of major estuarine fish kills. Nature 358:407-410. [DOI] [PubMed] [Google Scholar]
- 14.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2001. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 67:191-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Nabawi, A., M. S. Quesenberry, K. Saito, E. Silbergeld, G. R. Vasta, and A. Eldefrawi. 2000. The N-methyl-d-aspartate neurotransmitter receptor is a mammalian brain target for the dinoflagellate Pfiesteria piscicida toxin. Toxicol. Appl. Pharmacol. 169:84-93. [DOI] [PubMed] [Google Scholar]
- 16.Glasgow, H. B., J. M. Burkholder, M. A. Mallin, N. J. Deamer-Melia, and R. E. Reed. 2001. Field ecology of toxic Pfiesteria complex species and a conservative analysis of their role in estuarine fish kills. Environ. Health Perspect. 109:715-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grattan, L. M., D. Oldach, T. M. Perl, M. H. Lowitt, D. L. Matuszak, C. Dickson, C. Parrott, R. C. Shoemaker, C. L. Kauffman, M. P. Wasserman, J. R. Hebel, P. Charache, and J. G. Morris, Jr. 1998. Learning and memory difficulties after environmental exposure to waterways containing toxin-producing Pfiesteria or Pfiesteria-like dinoflagellates Lancet 352:532-539. [DOI] [PubMed] [Google Scholar]
- 18.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 26-60. In W. L. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum Press, New York, N.Y.
- 19.Harvell, C. D., K. Kim, J. M. Burkholder, R. R. Colwell, P. R. Epstein, D. J. Grimes, E. E. Hofmann, L. E. K., A. D. Osterhaus, R. M. Overstreet, J. W. Porter, G. W. Smith, and G. R. Vasta. 1999. Emerging marine diseases—climate links and anthropogenic factors. Science 285:1505-1510. [DOI] [PubMed] [Google Scholar]
- 20.Iwasaki, H. 1984. Growth physiology of red-tide microorganisms. Microbiol. Sci. 1:179-182. [PubMed] [Google Scholar]
- 21.Jeong, H. J., S. K. Kim, J. S. Kim, S. T. Kim, Y. D. Yoo, and J. Y. Yoon. 2001. Growth and grazing rates of the heterotrophic dinoflagellate Polykrikos kofoidii on red-tide and toxic dinoflagellates. J. Eukaryot. Microbiol. 48:298-308. [DOI] [PubMed] [Google Scholar]
- 22.Kane, A. S., D. Oldach, and R. Reimschuessel. 1998. Fish lesions in the Chesapeake Bay: Pfiesteria-like dinoflagellates and other etiologies. Maryland Med. J. 47:106-112. [PubMed] [Google Scholar]
- 23.Kiryu, Y., J. D. Shields, W. K. Vogelbein, H. Kator, and V. Blazer. 2003. Dose response and pathogenicity of secondary zoospores of the oomycete, Aphanomyces invadans, to Atlantic menhaden, Brevoortia tyrannus. Dis. Aquat. Organisms 54:135-146. [DOI] [PubMed] [Google Scholar]
- 24.Litaker, R. W., M. W. Vandersea, S. R. Kibler, V. J. Madden, E. J. Noga, and P. A. Tester. 2002. Life cycle of the heterotrophic dinoflagellate Pfiesteria piscicida (Dinophyceae). J. Phycol. 38:442-463. [Google Scholar]
- 25.Marchesi, J. R., T. Sato, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiom, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rrnA. Appl. Environ. Microbiol. 64:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marshall, H. M., A. S. Gordon, D. W. Seaborn, B. Dyer, W. M. Dunstan, and A. M. Seaborn. 2000. Comparative culture and toxicity studies between the toxic dinoflagellate Pfiesteria piscicida and a morphologically similar cryptoperidiniopsoid dinoflagellate. [DOI] [PubMed]
- 27.Moeller, P. D., S. L. Morton, B. A. Mitchell, S. K. Sivertsen, E. R. Fairey, T. M. Mikulski, H. B. Glasgow, N. J. Deamer-Melia, J. M. Burkholder, and J. S. Ramsdell. 2001. Current progress in isolation and characterization of toxins isolated from Pfiesteria piscicida. Environ. Health Perspect. 109:739-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noga, E. J. 2000. Skin ulcers in fish: Pfiesteria and other etiologies. Toxicol. Pathol. 28:807-823. [DOI] [PubMed] [Google Scholar]
- 29.Noga, E. J., S. A. Smith, J. M. Burkholder, C. H. Hobbs, and R. A. Bullis. 1993. A new ichthyotoxic dinoflagellate: cause of acute mortality in aquarium fishes. Vet. Res. 133:96-97. [DOI] [PubMed] [Google Scholar]
- 30.Parrow, M., and J. M. Burkholder. 2003. Reproduction and sexuality in Pfiesteria shumwayae (Dinophyceae). J. Phycol. 39:697-711. [Google Scholar]
- 31.Parrow, M., J. M. Burkholder, N. J. Deamer, and C. Zhang. 2002. Vegetative and sexual reproduction in Pfiesteria spp. (Dinophyceae) cultured with algal prey, and inferences for their classification. Harmful Algae 1:5-33. [Google Scholar]
- 32.Peglar, M. T., L. A. Amaral Zettler, O. R. Anderson, T. A. Nerad, P. M. Gillevet, T. E. Mullen, S. Frasca, J. D. Silberman, C. J. O'Kelly, and M. L. Sogin. 2003. Two new small-subunit ribosomal RNA gene lineages within the subclass gymnamoebia. J. Eukaryot. Microbiol. 50:224-232. [DOI] [PubMed] [Google Scholar]
- 33.Quesenberry, M. S., K. Saito, D. N. Krupatkina, J. A. F. Robledo, T. Drgon, W. T. Pecher, N. O'Leary, M. Alavi, T. Miller, R. E. Schneider, R. Belas, J. R. Deeds, A. R. Place, Y. Zohar, and G. R. Vasta. 2002. Bioassay for ichthyocidal activity of Pfiesteria piscicida: characterization of a culture flask assay format. J. Appl. Phycol. 14:241-254. [Google Scholar]
- 34.Quilliam, M. A. 1999. Phycotoxins. J. AOAC Int. 82:773-781. [PubMed] [Google Scholar]
- 35.Ritchie, N. J., M. E. Schutter, R. P. Dick, and D. D. Myrold. 2000. Use of length heterogeneity PCR and fatty acid methyl ester profiles to characterize microbial communities in soil. Appl. Environ. Microbiol. 66:1668-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogers, H. S., and L. Backer. 2001. Fish bioassay and toxin induction experiments for research on Pfiesteria piscicida and other toxic dinoflagellates: workshop summary. Environ. Health Perspect. 109:769-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito, K., T. Drgon, J. A. F. Robledo, D. N. Krupatkina, and G. R. Vasta. 2002. Characterization of the rRNA locus of Pfiesteria piscicida and development of standard and quantitative PCR-based detection assays targeted to the nontranscribed spacer. Appl. Environ. Microbiol. 68:5394-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silbergeld, E. K., L. Grattan, D. Oldach, and J. G. Morris. 2000. Pfiesteria: harmful algal blooms as indicators of human:ecosystem interactions. Environ. Res. 82:97-105. [DOI] [PubMed] [Google Scholar]
- 39.Stoecker, D. K., and D. E. Gustafson. 2002. Predicting grazing mortality of an estuarine dinoflagellate, Pfiesteria piscicida. Mar. Ecol. Prog. Ser. 233:31-38. [Google Scholar]
- 40.Stoecker, D. K., M. W. Parrow, J. M. Burkholder, and H. B. Galsgow. 2002. Grazing by microzooplankton on Pfiesteria piscicida cultures with different histories of toxicity. Aquat. Microb. Ecol. 28:79-85. [Google Scholar]
- 41.Suzuki, M., M. S. Rappe, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogelbein, W. K., V. J. Lovko, J. D. Shields, K. S. Reece, P. L. Mason, L. W. Haas, and C. C. Walker. 2002. Pfiesteria shumwayae kills fish by micropredation not exotoxin secretion. Nature 418:967-970. [DOI] [PubMed] [Google Scholar]
- 43.Vogelbein, W. K., J. D. Shields, L. W. Haas, K. S. Reece, and D. E. Zwerner. 2001. Skin ulcers in estuarine fishes: a comparative pathological evaluation of wild and laboratory-exposed fish. Environ. Health Perspect. 109:687-693. [DOI] [PMC free article] [PubMed] [Google Scholar]