Abstract
A new plasmid for the overexpression of His-tagged thermozymes in Thermus thermophilus was developed. With this plasmid, soluble and active histidine-tagged DNA polymerase from T. thermophilus was overproduced in larger amounts in the thermophile than in Escherichia coli. The protein purified from the thermophile was active in PCR.
The enzymes from thermophilic microorganisms, or thermozymes, are usually produced in surrogate mesophilic hosts (1). However, several thermozymes which require either cofactors, appropriate posttranslational processing, or specific components during multisubunit assembly cannot be overproduced in an active form in such mesophilic systems (1-3, 5, 10). On the other hand, contamination with material from the host, like the residual amounts of DNA present in most commercial thermostable DNA polymerases, leads to false-positive cases in specific analytical applications. Therefore, the use of homologous thermophilic host systems for the expression of such thermozymes appears to be the most straightforward solution to these problems (9, 11, 14).
Thermus spp. form one of the most widely distributed genera of thermophilic bacteria, and many of its isolates carry genes encoding enzymes of great biotechnological potential (4, 10). Due to the existence of well-adapted laboratory strains that are readily amenable to genetic manipulation, this genus has become the model of choice for the development of expression systems for thermozymes (6-9, 11, 14). Cloning and expression vectors have been described and essentially used in the T. thermophilus HB27 strain, which has been sequenced recently (4). However, moderate, 6- to 10-fold overexpression levels have been obtained in most cases (11). As an exception, the pMKE1 vector allowed up to 200-fold overexpression of a thermostable β-galactosidase reporter in the laboratory strain HB27::nar (9) and approximately 100-fold overexpression of an Mn-dependent catalase (5). Despite these two orders of magnitude of overexpression, the overproduced proteins were almost undetectable by Coomassie blue staining, thus prompting us to develop more effective expression systems.
We describe a new plasmid that uses a modified version of the promoter from the respiratory nitrate reductase (narp) to overproduce active DNA polymerase from T. thermophilus in larger amounts than those obtained with T7-dependent expression systems of E. coli.
Plasmid construction.
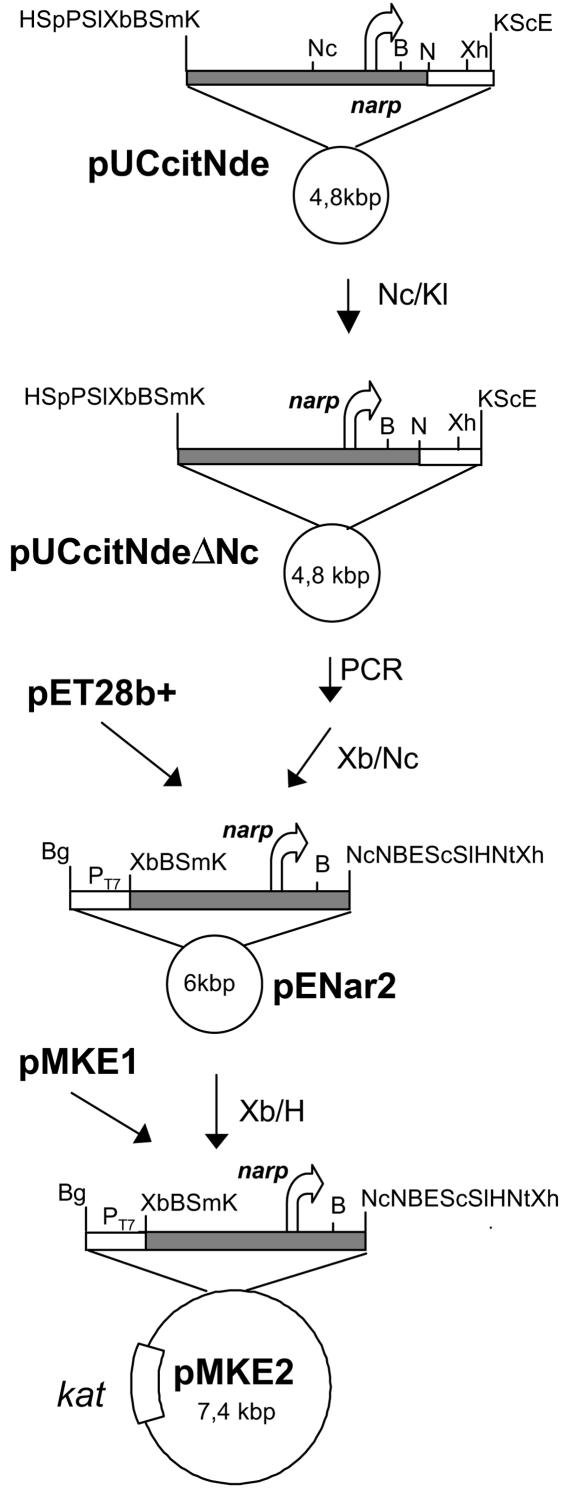
As illustrated in Fig. 1, the pMKE2 plasmid was constructed in three steps. The first step was designed to eliminate an NcoI restriction site located inside the narp promoter. This produced a 4-bp insertion. In the second step, the promoter was amplified (primers, 5′TCGCCATGGTCACCTCCGGC3′ and 5′GAAACAGCTATGACCATG3′) with the concomitant inclusion of an ATG start codon inside a new NcoI restriction site (underlined). The modified narp promoter was cloned into the XbaI-NcoI sites of plasmid pET28b(+) to yield pENar2. In the final step, the XbaI-HindIII DNA fragment from pMKE1 (9) was replaced by a similar DNA fragment from plasmid pENar2, thereby giving rise to the plasmid pMKE2.
FIG. 1.
Construction of plasmid pMKE2. The scheme for the construction of the expression plasmid pMKE2 is illustrated. Abbreviations for restriction enzymes sites are the following: B, BamHI; Bg, BglII; E, EcoRI; K, KpnI; H, HindIII; N, NdeI; Nc, NcoI; Nt, NotI; P, PstI; Sm, SmaI; Xb, XbaI; Xh, XhoI. Other symbols: Kl, Klenow fragment of DNA polymerase I of E. coli; kat, gene coding for thermostable resistance to kanamycin. Arrows indicate the transcription direction of the narp promoter (the sequence of which is shaded gray).
Expression of Tth polymerase.
The coding region of the DNA polymerase I gene from T. thermophilus (Tth) (Biotools B & M) was cloned into pMKE2, and the resulting plasmid (pMKETth) was used to transform T. thermophilus HB27::nar. One transformant colony was grown at 70°C in Thermus broth medium (12) under aerobic conditions (shaken at 150 rpm) prior to the addition of 40 mM potassium nitrate and the stopping of the shaker (time zero). Due to the low solubility of oxygen at this temperature and also its consumption, the culture rapidly became anaerobic, so the cells immediately started to transcribe the narp promoter. As shown in Fig. 2A, after 2 h a protein of the expected size for the His-Tth fusion (96 kDa) was detected in the soluble fraction of the cells. Two hours later, this protein accumulated to the point where it became the major component of the fraction.
FIG. 2.
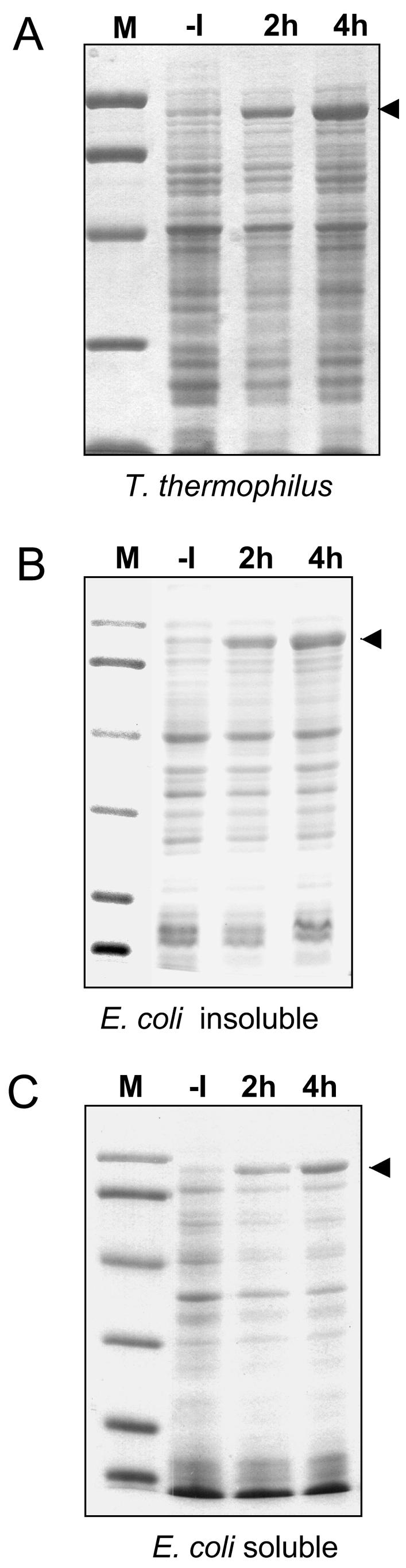
Expression of the His-tagged DNA polymerase of T. thermophilus and E. coli. (A) A culture of T. thermophilus HB27::nar carrying plasmid pMKE2Tth was subjected to inducing conditions by adding nitrate and simultaneously stopping shaking. Samples corresponding to identical amounts of soluble fractions were subsequently analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie blue. (B and C) A growing culture of E. coli BL21DE3 transformed with plasmid pET28Tth was treated with IPTG (1 mM), and the protein patterns of the particulate (B) and the soluble (C) fractions were analyzed by SDS-PAGE. Lanes: −I, before induction; 2 h and 4 h, 2 and 4 h after induction, respectively. The size markers (M) correspond to 97.4, 66.2, 45, 31, and 21.5 kDa. Arrowheads indicate the His-Tth polymerase.
To compare these results with those of expression systems of E. coli, the Tth gene was cloned into the pET28b+ plasmid (Novagen) to render pET28Tth. Interestingly, this plasmid was toxic at 37°C, but not at 30°C, for all strains of E. coli assayed. A colony of the strain BL21DE3 (13) transformed with pET28Tth was used to check the expression of the His-Tth polymerase 2 and 4 h after the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). As shown in Fig. 2B, most of the His-Tth polymerase accumulated in the particulate fraction of E. coli, whereas a minor fraction did so in the soluble fraction (Fig. 2C).
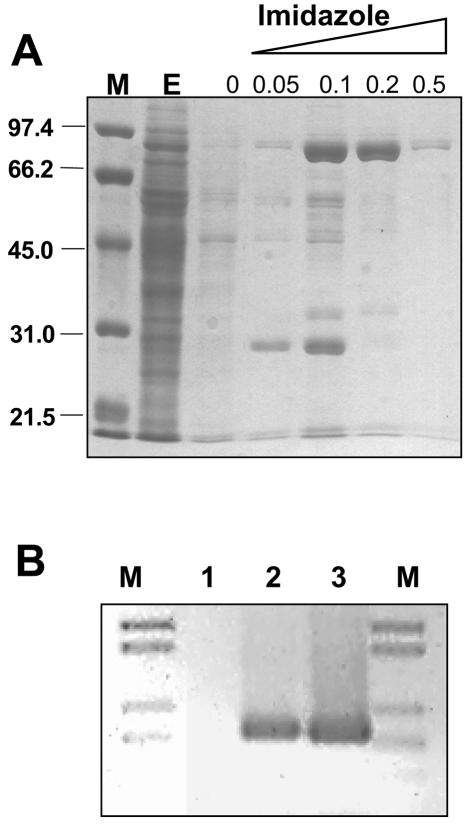
The enzyme overproduced in T. thermophilus was purified by affinity chromatography. As illustrated in Fig. 3A, most of the protein was bound to a nickel-nitrilotriacetic acid support, being further eluted with imidazole. A highly purified fraction was obtained and kept at −20°C in storage buffer (Tris, 20 mM; KCl, 50 mM; Nonidet P-40, 0.25%; glycerol, 40%; pH 8.0) until use. Figure 3B shows that this enzyme was as active in PCR as the commercial enzyme produced in E. coli.
FIG. 3.
Purification and activity of the His-Tth polymerase overexpressed in T. thermophilus. (A) A sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel is shown with fractions corresponding to an affinity Ni-agarose column in which the soluble fraction corresponding to the 4-h lane of Fig. 2A was assayed. The unbound material (E) and fractions corresponding to washing steps with the indicated concentrations of imidazole are shown. Lane M, protein markers of the labeled sizes (in kilodaltons). (B) Amplification of the kat gene with okat3 and okat4 primers. Lane 1, negative control without enzyme; lane 2, commercial Tth DNA polymerase; lane 3, His-tagged Tth DNA polymerase purified from T. thermophilus.
Concluding remarks.
The modification of the narp promoter resulted in unprecedentedly high levels of overexpression in T. thermophilus. There is presently no clear explanation for such an increase in pMKE2 compared to levels of pMKE1 (9), but it could be related to the binding of transcriptional activators close to the eliminated NcoI site. The involvement of this region in narp expression has been demonstrated by progressive deletions, which showed that regions upstream of this NcoI site are required for transcription (data not shown). This requirement for far-upstream regions is analogous to the requirement for specific heptameric sequences located upstream of the promoter for nitrate reductase A of E. coli, to which the phosphorylated transcription activator NarL binds (15). Thus, it is reasonable to speculate that either the fortuitous generation of a new site for an equivalent transcription factor in the thermophile or, more likely, a better architecture of the nucleoprotein formed around this NcoI site could be the basis for such an increase of expression. Of course, alternative explanations, such as higher mRNA stability or differences in plasmid copy number, also could contribute to this expression increase. However, as the leader RNA (91 bases long) of native nar-mRNA present in the construction is quite stabilizing (with a 15-min half-life) (16), and as the copy number per cell of pMKE2 was similar to that of pMKE1 (results not shown), these alternative explanations seem unlikely.
Whatever the reason underlying the increase in transcription from the modified narp promoter, it may be concluded that this new expression system represents a competitive route for the direct overexpression of soluble enzymes in T. thermophilus, even compared to the high-level overexpression systems of E. coli.
Acknowledgments
This work was supported by project BIO2001-1627 of the Ministerio de Ciencia y Tecnología, Spain. An institutional grant from the Fundación Ramón Areces to the Centro de Biologia Molecular “Severo Ochoa” is acknowledged. R. Moreno was in receipt of a fellowship from Biotools, B & M.
REFERENCES
- 1.Adams, M. W., and R. M. Kelly. 1998. Finding and using hyperthermophilic enzymes. Trends Biotechnol. 16:329-332. [DOI] [PubMed] [Google Scholar]
- 2.Coolbear, T., R. M. Daniel, and H. W. Morgan. 1992. The enzymes from extreme thermophiles: bacterial sources, thermostabilities and industrial relevance. Adv. Biochem. Eng. Biotechnol. 45:57-98. [DOI] [PubMed] [Google Scholar]
- 3.Fernández-Herrero, L. A., M. A. Badet-Denisot, B. Badet, and J. Berenguer. 1995. glmS of Thermus thermophilus HB8: an essential gene for cell-wall synthesis identified immediately upstream of the S-layer gene. Mol. Microbiol. 17:1-12. [DOI] [PubMed] [Google Scholar]
- 4.Henne, A., H. Bruggemann, C. Raasch, A. Wiezer, T. Hartsch, H. Liesegang, A. Johann, T. Lienard, O. Gohl, R. Martinez-Arias, C. Jacobi, V. Starkuviene, S. Schlenczeck, S. Dencker, R. Huber, H. P. Klenk, W. Kramer, R. Merkl, G. Gottschalk, and H. J. Fritz. 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 22:547-553. [DOI] [PubMed] [Google Scholar]
- 5.Hidalgo, A., L. Betancor, R. Moreno, O. Zafra, F. Cava, R. Fernández-Lafuente, J. M. Guisán, and J. Berenguer. 2004. Thermus thermophilus as a cell factory for the production of a thermophilic Mn-dependent catalase which fails to be synthesized in an active form in Escherichia coli. Appl. Environ. Microbiol. 70:3839-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koyama, Y., Y. Arikawa, and K. Furukawa. 1990. A plasmid vector for an extreme thermophile, Thermus thermophilus. FEMS Microbiol. Lett. 72:97-102. [DOI] [PubMed] [Google Scholar]
- 7.Lasa, I., M. de Grado, M. A. de Pedro, and J. Berenguer. 1992. Development of Thermus-Escherichia shuttle vectors and their use for expression of the Clostridium thermocellum celA gene in Thermus thermophilus. J. Bacteriol. 174:6424-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mather, M. W., and J. A. Fee. 1992. Development of plasmid cloning vectors for Thermus thermophilus HB8: expression of a heterologous, plasmid-borne kanamycin nucleotidyltransferase gene. Appl. Environ. Microbiol. 58:421-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moreno, R., O. Zafra, F. Cava, and J. Berenguer. 2003. Development of a gene expression vector for Thermus thermophilus based on the promoter of the respiratory nitrate reductase. Plasmid 49:2-8. [DOI] [PubMed] [Google Scholar]
- 10.Pantazaki, A. A., A. A. Pritsa, and D. A. Kyriakidis. 2002. Biotechnologically relevant enzymes from Thermus thermophilus. Appl. Microbiol. Biotechnol. 58:1-12. [DOI] [PubMed] [Google Scholar]
- 11.Park, H. S., K. J. Kayser, J. H. Kwak, and J. J. Kilbane II. 2004. Heterologous gene expression in Thermus thermophilus: beta-galactosidase, dibenzothiophene monooxygenase, PNB carboxy esterase, 2-aminobiphenyl-2,3-diol dioxygenase, and chloramphenicol acetyl transferase. J. Ind. Microbiol. Biotechnol. 31:189-197. [DOI] [PubMed] [Google Scholar]
- 12.Ramírez-Arcos, S., L. A. Fernández-Herrero, I. Marín, and J. Berenguer. 1998. Anaerobic growth, a property horizontally transferred by an Hfr-like mechanism among extreme thermophiles. J. Bacteriol. 180:3137-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg, A. H., B. N. Lada, D. Chui, S.-W. Lin, J. J. Dunn, and W. Studier. 1987. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene 56:125-135. [DOI] [PubMed] [Google Scholar]
- 14.Tamakoshi, M., M. Uchida, K. Tanabe, S. Fukuyama, A. Yamagishi, and T. Oshima. 1997. A new Thermus-Escherichia coli shuttle integration vector system. J. Bacteriol. 179:4811-4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Unden, G., and J. Bongaerts. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320:217-234. [DOI] [PubMed] [Google Scholar]
- 16.Zafra, O. 2004. Implicación de un citocromo c en la biosíntesis de la nitrato reductasa respiratoria de Thermus thermophilus. Ph.D. thesis. Universidad Autónoma, Madrid, Spain.