ABSTRACT
Giardia lamblia is the most frequently identified protozoan cause of intestinal infection. Over 200 million people are estimated to have acute or chronic giardiasis, with infection rates approaching 90% in areas where Giardia is endemic. Despite its significance in global health, the mechanisms of pathogenesis associated with giardiasis remain unclear, as the parasite neither produces a known toxin nor induces a robust inflammatory response. Giardia colonization and proliferation in the small intestine of the host may, however, disrupt the ecological homeostasis of gastrointestinal commensal microbes and contribute to diarrheal disease associated with giardiasis. To evaluate the impact of Giardia infection on the host microbiota, we used culture-independent methods to quantify shifts in the diversity of commensal microbes throughout the gastrointestinal tract in mice infected with Giardia. We discovered that Giardia's colonization of the small intestine causes a systemic dysbiosis of aerobic and anaerobic commensal bacteria. Specifically, Giardia colonization is typified by both expansions in aerobic Proteobacteria and decreases in anaerobic Firmicutes and Melainabacteria in the murine foregut and hindgut. Based on these shifts, we created a quantitative index of murine Giardia-induced microbial dysbiosis. This index increased at all gut regions during the duration of infection, including both the proximal small intestine and the colon. Giardiasis could be an ecological disease, and the observed dysbiosis may be mediated directly via the parasite's unique anaerobic fermentative metabolism or indirectly via parasite induction of gut inflammation. This systemic alteration of murine gut commensal diversity may be the cause or the consequence of inflammatory and metabolic changes throughout the gut. Shifts in the commensal microbiota may explain observed variations in giardiasis between hosts with respect to host pathology, degree of parasite colonization, infection initiation, and eventual clearance.
KEYWORDS: Giardia, microbiome, parasite, pathogenesis
INTRODUCTION
Giardia lamblia is a microaerophilic protozoan parasite of humans and animals that causes significant morbidity and diarrheal disease worldwide (1, 2). Giardiasis is a zoonotic disease with diverse animal reservoirs. Parasites infect and complete their life cycle in mammalian hosts, and infected animals shed Giardia cysts into water supplies. Over 200 million people are estimated to have acute or chronic giardiasis, and rates of giardiasis approach 90% in areas where it is endemic (3, 4). When prevalent, giardiasis has been implicated as a primary cause of growth restriction for children, resulting in long-term consequences, such as stunting, failure to thrive, malnutrition, and cognitive disabilities (1, 2, 5). In addition, Giardia has been associated with substantial postclearance irritable bowel symptoms in both children and adults (1, 6–8). The significant and adverse impact of giardiasis on global human health contrasts with a considerable lack of research efforts targeting prevention, treatment, and increased understanding of the basic biology of Giardia (9).
Common symptoms of acute and chronic giardiasis include abdominal cramps, gas, nausea, and weight loss. Severe giardiasis results in malabsorptive diarrhea with fatty, bulky stools. At the cellular level, parasite colonization of the gut is associated with malabsorption of glucose, salts, and water, disruption of intestinal barrier function, induction of enterocyte apoptosis, inhibition of brush border disaccharidases and lactases, loss of mucosal surface due to increased crypt:villus ratios and shortened microvilli, and manipulation of host immune responses via arginine/nitric oxide limitation (1, 11–14).
Following ingestion by a mammalian host, Giardia cysts transform into motile trophozoites as they pass into the gastrointestinal tract. Trophozoites navigate the lumen of the small intestine and attach to the microvilli via the ventral disc, but they do not invade the epithelium (11). Attachment to the gut epithelium allows the parasite to resist peristaltic flow and to proliferate in this low-oxygen, nutrient-rich environment. Trophozoites are triggered to transform into cysts, and these cysts eventually exit the host and are disseminated in feces into the environment. Despite its global importance as a primary cause of diarrheal disease, the mechanisms by which Giardia colonization of the gastrointestinal tract induces diarrheal disease remain elusive (4). Giardia produces no known toxin, and parasite colonization does not induce a robust inflammatory reaction (4, 15). Symptoms are generally believed to be the consequence of host tissue damage caused by the direct contact of the parasite with the intestinal villi (12). Paradoxically, parasite colonization is often associated with histologically normal tissue (16), and giardiasis may frequently by asymptomatic (1).
An ecological perspective on giardiasis could inform our understanding of Giardia's pathogenesis (17). Commensal microbes in the mammalian gut benefit the host by breaking down complex substrates, supplying essential nutrients, and defending against opportunistic pathogens (1). Commensal microbes from all three domains of life colonize the gastrointestinal tract of mammals and account for about 109 cells/g in the small intestine and 1012 cells/g of luminal contents in the large intestine (1). The maintenance of physiologically diverse commensal microbes by the host is critical for overall stability of this ecosystem (18). Physiological diversity can confer resilience and flexibility of commensal microbial responses to various external stresses, both promoting and modulating the development of the immune system. Commensal gut microbial communities are known to both limit and exacerbate pathogen colonization (19–22), and thus the disruption of the gut ecosystem may impact Giardia colonization and consequent symptoms of diarrheal disease. The histopathology of giardiasis has been noted to be less severe in germfree mice than in conventional mice, supporting the idea that commensal microbes can aggravate parasite infection or symptoms (23).
Parasite colonization within the small intestine occurs in an environment already inhabited by diverse commensal bacteria, yet positive or negative interactions of parasites with the commensal intestinal microbiota that occur during infection have largely been ignored (4). There is evidence that interactions between Giardia and these commensal microbes could contribute to variations in pathogenesis and adverse symptoms associated with giardiasis (24, 25). This idea is supported by evidence of a direct relationship between Giardia infection and intestinal bacterial overgrowth (26, 27). Cultures of jejunal juice from patients with active giardiasis revealed increased total bacterial load, increased abundance of Enterobacteriacae, and perturbed bile acid homeostasis (27, 28). Giardia infection also causes increased bacterial loads midinfection and increased bacterial invasiveness postclearance as a sequela to intestinal barrier disruption (29).
Viewed within the context of invasion biology, Giardia colonization of the mammalian host gut is analogous to an exotic invading species that causes cascading effects on the resident community (30). In conjunction with direct effects on host tissues, Giardia could cause disruptions to the ecological health of commensal microbes in the gastrointestinal tract by altering the microbial composition, metabolic capacities, or chemical homeostasis in the gastrointestinal lumen. Other gut ecologically mediated diseases include infectious diseases with known bacterial pathogens (e.g., Clostridium difficile or Salmonella enterica serovar Typhimurium) and immune-mediated diseases wherein the entirety of the gut community appears perturbed in the absence of a dominant pathogenic species (e.g., Crohn's disease and irritable bowel disease [IBD]) (22, 31–34). Similarly, the eukaryotic apicomplexan parasite Toxoplasma gondii has been proposed to act as a molecular adjuvant to the small intestinal microbiota, inducing acute ileitis (35). T. gondii infection induces a rapid overgrowth of Proteobacteria within the small intestine, aggravating immunopathology and promoting an antiparasitic immune response (36, 37).
To evaluate potential dysregulation of commensal gut microbial ecology associated with giardiasis, we infected mice with Giardia and used standard cultivation-independent methods (38–43) to describe and quantify shifts in microbial diversity throughout the gastrointestinal tract. We demonstrate that Giardia's colonization of the small intestine causes a systemic imbalance, or dysbiosis, throughout the murine gut that persists during infection. We propose that Giardia infection disrupts commensal gut microbial ecology and metabolism in the host. This dysbiosis may be mediated directly via the parasite's unique anaerobic metabolism and/or indirectly via parasite induction of host responses. This systemic alteration of murine gut ecology could also explain observed variation in giardiasis with respect to host pathology, degree of parasite colonization, infection initiation, and eventual clearance.
RESULTS
Giardia colonization is correlated with alterations in host commensal diversity in both antibiotic-treated and antibiotic-naive mice.
To quantify how Giardia infection alters the diversity and abundance of host-associated gut microbiota, we compared the gut microbiota of mice infected with Giardia to the microbiota of uninfected mice gavaged with a saline vehicle control (see Materials and Methods). Antibiotic treatment is generally required for robust experimental Giardia infections in mice (24, 44). To investigate potential colonization resistance, we compared infections and commensal diversity in both antibiotic-naive and antibiotic-treated animals. Giardia lamblia strain GS (assemblage B) was used for all infections, as this strain colonizes antibiotic-free mice more readily than the assemblage A strain WBC6 (24, 45, 46) (Fig. 1). Microbial diversity was quantified from a total of 192 samples from the intestinal tracts of 32 animals in 8 treatment groups, sampled from 6 sites (luminal and mucosal sampling of proximal and distal small intestine, bulk sampling of cecum and colon).
FIG 1.
Sampling scheme for querying murine gut ecological shifts during giardiasis with and without antibiotic treatment. Two cohorts (n = 16) of sibling mice, one not treated with antibiotics and one treated with antibiotics, were used to test the potential effects of Giardia colonization on the microbial diversity of the murine gut. Study animals were infected with Giardia lamblia GS trophozoites (n = 12 animals per treatment group) or a saline vehicle control (uninfected [U]; n = 4 per treatment group) by gavage. Cohorts of infected animals were sacrificed at 3, 7, and 14 days postgavage (n = 4 per day). Gut intestinal tracts for each treatment group and for each day postinoculation were dissected for subsequent diversity analyses (pSI, dSI, CEC, and COL). The total number of reads per cohort (n = 4) are shown for each experimental condition. Luminal and mucosal reads were pooled for most analyses (see Materials and Methods for details). The total reads per animal within each cohort under each experimental condition are summarized in Table S1 in the supplemental material.
For diversity analyses, total community DNA was extracted from each sample and amplified using universal small-subunit (SSU) rDNA primers (43). Amplified SSU genes were sequenced using high-throughput methods to generate 2,002,493 paired-end Illumina reads. A total of 12,171 individual operational taxonomic units (OTUs) were called at the 97% identity threshold from all samples (Fig. 1; see also Table S1 in the supplemental material). The total numbers of sequence reads for each anatomical site ranged from about 6,000 to over 60,000 (Fig. 1). Both antibiotic-treated and -naive mice had a median unique OTU count of 468.
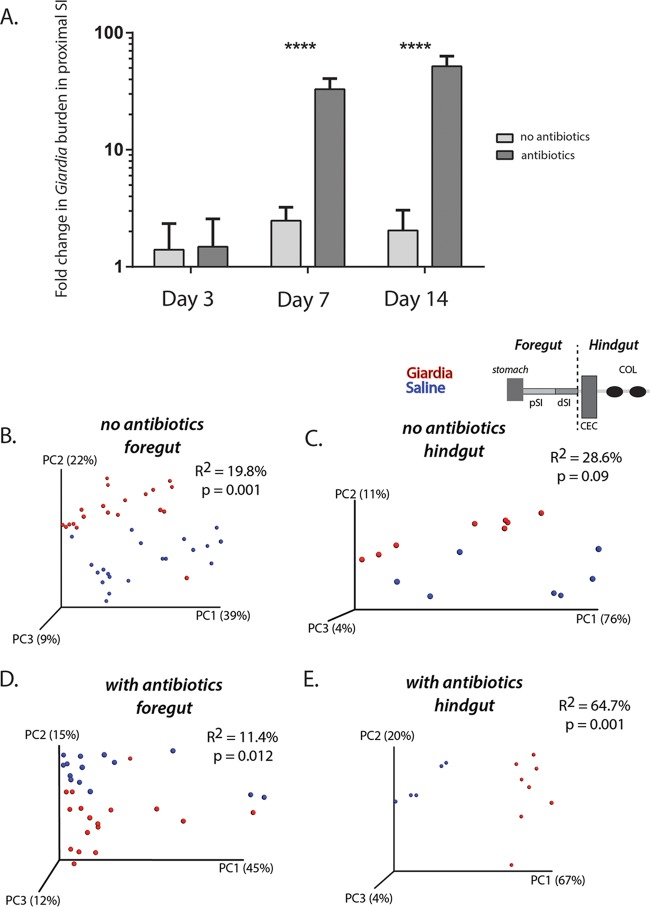
To quantify the degree of parasite infection (or Giardia burden) in the antibiotic-treated and untreated cohorts, we used Giardia-specific quantitative PCR (QPCR) of a single-copy intragenic region (GS-1-318). Proximal small intestinal samples (3-cm segments located 7 cm distal to the pylorus) were collected from animals in both treatment groups on day 3, day 7, and day 14, and the Giardia burden was assessed using QPCR. Without antibiotic pretreatment, we noted a moderate yet significant infection density that peaked on day 7 (Fig. 2A). With antibiotic pretreatment, however, we saw a significant 80-fold increase in Giardia abundance relative to that in untreated mice on day 7 and a further increase in parasite abundance by day 14 postinfection, when the Giardia burden in untreated mice had declined.
FIG 2.
Giardia infection significantly alters foregut and hindgut microbial diversity in antibiotic-treated and -naive mice. (A) Comparisons of the fold change in Giardia burden in pSI samples based on QPCR results between antibiotic-treated and nontreated animals are presented for days 3, 7, and 14 post-Giardia gavage. Asterisks indicate significant differences as assessed with Sidek's multiple comparison test. (B to E) Weighted Unifrac comparisons of beta diversity are shown using PCoA plots to show the variance in the diversity of all foregut samples and hindgut samples with and without antibiotic treatment. Samples with a saline vehicle control and samples after 2 weeks of Giardia infection are shown. Significance (P value) and the strength of explained variation (R2) were assessed with the adonis program.
To investigate potential colonization resistance in the murine giardiasis model, we infected both antibiotic-treated and antibiotic-naive mice with Giardia. Colonization occurred in both treatment groups, although the overall parasite burden was lower without antibiotic treatment (Fig. 2). In both treatment groups, the commensal diversity was significantly altered throughout the gastrointestinal tract in a similar manner at day 7 postinfection (Fig. 2; Fig. S1, S5, and S6). Specifically, with respect to overall microbial diversity for all sequences in the foregut and hindgut, we confirmed that Giardia infection resulted in significant differences in the variants of foregut and hindgut sequences, in both nontreated and antibiotic-treated animals (Fig. 2B to E; Fig. S5 and S6).
Overall species richness and evenness are not significantly altered within each anatomical site during giardiasis.
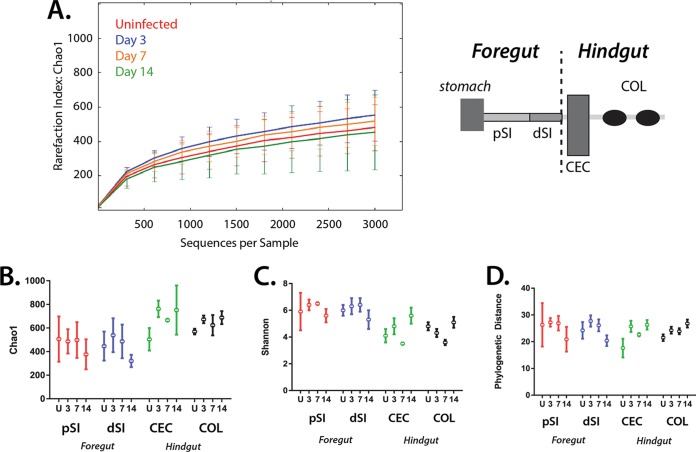
To evaluate the overall dynamics of ecological shifts in the gut microbiota during giardiasis, we first analyzed species richness and evenness (alpha diversity) using QIIME (Fig. 3; Fig. S1). Sampling accuracy for giardiasis samples with antibiotic treatment (Fig. 3A) and without (Fig. S1A) was evaluated for all anatomical samples at each time point postinfection by using rarefaction of Chao1 estimates (38–40). For both treatment groups, the Chao1 result for all samples (all anatomical sites) by day was estimated to be 3,000 OTUs.
FIG 3.
Overall alpha diversity by intestinal site is not significantly changed during giardiasis. (A) Sampling adequacy (alpha diversity) was assessed using rarefaction plots to compare all samples using the Chao1 estimate during the course of giardiasis (uninfected and day 3, day 7, and day 14) with antibiotic treatment (see Fig. S1 for results with no antibiotic treatment). (B to D) Alpha diversity measures of species richness, evenness, and phylogenetic distance, summarizing microbial diversity in regions of the murine gut (pSI, dSI, CEC, and COL) in control, antibiotic-treated, noninfected cohorts (U), and in infected cohorts (days 3, 7, and 14). For these analyses, the OTU tables were rarefied to 3,000 reads/sample and analyzed using QIIME to compare the relative read abundances. Chao 1 estimates of species richness for both antibiotic-treated groups are shown in panel B; increased Chao1 values correspond to increased numbers of unique taxa. Species evenness was also estimated using the Shannon index (C), where a high Shannon value indicated equal distribution between species and a low Shannon value indicated dominance by certain taxa. Lastly, the phylogenetic distance (D) was used to estimate the evolutionary relationships of the community; a higher phylogenetic distance value corresponds to a more diverse community. Error bars correspond to variations between gut samples of each of the four animals in one experimental group.
To compare alpha diversity between sites, four pooled anatomical samples were compared: proximal and distal small intestine (pSI and dSI), cecum (CEC), and colon (COL) (Fig. 3B and 2D). Species richness was similar for all samples based on Chao1 and observed OTU calculations. For any day of infection, regardless of antibiotic treatment, the proximal and distal small intestine samples were slightly less rich in unique OTUs than the cecal and colonic samples (Fig. 3B; Fig. S1B). This trend was more pronounced in the antibiotic-naive cohort (Fig. S1B).
To assess taxonomic evenness of the gut microbiota, we performed estimates of the Shannon index for each experimental condition and anatomical site (Fig. 3C; Fig. S1C). Higher Shannon index values imply an even distribution of abundances, in contrast to low Shannon values, which suggest that certain community members predominate. We found that antibiotic treatment increased the Shannon index in the proximal and distal small intestine compared to the index in antibiotic-naive mice, but it did not affect the cecal or colonic samples. Over the time of infection, microbiota diversity at all gut sites remained even with respect to OTU abundances. During giardiasis, the antibiotic-treated cohort exhibited more fluctuation than the nontreated cohort (Fig. 3C; Fig. S1C).
Lastly, we quantified phylogenetic distances between taxa during giardiasis in the four pooled anatomical samples (Fig. 3D; Fig. S1D). For antibiotic-treated mice, we observed that phylogenetic distances remained consistent over the time course of infection and between body sites. When individual antibiotic-treated mice were compared, the proximal and distal small intestines exhibited increased variability in phylogenetic distances, as indicated by wider error bars, compared to distances for the cecum and colon samples. Comparing phylogenetic distances between all gut sites and time points, we found that antibiotic-treated mice had higher estimated phylogenetic distances between taxa than nontreated animals.
Giardia infection is associated with a significant dysbiosis within the murine foregut and hindgut.
While Giardia trophozoites preferentially colonize the most proximal small intestine (Fig. 2A) (47–49), they are also found in the distal small intestine, as well as the cecum and large intestine (47–49). Within the antibiotic-treated proximal small intestine, we found that Giardia infection substantially changed the diversity of the host microbiota during the 14-day course of infection (Fig. S2A). Although mucosal and luminal communities are generally distinct in the large intestine (50), luminal/mucosal proximal and distal intestinal samples were not significantly clustered, based on principal-coordinate analysis (PCoA) (Fig. S3B) and were pooled for subsequent taxa-specific analyses. Total cecum and colonic samples did not significantly cluster according to PCoA during giardiasis (Fig. S3C) and were also pooled for subsequent analysis.
We used weighted UniFrac analysis to evaluate whether giardiasis is a predominant contributor to changes in murine gut microbial diversity (Fig. S2). By comparing sequences from uninfected animals to those from animals 14 days post-Giardia infection, we found that sequences from any small intestinal site clustered together based on giardiasis status, rather than with respect to small intestinal subsite (Fig. S2D).
Antibiotic treatment by itself changes the gut microbial ecosystem. Without antibiotics, we found that the proximal small intestinal microbiome was dominated by the Proteobacteria and Firmicutes bacterial phyla and that antibiotic treatment supported a larger population of Bacteroidetes and Melainabacteria than in naive individuals (Fig. 4). While we noted these significant differences in antibiotic versus non-antibiotic-treated samples, we highlight our subsequent analyses of the treatment group with antibiotics, as this group had significantly higher Giardia abundance, particularly by day 14 postinfection (Fig. 2). Specifically, we focused our attention on comparative analyses of antibiotic-treated animals with and without giardiasis by anatomic site. All cognate analyses for animals not treated with antibiotics are presented in the supplemental figures (Fig. S4 to S7).
FIG 4.
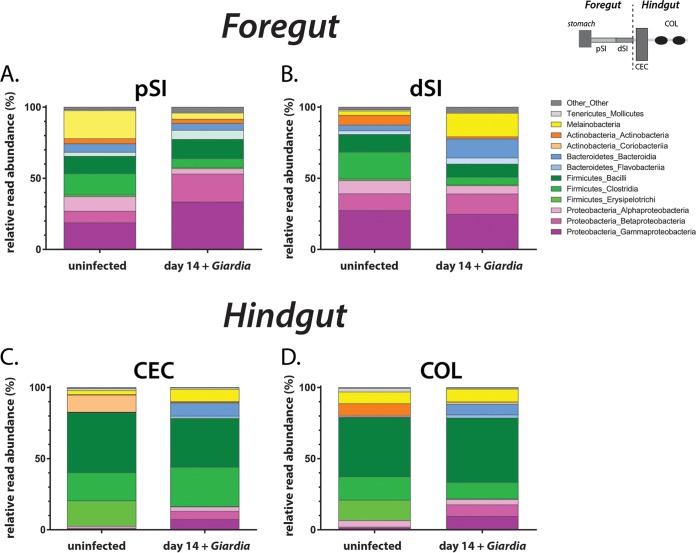
Significant changes in gut microbiome diversity in the proximal small intestine and colon during giardiasis. At 14 days postinfection, the relative read abundance (as a percentage) of the most abundant bacterial divisions and phyla are shown for the mouse foregut (pSI [A] and dSI [B]) and the mouse hindgut (CEC [C] and COL [D]). Each anatomic section was compared to the cognate uninfected sample.
Increased Proteobacteria diversity and decreased Firmicutes diversity exemplify the primary ecological changes during giardiasis.
In the pSI of antibiotic-treated animals, the diversity of several taxonomic groups was substantially altered during giardiasis (Fig. 4A). Specifically, Proteobacteria expanded from 33% to 56% of total reads after 14 days of infection. The expansion of both Betaproteobacteria and Gammaproteobacteria diversity accounted for over 80% of changes in unique Proteobacteria OTUs. In contrast, the overall percentage of Firmicutes reads decreased from 28% to 20% following infection. The significant decrease in Clostridia taxa during giardiasis accounted for over 80% of the changes in Firmicutes OTUs. Lastly, Giardia infection reduced diversity of Melainabacteria from almost 20% to 4% with respect to total reads in the proximal small intestine. We also saw alterations in the diversity of commensal microbes in the dSI (Fig. 4B), with increases in the diversity of both Melainabacteria and Firmicutes.
We observed similar trends in changing diversity in the foregut (pSI and dSI) within the antibiotic-naive mice during giardiasis (Fig. S4 and S5). With respect to the proximal small intestine, the overall diversity of Proteobacteria expanded from 14% to 21% of total reads, with the Betaproteobacteria in particular increasing from just over 1% of reads to almost 8% during giardiasis. Infection induced a decrease in the Clostridia from 25% to 8% of total reads. The Melainabacteria decreased from 4% to 1% of total reads. Clustering by treatment is evident in PCoA plots of antibiotic-treated and -naive mice, and adonis analysis showed a similar impact of Giardia infection, accounting for 13% of total variance in both antibiotic-treated and -naive groups (Fig. S5). In the dSI (Fig. S4B), we observed slight decreases in the diversity of both Melainabacteria and Firmicutes.
Due to host peristalsis and inflammatory responses occurring along the entire gastrointestinal tract, we predicted parasite colonization of the foregut could induce dysbiosis in the hindgut, far from the site of infection. We discovered that hindgut samples also had significant overall changes in bacterial diversity following 14 days of giardiasis in both antibiotic-treated and nontreated animals (Fig. 4; Fig. S3 to S5). During giardiasis, Firmicutes diversity in the cecum decreased as Proteobacteria diversity increased. Similarly, large intestine microbiota diversity was dominated by Firmicutes taxa, with an expansion of Proteobacteria in antibiotic-treated animals (Fig. 4D). Melainabacteria accounted for 8% of reads and thus were unchanged during infection. During giardiasis without antibiotic treatment, both the cecum and large intestine samples were composed predominantly of Firmicutes and Bacteroidetes, which together accounted for about 94% of reads regardless of infection (Fig. S4).
What are the primary bacterial taxa, or “indicator” taxa that shift in abundance during giardiasis? We used several multivariate statistical methods (DESeq2 with analysis of variance [ANOVA]; QIIME 1.9) to query microbial taxa that significantly changed after 2 weeks of giardiasis (51, 52). The OTUs with significant variations in relative abundance during infection ranged between 20 and 50% of total reads, depending on the day, site, and condition analyzed. Overall, Clostridiaceae and Proteobacteraciae were the taxa with the most dramatic changes in diversity throughout both the foregut and hindgut during giardiasis (Fig. 5).
FIG 5.
Significant shifts in beta diversity in both the murine foregut and hindgut during giardiasis. The relative abundance levels of bacterial taxa that were significantly enriched or depleted after 14 days of Giardia infection were plotted by comparing the fold changes in uninfected versus infected animals, using the same anatomical sample. The taxonomy of each species is indicated. Abbreviations: aProt, Alphaproteobacteria; bProt, Betaproteobacteria; gProt, Gammaproteobacteria; BacF, bacilli (Firmicutes); EryF, Erysipelotrichi (Firmicutes); ClosF, Clostridia (Firmicutes). Melainabacteria were categorized as Cyanobacteria-chloroplasts in QIIME. Underlined taxa were used to calculate the murine giardiasis dysbiosis index (see Fig. 7 and also Fig. S7).
To summarize overall shifts in microbial diversity by site during giardiasis, we plotted the overall abundance of each identified taxonomic group versus the fold change between infected and uninfected samples (Fig. 5). In antibiotic-treated animals, the taxa with increased diversity in both the foregut and hindgut included Rhodocyclaceae (most notably Dechloromonas and Zoogloea spp.), Moraxellaceae, due to enrichment of Acinetobacter spp., Flavobacteriales (Chryseobacterium and Cloacibacterium spp.), and Comomonadaceae, due to the enrichment in unclassified species (Fig. 5). Bacterial OTUs that were depleted across all regions of the gut included Firmicutes and the families Erysipelotrichaceae (including Allobaculum spp.) and Clostridiaceae (including Clostridium spp.). Melainabacteria were enriched in the proximal small intestine and depleted in the distal small intestine and hindgut. Other taxa (Lactobacillaceae, Lachnospiraceae, Xanthomonadaceae, Enterobacteriaceae, Pseudomonadaceae, Actinomycetales, Ruminococcaceae, Sphingomonadaceae, Bacteroidales) were either enriched or depleted in either the foregut or the hindgut. Based on these patterns of diversity during giardiasis, we defined taxa with significant diversity changes across all gut sites as “giardiasis indicator taxa” (underlined groups in Fig. 5).
We also summarized alterations in microbial taxa during giardiasis in non-antibiotic-treated animals (Fig. S6). Rhodocyclaceae OTUs were enriched in the proximal small intestine, and Moraxellaceae, Flavobacteriales, Comononadaceae, and Bacteroidales were enriched throughout the small intestine (Fig. 6). In the hindgut, only Bacteroidales were enriched. No Proteobacteria were found in the hindgut of non-antibiotic-treated animals. Clostridiacae were depleted across the intestinal tract of non-antibiotic-treated animals.
FIG 6.
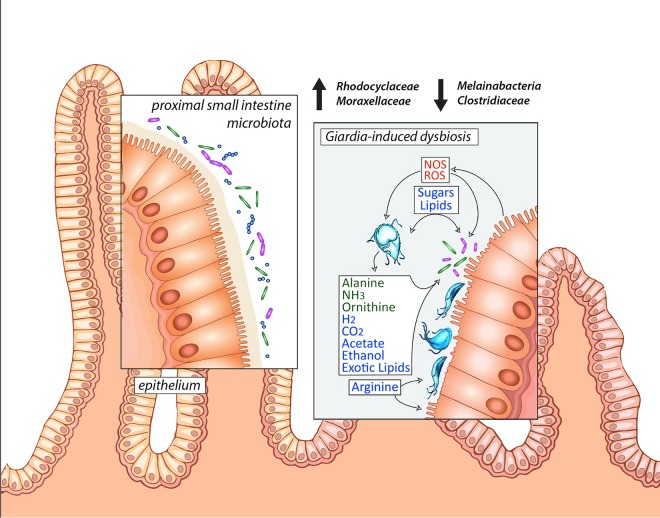
Model of Giardia-induced dysbiosis in the proximal small intestine with implications for disease symptoms. Hypothesized metabolic interactions between fermentative Giardia trophozoites with both the host commensal microbiota and host epithelium of the proximal small intestine, the primary site of parasite colonization. Reactive oxygen and nitrogen species (ROS and NOS) are induced by inflammatory responses. Trophozoites ferment sugars and amino acids and can excrete various waste products, depending on the oxygen tension in the surrounding lumen. Primary shifts in diversity of the commensal Rhodocyclaceae and Moraxellaceae and the Clostridiales (as well as Melainabacteria) are indicated and are also summarized in our Giardia microbial dysbiosis index (see Fig. 7).
Because the bacterial families Rhodocylaceae and Moraxellaceae were enriched and Clostridiales were depleted throughout the course of infection, we created a murine giardiasis dysbiosis index (MGDI) to summarize shifts in the microbiota during giardiasis. The MGDI combines the relative abundance of Rhodocylcaceae and Moraxellaceae in the numerator and Clostridiales in the denominator, such that a higher MGDI indicates depleted Clostridiales and enriched Rhodocyclaceae and Moraxellaceae. Using this measure, the MGDI steadily rose at all body sites during the duration of infection (Fig. 7A). Plotting samples with infection day as an explicit axis showed that elevated MGDI values were only associated with more-prolonged infection (Fig. 7B).
FIG 7.
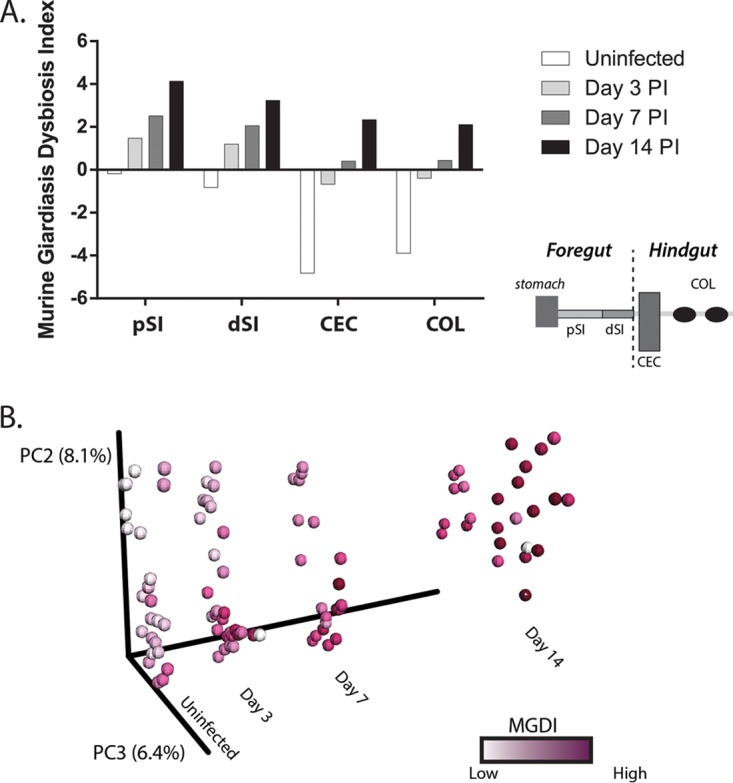
The murine giardiasis dysbiosis index defines microbial community shifts during the time course of infection in antibiotic-treated animals. The murine giardiasis dysbiosis index was calculated for each sample by dividing the sum of the abundances of the Rhodocylaceae and Moraxellaceae families by the abundance of Clostridiaceae (see Materials and Methods). The average MGDI per body site over the infection time course is presented. (A) The MGDI increased during infection for all body sites analyzed. (B) PCoA revealed the infection time course as an explicit axis. Samples are shaded according to their calculated MGDI, with darker shading intensities representing higher MGDI values.
The MGDI similarly defined microbial shifts associated with infection duration in non-antibiotic-treated animals (Fig. S7). This shift was magnified in the foregut, yet an increased MGDI was also seen in the hindgut. The commensal taxa characterizing the cecum and feces clustered distinctly from those in the small intestine, and this was reflected in substantially lower MGDI values in these anatomical regions.
DISCUSSION
This is the first high-throughput, culture-independent assessment of the commensal microbiota during Giardia infection. Giardiasis perturbs the diversity and abundance of host commensal microbial communities in both the foregut, where parasites colonize, and also in the hindgut (Fig. 4 and 5; Fig. S3 to S6 in the supplemental material). This systemic alteration of the murine commensal gut ecology during giardiasis likely contributes to the variation in infection initiation, degree of colonization, induction of pathology in the host, and/or eventual clearance of the parasite (Fig. 6) (24, 25).
Summarizing commensal microbial shifts using our murine giardiasis dysbiosis index.
Based on our observations, we developed the MGDI as a simplified and quantitative metric of the changing community composition during giardiasis (Fig. 7; Fig. S7). Like other indexes of microbial dysbioses (33), the MGDI summarizes primary ecological shifts in taxon abundance (e.g., the enrichment of Rhodocylaceae and Moraxellaceae and depletion of Clostridiales) and the relative abundance levels of enriched and depleted microbes. Using this measure, the MGDI steadily increased at all gut regions during the duration of infection, regardless of antibiotic pretreatment (Fig. 7; Fig. S6). Use of this or a similar dysbiosis index, if validated in human samples, could aid in evaluating gut commensal health in infected patients and monitoring long-term functional gastrointestinal consequences and efficacy of therapies, such as fecal microbial transplant (53).
Giardiasis symptoms may be associated with the ecological shifts induced by parasite metabolism.
Giardia colonization likely alters the local gut chemistry and metabolite availability, changing the gut ecology. Giardia trophozoites are microaerophilic (54), and the end products of glycolysis and pyruvate metabolism—acetate, ethanol, alanine, carbon dioxide, and hydrogen—vary according to the local oxygen tension (55–61). We found that Giardia infection enriches metabolically flexible taxa known to thrive with increased oxygen tension, lipid availability, and competition for arginine. In this way, Giardia acts an ecosystem engineer (62), indirectly modulating the availability of resources to other species in the same environment and excreting novel waste products, such as alanine (56) and exotic lipids (63), that can be metabolized by commensal bacteria.
Even in the absence of histopathologic evidence of inflammation, giardiasis is associated with redox perturbations (15). Increased diversity and abundance of facultative anaerobes, such as various members of the Gammaproteobacteria and Betaproteobacteria, can accompany the induction of inflammatory states, and as such, their growth is a sensitive indicator of redox potential in the gastrointestinal environment (64). Alterations in luminal redox potential during giardiasis could be linked to lower levels of anaerobic metabolism by the host microbiota (Fig. 6). Giardia is proposed to reduce epithelial cell inflammatory responses through decreased host nitric oxide synthesis and reductions in interleukin-8 (IL-8) (14, 65). Giardia-mediated dysbiosis may be caused indirectly by microinflammation in the gastrointestinal environment, as has been noted with other opportunistic gut pathogens (22, 33, 66). Inflammation throughout the gut is associated with an overall increased oxygen tension in this generally low-oxygen environment, however, and we suggest that alterations in the redox potential during giardiasis are linked to markedly lower levels of anaerobic metabolism by the host microbiota (Fig. 6).
The increased abundance of facultative aerobes and strict aerobes we observed during infection could be explained by low-level inflammation associated with giardiasis (67). However, the Gammaproteobacteria taxa enriched in the proximal and distal small intestine during giardiasis are not the members of the Enterobacteriaceae that are often noted to bloom during gut inflammation (31, 68). While the Enterobacteriaceae decreased in diversity, the diversity of the strictly aerobic Moraxellaceae and the Betaproteobacteria Rhodocyclaceae and Comomonadaceae increased. These taxa are generally metabolically flexible with respect to respiratory potential and carbon/nitrogen metabolism, and they often metabolize complex carbon polymers, including hydrocarbons and lipids (69). Concomitantly, giardiasis-mediated inflammation in the gut resulted in decreased diversity and abundance of obligate anaerobes, such as the Firmicutes (e.g., Lactobacillaceae, Eryipelotichaeae, Ruminococcus, and Clostridia) (33). We also saw decreases in the obligately anaerobic Melainabacteria, members of the cyanobacteria that do not perform photosynthesis and instead obtain energy by fermentation, producing hydrogen as a waste product. In the human gut, they also synthesize some B and K vitamins, implying that commensal Melainabacteria are beneficial to the mammalian host (70, 71).
The lipid concentrations in luminal contents and feces are markedly increased in giardiasis (11, 72, 73). Novel dietary lipids have been shown to notably change microbial community makeup (74), and the microbial metabolism of giardial lipids may play an important role in the commensal diversity shifts in the gut. Giardia directly perturbs host lipid metabolism by scavenging lipids and by excreting novel end products of lipid metabolism (69). Encysting or colonizing Giardia may contribute to shifts in the local gut microbiota (1) by both excreting novel lipids and influencing the bioavailability of bile acids (63, 72, 75, 76). By creating a lipid-rich environment in the gut lumen, Giardia colonization may enrich for lipid specialists within the commensal microbiota. The taxa enriched during giardiasis are similar to taxa dominating the microbiome of other hydrocarbon-enriched environments (77–80). Acinetobacter spp., which are Gammaproteobacteria within the Moraxellaceae family, are notably enriched during giardiasis. Acinetobacter species, such as Acinetobacter calcoaceticus, grow using diverse carbon sources, including acetate, ethanol, and exotic hydrocarbons, and have been promoted to degrade these compounds in contaminated environments (81, 82). It is possible that during giardiasis, Acinetobacter spp. may metabolize these similar lipids as carbon sources. Commensal metabolism of exotic lipids induced during giardiasis could also contribute to this dysbiosis.
Giardia arginine metabolism could also directly impact commensal microbial metabolism. Trophozoites use the arginine dihydrolase pathway (ADiHP) for energy production, yielding ATP through the conversion of arginine to ammonia and citrulline, with the substrate-level phosphorylation of citrulline yielding ornithine and carbamoylphosphate, resulting in NH3 and CO2 as end products (60). Encysting trophozoites are thought to increase ATP production via AdiHP, as glucose catabolized for energy is diverted to synthesize N-acetylgalactosamine, a component of the cyst wall. Competition for arginine between the parasite and host could directly influence the observed severe malnutrition and increased susceptibility to villus shortening (4, 83). The enrichment of denitrifiers, such as the Betaproteobacteria (Rhodocyclaecae) (Fig. 5), that we observed during giardiasis may be related to the metabolism of both vegetative and encysting trophozoites through the excretion of ornithine and ammonium via the arginine dihydrolase pathway. Many of these organisms are noted for their broad metabolic capacities, and some, such as Zoogloea and Dechloromonas spp., both denitrify and degrade hydrocarbons (84). Other Rhodocyclaecae use organic acids, alcohols, or lipids, such as cholesterol, for growth (85, 86). Further profiling of metabolic processes of the Giardia-infected gut via metabolomics methods could illuminate possible roles of microbial metabolic by-products during giardiasis.
Host commensal microbes may limit the initiation or the degree of Giardia colonization.
By a general mechanism known as colonization resistance (87–89), members of the gut microbiota are thought to antagonize pathogens. Production of bacteriocidins or peroxidases can inhibit or kill pathogenic bacteria. For instance, Enterococcus, one of the major constituents of the small intestine, produces superoxide radicals, possibly as a defense mechanism against competitors (90). Understanding how the host gut microbiome limits the initiation of infections could lead to novel therapeutic mechanisms for reducing the severity or duration of symptoms, including treatment of at-risk children with pre- or probiotics.
Commensal microbiota could limit the initiation or the degree of giardiasis in mice; this may also be reflected in giardiasis in humans. Though convenient for immunologic and genetic manipulations, mice are not a natural host for human strains of Giardia lamblia. Reproducible and robust infections of mice with the human-adapted assemblages of Giardia lamblia WB (assemblage A) or GS (assemblage B) require pretreatment of study animals with a cocktail of antibiotics (45). Singer and Nash implicated commensal gastrointestinal bacteria in Giardia's pathogenesis when they observed that mice of the same genotype, from two different vendors, were differentially susceptible to infection with strain GS, that the susceptibility was sensitive to antibiotics, and it was transferrable by cohousing (24). In fact, host commensal bacteria may have a greater influence over infection susceptibility than the absence of CD4+ T cells (91). More recently, Solaymani-Mohammadi and Singer used a cocktail of antibiotics to successfully infect mice with strain WB (45). Lactobacillus probiotic treatment has been reported to reduce the severity and duration of murine giardiasis (92, 93). In humans, symptoms of cramping and steatorrhea do not resolve with antibiotics (94), yet fecal bacteriotherapy has been reported to relieve postgiardiasis gastrointestinal symptoms for over a year after infection (53).
Immunity to Giardia has been shown to depend significantly on intestinal production of IL-17 (reviewed in reference 95). This is supported by findings that the presence of segmented filamentous bacteria in mice from Taconic Farms, which are resistant to Giardia colonization, has been shown to drive intestinal IL-17 production (96). Thus, modulation of baseline host immunity is a potential mechanism for the commensal microbiota to impact Giardia colonization. However, because our analyses were performed using mice from Jackson Laboratories, which lack SFB, our data do not support or refute this particular model.
The intestinal microbiota are known to modulate development of other host immune responses. We recently reported that antibiotic treatment of mice reduced the activation of CD8+ T cells in the intestinal lamina propria and intraepithelial lymphocytes expressing the γδ T cell receptor (97). CD8+ T cells have previously been shown to mediate microvillous shortening and reduce levels of disaccharidase activity in the small intestine (44, 98). Indeed, antibiotic treatment blunted the sucrose deficiency normally seen in the GS model of infection. Unpublished data (J. G. Maloney and S. M. Singer) further suggest that antibiotic treatment may reduce the accumulation of macrophages in the intestinal lamina propria, which is seen in this model. It is unclear if these altered immune responses reflect changes in the full ecosystem of microbes or reflect changes in abundance of a few select species.
Direct and indirect parasite-commensal interactions, as well as host-parasite interactions, can be dysregulated during giardiasis through host-mediated inflammatory responses and microbially mediated metabolic responses. The specific ecological mechanisms by which Giardia's interactions with the commensal microbiota contribute to parasite colonization, infection initiation, pathology, and parasite clearance remain to be explored. A mechanistic understanding of ecological shifts induced during giardiasis could lead to both preventative and therapeutic strategies for treatment. Host-associated microbiota are known to restrict pathogenic colonization (19, 20) and exacerbate or support invasion and pathogenesis (21, 22); thus, they could be used therapeutically. While Lactobacillus probiotic treatment is reported to reduce the severity and duration of murine giardiasis (92, 93), other probiotic treatments remain unexplored. A clearer understanding of parasite-induced ecological shifts could lead to improved treatments that reduce the severity or duration of symptoms during acute or chronic infection.
MATERIALS AND METHODS
Giardia growth conditions and infections.
Giardia lamblia strain GS/M/H7 (ATCC 50801) was cultured in modified TYI-S-33 medium supplemented with bovine bile and 5% adult and 5% fetal bovine serum (48) in sterile 13-ml screw-cap disposable tubes (BD Falcon) and incubated upright at 37°C without shaking.
For infections, 8-week-old C57BL/6J female mice (Jackson Laboratories) were kept under specific-pathogen-free conditions. Mice were kept as cage mates, with four mice per cage. The experimental design included two cohorts of sibling mice (n = 32). Prior to inoculation, one cohort of 16 mice was treated for 4 days with antibiotics (0.5 mg/ml vancomycin, 1 mg/ml neomycin, 1 mg/ml ampicillin) ad libitum in their drinking water to promote parasite colonization (45). Antibiotics were maintained in drinking water throughout the infection. Sixteen control mice were not given antibiotics. Study animals in each cohort were gavaged with 1 million Giardia lamblia GS (n = 12) trophozoites or a saline vehicle control (n = 4 per antibiotic group). Mice in replicates of four were sacrificed at 3, 7, and 14 days postinoculation. Infection dates were staggered such that all sacrifices occurred within a 3-day window. All animal studies were performed with IACUC approval at Georgetown University (Steven Singer, Principal Investigator).
Anatomical sampling of the murine gastrointestinal tract for microbial community analyses.
For comparisons of microbial diversity in various regions of the murine gut during giardiasis, uninfected and Giardia-infected mice were sacrificed at 3, 7, and 14 days after inoculation with Giardia. Anatomical sampling of the gut yielded six community DNA samples: mucosal and luminal proximal small intestine, mucosal and luminal distal small intestine, cecal contents, and colonic contents (Fig. 2). The pSI was sampled as a 3-cm segment 3 cm distal to the pylorus, and the dSI was sampled as a 3-cm segment 10 cm distal to the pylorus. For the proximal and distal small intestinal samples, the luminal and mucosal contents were extracted separately. Luminal contents were isolated by gentle pressure, and the mucosa was removed by gentle scraping with a sterile glass slide. The cecum was opened with a sterile blade, and 200 μl of cecal contents was removed. Colonic contents were sampled by excising the colon and removing two fecal pellets. Pelleted colonic samples from cage mate mice exposed to the same diet were similar in weight and size. No samples were taken from the stomach. All samples were immediately flash-frozen in liquid nitrogen and stored at −80C for subsequent microbial diversity analyses.
Total gut microbial community DNA extraction.
Total community DNA was extracted from 192 gastrointestinal tract samples from the 32 mice using standard methods with mechanical disruption (41) for paired-end sequencing with the llumina MiSeq system. Small subunit rDNA primers (515F and 806R) were used to amplify total community DNA according to protocols developed at the Earth Microbiome Project (42, 43). Small subunit rDNA amplicon sequencing was performed at Argonne National Laboratories using protocols from the Earth Microbiome Project (43).
Amplicon assembly and identification of OTUs.
More than 2 million total reads were used for analysis after sequence quality filtering and assembly of paired-end reads. Default parameters were used to pair forward and reverse reads via SeqPrep (https://github.com/jstjohn/SeqPrep) and call OTUs with open-reference picking at 97% confidence via the GreenGenes 13_8 database. Singleton OTUs were discarded. Analyses of alpha and beta diversities were performed in a QIIME pipeline (99) using default parameters for OTU tables rarefied to 3,000 sequences/sample. OTU tables were filtered to reflect only antibiotic-treated, antibiotic-naive, foregut, hindgut, or any other subclassifications as needed for diversity analyses. Samples with less than 3,000 reads were discarded.
Analyses of gut microbiome diversity.
All microbial diversity metrics were calculated using rarefied OTU tables at 3,000 sequences/sample, with the exception of DESeq (used for unrarefied OTU tables with a 3,000 sequences/sample minimum count) (51). Alpha diversity for each sample was evaluated using Chao1, Shannon, and whole tree phylogenetic distances. PCoA plots were visualized using EMPeror (100), and both unweighted and weighted UniFrac samples were used for further analyses (101, 102). adonis analysis was performed on unweighted and weighted Unifrac matrices to calculate P values. All diversity metrics were calculated using weighted UniFrac. Similar trends were seen using both weighted and unweighted algorithms (unweighted R2 values are presented in Table S2 in the supplemental material). Mucosal and luminal samples from the same small intestinal site were combined unless otherwise indicated. For overall summaries of microbial diversity, proximal and distal small intestine samples were combined and designated foregut samples, whereas combined cecum and colon samples were designated hindgut samples.
Taxa with significant variation (e.g., dynamic taxa) were identified between anatomical sites in experimental treatments using a combination of group_significance.py script to perform a Kruskal-Wallis test on rarefied OTU tables and differential_abundance.py on unrarefied OTU tables to implement the DESeq algorithm (51, 52). Giardiasis indicator taxa were defined as taxonomic groupings identified through both DESeq and Kruskal-Wallis analyses that had consistent variation in diversity (corrected P values of <0.05) and fold changes in relative read abundance in infected animals were calculated relative to relative read abundance in uninfected animals. Lastly, the MGDI was used for calculate_taxonomy_ratios.py in QIIME, specifying increased Moraxellaceae and Rhodocyclaceae with decreased Clostridiales. PCoA plots with explicit axes were created using the Make_emporer.py script in QIIME.
Quantitation of parasite density (burden) using QPCR.
For the QPCR-based analysis of microbial abundance, the gastrointestinal tracts of euthanized mice were isolated and frozen as described above. Specifically, a 3-cm segment located 7 cm distal to the pylorus (proximal small intestine) was used in its entirety for Giardia quantification. QPCR of an intragenic region unique to chromosome 1 was performed with Giardia-specific primers GS-1-318F, GCAGAAACAGTGCTTTGAGG; and GS-1-318R, TTGTTTACGGCAAGGAAATG. DNA was extracted using phenol-chloroform and bead beating (103). The single-copy, stably expressed murine nidogen-1 (nid1) gene was used as an internal control to quantify the contribution of murine DNA to intestinal segments by using the primers nido.F, CCAGCCACAGAATACCATCC, and nido.R, GGACATACTCTGCTGCCATC (104). The differential counts to threshold (ΔCT) between nid1 and GS-1-318 was calculated and the ΔCT was normalized against murine contribution. Standard curves for QPCR were created using known concentrations of Giardia GS DNA. The averaged CT of three technical replicates was utilized to extrapolate the concentration of Giardia per sample. The fold change was calculated between averaged negative samples and each infected sample.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant R01AI077571 to S.C.D. and American Association of Immunologists (AAI) Careers in Immunology fellowship, NIH AI094492, and NIH grant AI109591 to S.M.S.
We graciously thank Sarah Guest for the schematic representation of the Giardia-induced dysbiosis, as well as Kari Hagen, Katherine Karberg, and Chris Nosala for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00948-16.
REFERENCES
- 1.Halliez MC, Buret AG. 2013. Extra-intestinal and long term consequences of Giardia duodenalis infections. World J Gastroenterol 19:8974–8985. doi: 10.3748/wjg.v19.i47.8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Mekhlafi HM, Al-Maktari MT, Jani R, Ahmed A, Anuar TS, Moktar N, Mahdy MA, Lim YA, Mahmud R, Surin J. 2013. Burden of Giardia duodenalis infection and its adverse effects on growth of schoolchildren in rural Malaysia. PLoS Negl Trop Dis 7:e2516. doi: 10.1371/journal.pntd.0002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furness BW, Beach MJ, Roberts JM. 2000. Giardiasis surveillance—United States, 1992-1997. MMWR CDC Surveill Summ 49:1–13. [PubMed] [Google Scholar]
- 4.Bartelt LA, Sartor RB. 2015. Advances in understanding Giardia: determinants and mechanisms of chronic sequelae. F1000Prime Rep 7:62. doi: 10.12703/P7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartelt LA, Roche J, Kolling G, Bolick D, Noronha F, Naylor C, Hoffman P, Warren C, Singer S, Guerrant R. 2013. Persistent G. lamblia impairs growth in a murine malnutrition model. J Clin Invest 123:2672–2684. doi: 10.1172/JCI67294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morch K, Hanevik K, Rortveit G, Wensaas KA, Eide GE, Hausken T, Langeland N. 2009. Severity of Giardia infection associated with post-infectious fatigue and abdominal symptoms two years after. BMC Infect Dis 9:206. doi: 10.1186/1471-2334-9-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morch K, Hanevik K, Rortveit G, Wensaas KA, Langeland N. 2009. High rate of fatigue and abdominal symptoms 2 years after an outbreak of giardiasis. Trans R Soc Trop Med Hyg 103:530–532. doi: 10.1016/j.trstmh.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Stark D, Barratt JL, van Hal S, Marriott D, Harkness J, Ellis JT. 2009. Clinical significance of enteric protozoa in the immunosuppressed human population. Clin Microbiol Rev 22:634–650. doi: 10.1128/CMR.00017-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savioli L, Smith H, Thompson A. 2006. Giardia and Cryptosporidium join the Neglected Diseases Initiative. Trends Parasitol 22:203–208. doi: 10.1016/j.pt.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 10.Reference deleted.
- 11.Adam RD. 2001. Biology of Giardia lamblia. Clin Microbiol Rev 14:447–475. doi: 10.1128/CMR.14.3.447-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ankarklev J, Jerlstrom-Hultqvist J, Ringqvist E, Troell K, Svard SG. 2010. Behind the smile: cell biology and disease mechanisms of Giardia species. Nat Rev Microbiol 8:413–422. [DOI] [PubMed] [Google Scholar]
- 13.Maloney J, Keselman A, Li E, Singer SM. 2015. Macrophages expressing arginase 1 and nitric oxide synthase 2 accumulate in the small intestine during Giardia lamblia infection. Microbes Infect 17:462–467. doi: 10.1016/j.micinf.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stadelmann B, Merino MC, Persson L, Svard SG. 2012. Arginine consumption by the intestinal parasite Giardia intestinalis reduces proliferation of intestinal epithelial cells. PLoS One 7:e45325. doi: 10.1371/journal.pone.0045325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotton JA, Amat CB, Buret AG. 2015. Disruptions of host immunity and inflammation by Giardia duodenalis: potential consequences for co-infections in the gastro-intestinal tract. Pathogens 4:764–792. doi: 10.3390/pathogens4040764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koot BGP, ten Kate FJ, Juffrie M, Rosalina I, Taminiau JJ, Benninga MA. 2009. Does Giardia lamblia cause villous atrophy in children? A retrospective cohort study of the histological abnormalities in giardiasis. J Pediatr Gastroenterol Nutr 49:304–308. doi: 10.1097/MPG.0b013e31818de3c4. [DOI] [PubMed] [Google Scholar]
- 17.Frank DN, Pace NR. 2008. Gastrointestinal microbiology enters the metagenomics era. Curr Opin Gastrenterol 24:4–10. doi: 10.1097/MOG.0b013e3282f2b0e8. [DOI] [PubMed] [Google Scholar]
- 18.McCann KS. 2000. The diversity-stability debate. Nature 405:228–233. doi: 10.1038/35012234. [DOI] [PubMed] [Google Scholar]
- 19.Swe PM, Zakrzewski M, Kelly A, Krause L, Fischer K. 2014. Scabies mites alter the skin microbiome and promote growth of opportunistic pathogens in a porcine model. PLoS Negl Trop Dis 8:e2897. doi: 10.1371/journal.pntd.0002897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsiao A, Ahmed AM, Subramanian S, Griffin NW, Drewry LL, Petri WA Jr, Haque R, Ahmed T, Gordon JI. 2014. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515:423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreira RB, Gill N, Willing BP, Antunes LC, Russell SL, Croxen MA, Finlay BB. 2011. The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLoS One 6:e20338. doi: 10.1371/journal.pone.0020338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torres MF, Uetanabaro AP, Costa AF, Alves CA, Farias LM, Bambirra EA, Penna FJ, Vieira EC, Nicoli JR. 2000. Influence of bacteria from the duodenal microbiota of patients with symptomatic giardiasis on the pathogenicity of Giardia duodenalis in gnotoxenic mice. J Med Microbiol 49:209–215. doi: 10.1099/0022-1317-49-3-209. [DOI] [PubMed] [Google Scholar]
- 24.Singer SM, Nash TE. 2000. The role of normal flora in Giardia lamblia infections in mice. J Infect Dis 181:1510–1512. doi: 10.1086/315409. [DOI] [PubMed] [Google Scholar]
- 25.Eckmann L. 2003. Mucosal defences against Giardia. Parasite Immunol 25:259–270. doi: 10.1046/j.1365-3024.2003.00634.x. [DOI] [PubMed] [Google Scholar]
- 26.Müller N, von Allmen N. 2005. Recent insights into the mucosal reactions associated with Giardia lamblia infections. Int J Parasitol 35:1339–1347. doi: 10.1016/j.ijpara.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 27.Tandon BN, Tandon RK, Satpathy BK, Shriniwas. 1977. Mechanism of malabsorption in giardiasis: a study of bacterial flora and bile salt deconjugation in upper jejunum. Gut 18:176–181. doi: 10.1136/gut.18.3.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomkins AM, Wright SG, Drasar BS, James WP. 1978. Bacterial colonization of jejunal mucosa in giardiasis. Trans R Soc Trop Med Hyg 72:33–36. [DOI] [PubMed] [Google Scholar]
- 29.Chen TL, Chen S, Wu HW, Lee TC, Lu YZ, Wu LL, Ni YH, Sun CH, Yu WH, Buret AG, Yu LC. 2013. Persistent gut barrier damage and commensal bacterial influx following eradication of Giardia infection in mice. Gut Pathog 5:26. doi: 10.1186/1757-4749-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooks J. 2002. Characterizing ecosystem-level consequences of biological invasions: the role of ecosystem engineers. Oikos 97:153–166. doi: 10.1034/j.1600-0706.2002.970201.x. [DOI] [Google Scholar]
- 31.Faber F, Baumler AJ. 2014. The impact of intestinal inflammation on the nutritional environment of the gut microbiota. Immunol Lett 162:48–53. doi: 10.1016/j.imlet.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weingarden A, Gonzalez A, Vazquez-Baeza Y, Weiss S, Humphry G, Berg-Lyons D, Knights D, Unno T, Bobr A, Kang J, Khoruts A, Knight R, Sadowsky MJ. 2015. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome 3:10. doi: 10.1186/s40168-015-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, Gonzalez A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. 2014. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hold GL, Smith M, Grange C, Watt ER, El-Omar EM, Mukhopadhya I. 2014. Role of the gut microbiota in inflammatory bowel disease pathogenesis: what have we learnt in the past 10 years? World J Gastroenterol 20:1192–1210. doi: 10.3748/wjg.v20.i5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molloy MJ, Grainger JR, Bouladoux N, Hand TW, Koo LY, Naik S, Quinones M, Dzutsev AK, Gao JL, Trinchieri G, Murphy PM, Belkaid Y. 2013. Intraluminal containment of commensal outgrowth in the gut during infection-induced dysbiosis. Cell Host Microbe 14:318–328. doi: 10.1016/j.chom.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benson A, Pifer R, Behrendt CL, Hooper LV, Yarovinsky F. 2009. Gut commensal bacteria direct a protective immune response against Toxoplasma gondii. Cell Host Microbe 6:187–196. doi: 10.1016/j.chom.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heimesaat MM, Bereswill S, Fischer A, Fuchs D, Struck D, Niebergall J, Jahn HK, Dunay IR, Moter A, Gescher DM, Schumann RR, Gobel UB, Liesenfeld O. 2006. Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J Immunol 177:8785–8795. doi: 10.4049/jimmunol.177.12.8785. [DOI] [PubMed] [Google Scholar]
- 38.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 39.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zoetendal EG, Heilig HG, Klaassens ES, Booijink CC, Kleerebezem M, Smidt H, de Vos WM. 2006. Isolation of DNA from bacterial samples of the human gastrointestinal tract. Nat Protoc 1:870–873. doi: 10.1038/nprot.2006.142. [DOI] [PubMed] [Google Scholar]
- 42.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilbert JA, Meyer F, Jansson J, Gordon J, Pace N, Tiedje J, Ley R, Fierer N, Field D, Kyrpides N, Glockner FO, Klenk HP, Wommack KE, Glass E, Docherty K, Gallery R, Stevens R, Knight R. 2010. The Earth Microbiome Project: meeting report of the 1 EMP Meeting on Sample Selection and Acquisition at Argonne National Laboratory, October 6 2010. Stand Genomic Sci 3:249–253. doi: 10.4056/aigs.1443528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solaymani-Mohammadi S, Singer SM. 2013. Regulation of intestinal epithelial cell cytoskeletal remodeling by cellular immunity following gut infection. Mucosal Immunol 6:369–378. doi: 10.1038/mi.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solaymani-Mohammadi S, Singer SM. 2011. Host immunity and pathogen strain contribute to intestinal disaccharidase impairment following gut infection. J Immunol 187:3769–3775. doi: 10.4049/jimmunol.1100606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts-Thomson IC, Stevens DP, Mahmoud AA, Warren KS. 1976. Giardiasis in the mouse: an animal model. Gastroenterology 71:57–61. [PubMed] [Google Scholar]
- 47.Gillin FD, Boucher SE, Reiner DS. 1987. Stimulation of in-vitro encystation of Giardia lamblia by small intestinal conditions. Clin Res 35:475A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lujan HD, Mowatt MR, Byrd LG, Nash TE. 1996. Cholesterol starvation induces differentiation of the intestinal parasite Giardia lamblia. Proc Natl Acad Sci U S A 93:7628–7633. doi: 10.1073/pnas.93.15.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gillon J, Al Thamery D, Ferguson A. 1982. Features of small intestinal pathology (epithelial cell kinetics, intraepithelial lymphocytes, disaccharidases) in a primary Giardia muris infection. Gut 23:498–506. doi: 10.1136/gut.23.6.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peterson DA, Frank DN, Pace NR, Gordon JI. 2008. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morken MH, Valeur J, Norin E, Midtvedt T, Nysaeter G, Berstad A. 2009. Antibiotic or bacterial therapy in post-giardiasis irritable bowel syndrome. Scand J Gastroenterol 44:1296–1303. doi: 10.3109/00365520903274401. [DOI] [PubMed] [Google Scholar]
- 54.Lloyd D. 2004. Anaerobic protists: some misconceptions and confusions. Microbiology 150:1115–1116. doi: 10.1099/mic.0.26802-0. [DOI] [PubMed] [Google Scholar]
- 55.Lindmark DG. 1980. Energy metabolism of the anaerobic protozoan Giardia lamblia. Mol Biochem Parasitol 1:1–12. doi: 10.1016/0166-6851(80)90037-7. [DOI] [PubMed] [Google Scholar]
- 56.Paget TA, Raynor MH, Shipp DW, Lloyd D. 1990. Giardia lamblia produces alanine anaerobically but not in the presence of oxygen. Mol Biochem Parasitol 42:63–67. doi: 10.1016/0166-6851(90)90113-Z. [DOI] [PubMed] [Google Scholar]
- 57.Schofield PJ, Costello M, Edwards MR, O'Sullivan WJ. 1990. The arginine dihydrolase pathway is present in Giardia intestinalis. Int J Parasitol 20:697–699. doi: 10.1016/0020-7519(90)90133-8. [DOI] [PubMed] [Google Scholar]
- 58.Schofield PJ, Edwards MR, Kranz P. 1991. Glucose metabolism in Giardia intestinalis. Mol Biochem Parasitol 45:39–47. doi: 10.1016/0166-6851(91)90025-2. [DOI] [PubMed] [Google Scholar]
- 59.Edwards MR, Schofield PJ, O'Sullivan WJ, Costello M. 1992. Arginine metabolism during culture of Giardia intestinalis. Mol Biochem Parasitol 53:97–103. doi: 10.1016/0166-6851(92)90011-8. [DOI] [PubMed] [Google Scholar]
- 60.Schofield PJ, Edwards MR, Matthews J, Wilson JR. 1992. The pathway of arginine catabolism in Giardia intestinalis. Mol Biochem Parasitol 51:29–36. doi: 10.1016/0166-6851(92)90197-R. [DOI] [PubMed] [Google Scholar]
- 61.Lloyd D, Harris JC, Maroulis S, Wadley R, Ralphs JR, Hann AC, Turner MP, Edwards MR. 2002. The “primitive” microaerophile Giardia intestinalis (syn. lamblia, duodenalis) has specialized membranes with electron transport and membrane-potential-generating functions. Microbiology 148:1349–1354. doi: 10.1099/00221287-148-5-1349. [DOI] [PubMed] [Google Scholar]
- 62.Jones C, Lawton J, Shachak M. 1994. Organisms as ecosystem engineers. Oikos 69:373–386. doi: 10.2307/3545850. [DOI] [Google Scholar]
- 63.Yichoy M, Duarte TT, De Chatterjee A, Mendez TL, Aguilera KY, Roy D, Roychowdhury S, Aley SB, Das S. 2011. Lipid metabolism in Giardia: a post-genomic perspective. Parasitology 138:267–278. doi: 10.1017/S0031182010001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rigottier-Gois L. 2013. Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J 7:1256–1261. doi: 10.1038/ismej.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mastronicola D, Falabella M, Forte E, Testa F, Sarti P, Giuffre A. 2015. Antioxidant defence systems in the protozoan pathogen Giardia intestinalis. Mol Biochem Parasitol 206:56–66. doi: 10.1016/j.molbiopara.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 66.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 67.Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, Thom SR, Bushman FD, Vinogradov SA, Wu GD. 2014. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 147:1055–1063.e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Winter SE, Baumler AJ. 2014. Dysbiosis in the inflamed intestine: chance favors the prepared microbe. Gut Microbes 5:71–73. doi: 10.4161/gmic.27129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mendez TL, De Chatterjee A, Duarte T, De Leon J, Robles-Martinez L, Das S. 2015. Sphingolipids, lipid rafts, and giardial encystation: the show must go on. Curr Trop Med Rep 2:136–143. doi: 10.1007/s40475-015-0052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wrighton KC, Castelle CJ, Wilkins MJ, Hug LA, Sharon I, Thomas BC, Handley KM, Mullin SW, Nicora CD, Singh A, Lipton MS, Long PE, Williams KH, Banfield JF. 2014. Metabolic interdependencies between phylogenetically novel fermenters and respiratory organisms in an unconfined aquifer. ISME J 8:1452–1463. doi: 10.1038/ismej.2013.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Di Rienzi SC, Sharon I, Wrighton KC, Koren O, Hug LA, Thomas BC, Goodrich JK, Bell JT, Spector TD, Banfield JF, Ley RE. 2013. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. eLife 2:e01102. doi: 10.7554/eLife.01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Das S, Stevens T, Castillo C, Villasenor A, Arredondo H, Reddy K. 2002. Lipid metabolism in mucous-dwelling amitochondriate protozoa. Int J Parasitol 32:655–675. doi: 10.1016/S0020-7519(02)00006-1. [DOI] [PubMed] [Google Scholar]
- 73.Katelaris P, Seow F, Ngu M. 1991. The effect of Giardia lamblia trophozoites on lipolysis in vitro. Parasitology 103:35–39. doi: 10.1017/S0031182000059266. [DOI] [PubMed] [Google Scholar]
- 74.Marques TM, Wall R, O'Sullivan O, Fitzgerald GF, Shanahan F, Quigley EM, Cotter PD, Cryan JF, Dinan TG, Ross RP, Stanton C. 2015. Dietary trans-10, cis-12-conjugated linoleic acid alters fatty acid metabolism and microbiota composition in mice. Br J Nutr 113:728–738. doi: 10.1017/S0007114514004206. [DOI] [PubMed] [Google Scholar]
- 75.Das S, Schteingart CD, Hofmann AF, Reiner DS, Aley SB, Gillin FD. 1997. Giardia lamblia: evidence for carrier-mediated uptake and release of conjugated bile acids. Exp Parasitol 87:133–141. doi: 10.1006/expr.1997.4197. [DOI] [PubMed] [Google Scholar]
- 76.Gillin FD, Gault MJ, Hofmann AF, Gurantz D, Sauch JF. 1986. Biliary lipids support serum-free growth of Giardia lamblia. Infect Immun 53:641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abbai NS, Pillay B. 2013. Analysis of hydrocarbon-contaminated groundwater metagenomes as revealed by high-throughput sequencing. Mol Biotechnol 54:900–912. doi: 10.1007/s12033-012-9639-z. [DOI] [PubMed] [Google Scholar]
- 78.Paisio CE, Quevedo MR, Talano MA, Gonzalez PS, Agostini E. 2014. Application of two bacterial strains for wastewater bioremediation and assessment of phenolics biodegradation. Environ Technol 35:1802–1810. doi: 10.1080/09593330.2014.882994. [DOI] [PubMed] [Google Scholar]
- 79.Chen Y, Li C, Zhou Z, Wen J, You X, Mao Y, Lu C, Huo G, Jia X. 2014. Enhanced biodegradation of alkane hydrocarbons and crude oil by mixed strains and bacterial community analysis. Appl Biochem Biotechnol 172:3433–3447. doi: 10.1007/s12010-014-0777-6. [DOI] [PubMed] [Google Scholar]
- 80.Singleton DR, Dickey AN, Scholl EH, Wright FA, Aitken MD. 2015. Complete genome sequence of a novel bacterium within the family Rhodocyclaceae that degrades polycyclic aromatic hydrocarbons. Genome Announc 3:e00251-15. doi: 10.1128/genomeA.00251-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abdel-El-Haleem D. 2003. Acinetobacter: environmental and biotechnological applications. Afr J Biotechnol 2:71–74. doi: 10.5897/AJB2003.000-1014. [DOI] [Google Scholar]
- 82.Khoramnia A, Ebrahimpour A, Beh BK, Lai OM. 2011. Production of a solvent, detergent, and thermotolerant lipase by a newly isolated Acinetobacter sp. in submerged and solid-state fermentations. J Biomed Biotechnol 2011:702179. doi: 10.1155/2011/702179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ventura LL, Oliveira DR, Viana JC, Santos JF, Caliari MV, Gomes MA. 2013. Impact of protein malnutrition on histological parameters of experimentally infected animals with Giardia lamblia. Exp Parasitol 133:391–395. doi: 10.1016/j.exppara.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 84.Farkas M, Tancsics A, Kriszt B, Benedek T, Toth EM, Keki Z, Veres PG, Szoboszlay S. 2015. Zoogloea oleivorans sp. nov., a floc-forming, petroleum hydrocarbon-degrading bacterium isolated from biofilm. Int J Syst Evol Microbiol 65:274–279. doi: 10.1099/ijs.0.068486-0. [DOI] [PubMed] [Google Scholar]
- 85.Smalley NE, Taipale S, Marco P, Doronina NV, Kyrpides N, Shapiro N, Woyke T, Kalyuzhnaya MG. 2015. Functional and genomic diversity of methylotrophic Rhodocyclaceae: description of the new species Methyloversatilis discipulorum sp. nov. Int J Syst Evol Microbiol 65:2227–2233. doi: 10.1099/ijs.0.000190. [DOI] [PubMed] [Google Scholar]
- 86.Chiang YR, Ismail W, Heintz D, Schaeffer C, Van Dorsselaer A, Fuchs G. 2008. Study of anoxic and oxic cholesterol metabolism by Sterolibacterium denitrificans. J Bacteriol 190:905–914. doi: 10.1128/JB.01525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sharma A, Dhayal D, Singh OP, Adak T, Bhatnagar RK. 2013. Gut microbes influence fitness and malaria transmission potential of Asian malaria vector Anopheles stephensi. Acta Trop 128:41–47. doi: 10.1016/j.actatropica.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 88.Flavia Nardy A, Freire-de-Lima CG, Morrot A. 2015. Immune evasion strategies of Trypanosoma cruzi. J Immunol Res 2015:178947. doi: 10.1155/2015/178947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mooney JP, Lokken KL, Byndloss MX, George MD, Velazquez EM, Faber F, Butler BP, Walker GT, Ali MM, Potts R, Tiffany C, Ahmer BM, Luckhart S, Tsolis RM. 2015. Inflammation-associated alterations to the intestinal microbiota reduce colonization resistance against non-typhoidal Salmonella during concurrent malaria parasite infection. Sci Rep 5:14603. doi: 10.1038/srep14603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang X, Huycke MM. 2007. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology 132:551–561. doi: 10.1053/j.gastro.2006.11.040. [DOI] [PubMed] [Google Scholar]
- 91.Singer SM, Nash TE. 2000. T-cell-dependent control of acute Giardia lamblia infections in mice. Infect Immun 68:170–175. doi: 10.1128/IAI.68.1.170-175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goyal N, Tiwari RP, Shukla G. 2011. Lactobacillus rhamnosus GG as an effective probiotic for murine giardiasis. Interdiscip Perspect Infect Dis 2011:795219. doi: 10.1155/2011/795219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shukla G, Sidhu RK. 2011. Lactobacillus casei as a probiotic in malnourished Giardia lamblia-infected mice: a biochemical and histopathological study. Can J Microbiol 135:127–135. doi: 10.1139/W10-110. [DOI] [PubMed] [Google Scholar]
- 94.Wright SG, Tomkins AM, Ridley DS. 1977. Giardiasis: clinical and therapeutic aspects. Gut 18:343–350. doi: 10.1136/gut.18.5.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Singer SM. 2015. Control of giardiasis by interleukin-17 in humans and mice—are the questions all answered? Clin Vaccine Immunol 23:2–5. doi: 10.1128/CVI.00648-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Keselman A, Li E, Maloney J, Singer SM. 2016. The microbiota contributes to CD8+ T cell activation and nutrient malabsorption following intestinal infection with Giardia duodenalis. Infect Immun 84:2853–2860. doi: 10.1128/IAI.00348-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scott KG, Yu LC, Buret AG. 2004. Role of CD8+ and CD4+ T lymphocytes in jejunal mucosal injury during murine giardiasis. Infect Immun 72:3536–3542. doi: 10.1128/IAI.72.6.3536-3542.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vazquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. 2013. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lozupone CA, Stombaugh J, Gonzalez A, Ackermann G, Wendel D, Vazquez-Baeza Y, Jansson JK, Gordon JI, Knight R. 2013. Meta-analyses of studies of the human microbiota. Genome Res 23:1704–1714. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wagner Mackenzie B, Waite DW, Taylor MW. 2015. Evaluating variation in human gut microbiota profiles due to DNA extraction method and inter-subject differences. Front Microbiol 6:130. doi: 10.3389/fmicb.2015.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, Zhu W, Sartor RB, Boedeker EC, Harpaz N, Pace NR, Li E. 2011. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis 17:179–184. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morrison TB, Ma Y, Weis JH, Weis JJ. 1999. Rapid and sensitive quantification of Borrelia burgdorferi-infected mouse tissues by continuous fluorescent monitoring of PCR. J Clin Microbiol 37:987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.