Abstract
We present a new approach for the detection and identification of enteroviruses concentrated and isolated from sewage. Samples were collected from two study sites located at Nicosia and Limassol sewage treatment plants in Cyprus. Viruses were adsorbed to cellulose nitrate membrane filters, cultured directly from the membrane filters by using the VIRADEN method, and identified by reverse transcription-PCR, followed by 5′ untranslated region (5′-UTR) restriction fragment length polymorphism (RFLP) analysis and partial sequencing of the VP1 protein coding region. Initial subgrouping based on the HpaII restriction profile showed that all of the isolates except one belonged to the same genetic subcluster. Partial VP1 sequencing revealed that most isolates belonged to serotypes coxsackie B4 (42.5%) and coxsackie Α9 (30%), whereas coxsackie B2 (17.5%) and coxsackie B1 (3%) isolates were less frequently observed. One poliovirus type 2 isolate (2.5%) of vaccine origin was also found. The HpaII digests predicted the genetic subcluster for all isolates. They also accurately differentiated the isolates as nonpolio or polio isolates. This approach seems to be very promising for environmental surveillance of enterovirus circulation and epidemiology, with all of the significant effects that this entails for public health. Partial VP1 sequencing is efficient for molecular serotyping of enteroviruses, while 5′-UTR RFLP analysis with HpaII can also be considered an asset for the initial subclassification of enterovirus isolates.
Approximately 140 types of viruses may contaminate water and wastewater, and enteroviruses are the most frequently detected viruses (30). The enterovirus genus, which is the most important genus of the Picornaviridae in terms of human pathogenicity (16, 36), contains 65 immunologically distinct human serotypes in five Human enterovirus species (Human enterovirus A to D and Poliovirus) (14). It consists of polioviruses (PV) (three serotypes), coxsackie A viruses (CAV) (23 serotypes), coxsackie B viruses (CBV) (six serotypes), echoviruses (28 serotypes), and enteroviruses 68 to 71 and 73. The clinical manifestations and diseases caused by human enteroviruses range widely from asymptomatic infections and the common cold to fatal cases of meningitis, encephalitis, and poliomyelitis. Central nervous system diseases are common manifestations of infections caused by several different types of enteroviruses, and aseptic meningitis is the most common such disease recorded (16). Other less common central nervous system diseases include encephalitis, Guillain-Barré syndrome, paralysis, cerebellar ataxia, and peripheral neuritis (16). Enterovirus infections of the myocardium are also extremely important, since they are the most common cause of acute myocarditis and they have also been implicated in dilated cardiomyopathy (especially CBV serotype 1 to 5 infections), which is one of the most common cardiac diseases requiring heart transplantation.
Enteroviruses are transmitted by a fecal-oral cycle, multiply in the gastrointestinal tract, and are finally excreted in large numbers into the environment through feces. Although these viruses are readily found in fecally contaminated waters, waterborne enterovirus infection has only occasionally been documented because many infections caused by these agents are subclinical (30).
Identification of enterovirus isolates does not have a significant impact on the clinical management of infected individuals. Nevertheless, it can contribute significantly to the identification of various epidemics and to the subsequent effective surveillance of populations by determining the source of infection, the correlation between enterovirus serotype or strain and clinical symptoms, the characteristics of particularly virulent viruses, the possible means of transmission, and the emergence of new strains or the reemergence of older strains. Moreover, a means for accurate distinction between polio and nonpolio isolates is essential for public health polio surveillance programs that aim to eradicate wild-type polioviruses.
Environmental surveillance has been successfully used in monitoring enteric virus circulation and in assessing the extent or duration of epidemic poliovirus infection in specific conditions (3). The methods used for identification of enteroviruses from raw sewage and sewage effluents have many limitations, mainly due to the nature of the samples examined. Raw sewage usually contains organic compounds at high concentrations, which may interfere with either the adsorption of the virus on a membrane filter or the cultivation of the virus in tissue cultures. Moreover, a large proportion of viruses found in raw sewage are associated with solids, which leads to major technical limitations due to incomplete virus elution. A simple, low-cost, efficient method for concentrating and counting cytopathogenic viruses present in wastewater after the viruses are adsorbed to cellulose nitrate membrane filters has recently been described (26). This method, which is referred as VIRADEN (an acronym derived from virus adsorption enumeration), is suitable for testing raw sewage and secondary sewage effluents in volumes large enough to find significant numbers of enteroviruses.
Typing of concentrated enteroviruses is based mainly on (i) serotyping with equine type-specific hyperimmune sera that have been mixed to obtain intersecting pools (16) and (ii) use of molecular methods based on reverse transcription PCR (RT-PCR) of RNA extracted from preparations showing cytopathic effects on cell monolayers and application of other molecular techniques (notably restriction fragment length polymorphism [RFLP] analysis and sequencing) (20, 29). However, serotyping by seroneutralization is time-consuming, labor-intensive, and costly, the antiserum supply is limited, and there is still the problem of untypeable enteroviruses.
A new approach for molecular serotyping of enteroviruses concentrated and isolated from sewage samples by the VIRADEN method was developed in the present study. We performed both RFLP-based molecular typing of the 5′ untranslated region (5′-UTR) of isolated enteroviruses in genetic clusters, using restriction endonuclease HpaII as previously described (35), and molecular serotyping of a portion of the VP1 gene, based on RT-PCR amplification and sequencing (22). The combination of these techniques was tested by using raw sewage collected at Nicosia and Limasol sewage treatment plants in Cyprus. This approach may be used as a rapid, low-cost, efficient method for environmental surveillance of circulating enteroviruses in sewage.
MATERIALS AND METHODS
Sampling sites.
Six sewage samples were collected between April and December 2003 from two study sites located at the Nicosia and Limassol sewage treatment plants in Cyprus. The Nicosia sewage treatment plant is a waste stabilization pond plant that receives about 13,000 m3 of sewage effluents per day. The Limasol sewage treatment plant is one of the most sophisticated activated sludge sewage treatment plants in Cyprus. It is relatively new (it was built in the mid-1990s), serves a total population of 80,000 people, and receives 15,000 m3 of sewage effluents per day on average.
Collection of sewage samples.
Trained local authorities collected samples in April, May, September, and December 2003. The existing sewage sample collection system was used for assessment of the microbiological and chemical quality of sewage effluents. One-liter grab samples were collected as recommended previously (3) during the peak morning flow. The samples were transported in clean, sterile, leak-proof containers at 4°C. Samples were transported and analyzed on the day of collection.
VIRADEN method. (i) Cells, media, and reagents.
The Buffalo green monkey (BGM) continuous cell line was used for propagation of viruses. Cells were grown in Eagle's minimum essential medium with Earle's salts (Gibco, Invitrogen, Paisley, United Kingdom) containing 5% fetal bovine serum, 2 mM l-glutamine, 26.8 mM NaHCO3, 100 U of penicillin per ml, and 100 μg of streptomycin per ml.
The overlay medium used for the standard plaque assay was medium 199 with Earle's salts (Gibco) supplemented with 2% fetal bovine serum, 26.8 mM NaHCO3, 100 U of penicillin per ml, and 100 mg of streptomycin per ml. The overlay medium was prepared as a 2× stock and was mixed before use with an equal volume of 2% purified agar (Oxoid, Basingstoke, Hampshire, United Kingdom).
Penicillin (100 U/ml), streptomycin (100 mg/ml), gentamicin (50 mg/ml), nystatin (50 mg/ml), and ceftazidime (20 mg/ml) were added to the growth and overlay media. All of the antibiotics except ceftazidime were obtained from Gibco. Ceftazidime (Ceftazidime Fortum 1g) was obtained from GlaxoWellcome (Greenford, United Kingdom). Ceftazidime was diluted in sterile distilled water to a concentration of 10,000 mg per ml and was stored at −20°C. Defrosted ceftazidime was used within 7 days.
(ii) Membrane prefilters and filters.
The suspended solids and the microbial load were removed from a sample by filtering it through 0.22-μm-pore-size, 33-mm-diameter, syringe-driven, hydrophilic polyester sulfonate filter units (SLGP 033 RS; Millipore Corp., Bedford, Mass.) as previously described (18, 19). Cellulose nitrate filters with a pore size of 3 μm and a diameter of 47 mm (Millipore) were used to concentrate viruses from the samples.
(iii) Virus adsorption on the membranes and detection by the VIRADEN method.
Viruses present in the filtered raw sewage samples were first adsorbed to the cellulose nitrate membrane filters by using the VIRADEN method as previously described (26). Briefly, a sample was amended by adding MgCl2 · 6H2O to a final MgCl2 concentration of 0.05 M. Then it was filtered through a 47-mm-diameter, 3-μm-pore-size cellulose nitrate membrane at a flow rate that did not exceed 200 ml per min. When the entire sample had been filtered, the membrane filter was washed by passing 100 ml of sterile 0.05 M MgCl2 through it. The volumes examined for each sample were 5, 10, and 20 ml. For such small sample volumes (up to 20 ml), amended sterile water (20 to 30 ml) was added to the funnel before the sample was poured in order to make the flow rate more realistic and to evenly disperse the viruses on the filter. The VIRADEN method can be used for raw sewage samples with volumes up to 20 ml without a significant loss of recovery (19).
Finally, the viruses adsorbed on a membrane filter were detected and counted on a BGM monolayer as follows. The growth medium in a 60-mm-diameter petri dish with a confluent monolayer was discarded. Then 100 μl of a suspension of BGM cells in Eagle's minimum essential medium with Earle's salts supplemented with antibiotics containing 1.75 × 107 ± 0.25 × 107 cells per ml was placed in the center of the petri dish. The membrane filter with the adsorbed viruses was placed upside down on top of the suspension and the cell monolayer. Five milliliters of overlay medium was then poured slowly onto the center of the membrane filter and spread all over the plate. The whole procedure was performed under aseptic conditions. The agar was allowed to set protected from light, and the petri dishes were incubated at 37°C in the presence of 5% CO2 at a relative humidity of more than 80% for 48 to 72 h. We did not use neutral red in our overlay medium since the plaques were visible without it. Addition of neutral red (1 ml of a 0.01% solution in 200 ml of overlay agar medium) could enhance plaque visibility, but care had to be taken to protect the cultures from direct light. After incubation, both the agar and the membrane were carefully detached with a spatula. The plate was examined carefully from the top in order to observe any cytopathogenic effects on the cell monolayer in the form of plaques. Virus material was picked from each plaque after resuspension in 3 to 4 μl of sterile distilled water by using a sterile pipette tip. It was then used to inoculate growing BGM cells in 24-well tissue culture trays. Mixing of material from different plaques was avoided. The final products, subcultures in liquid medium, were kept at −20°C until they were examined further.
RNA extraction.
Viral RNA extraction was carried out by the method described by Casas et al. (6), and it was applied directly to the VIRADEN final products. The RNA extraction technique involved the use of a lysis buffer, which contained the chaotropic agent guanidinium thiocyanate acid (GuSCN) buffer and did not contain organic solvents. Specifically, 100 μl of a VIRADEN final product was lysed in 400 μl of GuSCN buffer. Following addition of 50 μg glycogen, the mixture was incubated for 20 min at room temperature. Then 500 μl of isopropyl alcohol (−20°C) was added to precipitate the nucleic acids, and the preparation was incubated on ice for 20 min. The mixture was centrifuged at 14,000 × g for 10 min at 4°C, the supernatant was discarded, and the pellet was washed with 1 ml of 70% ethanol. The centrifugation at 14,000 × g for 10 min at 4°C was repeated, and the supernatant was discarded again. The pellet was left to dry at 70°C for 5 min and was finally dissolved in 50 μl of double-distilled H2O. The efficiency of the RNA extraction procedure used and the presence of inhibitors of RNA amplification in each specimen were evaluated with α-tubulin primers in a separate PCR assay (15). Moreover, poliovirus Sabin type 1 was used as a positive control (33).
RT-PCR.
Isolated RNA was converted to cDNA by reverse transcription; 40 U of RNase inhibitor (Promega Corporation, Madison, Wis.), 2 μl of d(N)9 primers (50 pmol/μl; Takara, Tokyo, Japan), and 5 μl of extracted RNA from each sample were initially mixed and heated at 70°C for 5 min. The tubes were immediately transferred to ice, and 5 μl of 5× RT buffer, 5 μl of a preparation containing each deoxynucleoside triphosphate at a concentration of 10 mM, 100 U of Moloney murine leukemia virus reverse transcriptase (Promega Corporation), and 6.5 μl of RNase-free water (Sigma Aldrich) were added to each tube; the total volume of the reaction mixture was 25 μl. This mixture was incubated at 37°C for 1 h, and heating at 95°C for 5 min was used to inactivate the Moloney murine leukemia virus reverse transcriptase. The cDNA produced was amplified by PCR by using a reaction mixture (50 μl/tube) containing 5 μl of 10× PCR buffer, 5 μl of a preparation containing each deoxynucleoside triphosphate at a concentration of 10 mM, 2 μl of 50 mM MgCl2 (resulting in a final MgCl2 concentration of 2 mM), 32.6 μl of RNase-free water, 2 U of Taq polymerase (BIOTAQ, Moscow, Russia), 5 μl of cDNA, and 2 μl of each of the two primer pairs (either UC53 and UG52 [20 pmol/tube] or 292 and 222 [50 pmol/tube]). Forty cycles of denaturation at 94°C for 10 s, annealing at 42°C for 30 s, and extension at 74°C for 10 s, followed by incubation for 15 min at 78°C in order to complete the extension of the primers, were performed with a Perkin-Elmer 9600 thermal cycler. Ten microliters of each amplified product was analyzed by agarose gel electrophoresis by using 2.5% agarose (ultrapure; electrophoresis grade; Gibco BRL) containing 1 μg of ethidium bromide per ml in Tris-boric acid-EDTA buffer. The amplicons were then visualized with a FOTO/PHORESIS I UV transilluminator (Fotodyne, Hartland, Wis.).
The antisense primer UC53 (5′-TTGTCACCATAACCAGCCA-3′; positions 583 to 601 in the genome of CAV9 reference strain Griggs) and the sense primer UG52 (5′-CAAGCACTTCTGTTTCCCCGG-3′; positions 167 to 187 in the genome of CAV9 reference strain Griggs) were selected to be homologous to the corresponding parts in the highly conserved 5′-UTR; they were synthesized by and purchased from Genosys Biotechnologies (Cambridge, United Kingdom). These primers yielded amplicons that were 435 bp long; they were adjusted to a concentration of 10 pmol/μl in sterile distilled water and were stored at −20°C (11, 32, 33, 34).
The sense primer 292 (5′-MIGCIGYIGARACNGG-3′; positions 2612 to 2627 in the genome of PV1 reference strain Mahoney) and the antisense primer 222 (5′-CICCIGGIGGIAYRWACAT-3′; positions 2969 to 2951 in the genome of PV1 reference strain Mahoney) were used to amplify a portion of the gene encoding VP1 capsid protein (23, 24). These primers yielded amplicons that were approximately 340 bp long; they were adjusted to a concentration of 50 pmol/μl in sterile distilled water and were stored at −20°C.
All procedures were carried out under conditions that minimized the risk of contamination from exogenous nucleic acid sources or carryover of amplification products during RT-PCR. There was physical separation of the pre- and post-PCR procedures in separate rooms, and sets of pipettes with plugged, aerosol-resistant tips were allocated for each step of the PCR (i.e., reaction mixture preparation, template addition, and amplified product electrophoretic analysis). Negative controls were used in each amplification assay and were always RT-PCR negative, which indicated the effectiveness of these preventative measures.
Restriction fragment length polymorphism analysis of UC53-UG52-produced RT-PCR amplicons of the VIRADEN isolates.
In a previous study it was shown that RFLP analysis of the 5′-UTR with the enzyme HpaII could be used to accurately classify both reference and wild-type strains into five different genetic subclusters with no intraserotypic variation in the HpaII-produced haplotypes (35). Therefore, an initial RFLP analysis of the UC53-UG52-produced RT-PCR amplicons with the restriction enzyme HpaII (New England Biolabs, Beverly, Mass.) was carried out as described previously. Briefly, 1 μl of the appropriate 10× buffer, 20 U of the restriction enzyme (2 μl), and 1 μl of distilled, RNase-free, sterile water (Sigma Aldrich) were added to 6 μl of each of the UC53-UG52-produced RT-PCR amplicons of the VIRADEN isolates to obtain a final volume of 10 μl. The samples were then incubated at 37°C for 2 h, and the products were subjected to electrophoresis in 3% agarose gels made from high-resolution agarose (Metaphor FMC Bioproducts, Rockland Maine) containing 1 μg of ethidium bromide per μl and visualized with a UV transilluminator. The results were analyzed by using the GelPro Analyzer software (Media Cybernetics, Silver Spring, Md.).
Sequence and phylogenetic analysis of the 292-222-produced RT-PCR amplicons of the VIRADEN isolates.
As there cannot be a direct correlation between the 5′-UTR and enterovirus serotype (35), the partial VP1 sequences of the environmental isolates were also obtained for serotypic identification of these isolates by using the model proposed by Oberste et al. (22, 24), since the VP1 gene contains important serotype-specific epitopes. The 292-222-produced amplification products of the VIRADEN isolates were extracted from the electrophoresis gels with a NucleoSpin extract isolation kit (Macherey-Nagel, Düren, Germany) and sequenced by Macrogen Inc. (Seoul, Korea). The serotypic identity of each isolate was deduced by comparison of the partial VP1 sequences with the sequences of the corresponding genomic regions of all human enteroviruses which are available in the GenBank database by using the program BLAST, version 2.2.8 (http://www.ncbi.nlm.nih.gov/BLAST) (1, 2).
A phylogenetic analysis was carried out by pairwise comparison of the partial VP1 sequences of the enterovirus isolates and the corresponding partial VP1 sequences of reference and wild-type human enterovirus strains of the same serotype, which are available from GenBank, by using ClustalW (www.ebi.ac.uk./ClustalW) (39). Construction of the phylogenetic tree was carried out with ClustalX (version 1.83) by using the alignment file obtained by analysis with ClustalW. By using this program the distances between all pairs of sequences were calculated, and this was followed by application of the neighbor-joining method to the distance matrix. Confidence values for the groups in the tree (bootstrap values on a scale from 1 to 1,000) were also calculated by using ClustalX. A dendrogram showing the phylogenetic relationships among the enterovirus isolates used in the present study and prototype strains was plotted in the PHYLIP format output by using the TreeView software (version 3.0), which was obtained from the website of the University of Glasgow (http://taxonomy.zoology.gla.ac.uk/rod/rod/html).
Nucleotide sequence accession numbers.
The accession numbers of all the virus sequences obtained in this and previous studies that were used for the phylogenetic comparison are shown in Tables 1 and 2.
TABLE 1.
Collection sites, dates, VIRADEN isolates, and GenBank accession numbers
| Location | Datea | VIRADEN isolate | VP1 sequence accession no. |
|---|---|---|---|
| Nicosia | April | A1 | AY563382 |
| A2 | AY563351 | ||
| A3 | AY563352 | ||
| A4 | AY563353 | ||
| A5 | AY563379 | ||
| A6 | AY563354 | ||
| Nicosia | May | B1 | AY563355 |
| B4 | AY563356 | ||
| B5 | AY563357 | ||
| B6 | AY563358 | ||
| B8 | AY563359 | ||
| B9 | AY563360 | ||
| B10 | AY563361 | ||
| Nicosia | September | LK1 | AY563362 |
| LK2 | AY563363 | ||
| LK5 | AY563364 | ||
| LK8 | AY563365 | ||
| LK11 | AY563366 | ||
| LK14 | AY563367 | ||
| Limassol | September | L1 | AY563380 |
| L2 | AY563381 | ||
| L3 | AY563326 | ||
| L4 | AY563329 | ||
| L5 | AY563330 | ||
| L6 | AY563331 | ||
| L7 | AY563368 | ||
| L8 | AY563369 | ||
| L9 | AY563332 | ||
| L10 | AY563333 | ||
| Nicosia | December | LE1 | AY563370 |
| LE2 | AY563371 | ||
| LE3 | AY563372 | ||
| LE4 | AY563373 | ||
| Limassol | December | LM1 | AY563374 |
| LM2 | AY563375 | ||
| LM3 | AY563376 | ||
| LM4 | AY563377 | ||
| LM5 | AY563378 | ||
| LM6 | AY563328 | ||
| LM7 | AY563327 |
Months in 2003.
TABLE 2.
Accession numbers of reference and wild-type human enterovirus strains used for the phylogenetic analysis
| Serotype | Strain | Accession no. |
|---|---|---|
| PV2 | Lansing | M12197 |
| Sabin | X00595 | |
| W-2 | D00625 | |
| MEF-1 | AY238473 | |
| EGY88-074 | AF448782 | |
| EGY93-034 | AF448783 | |
| P2S/Mog65-3 | AY278549 | |
| P2S/Mog65-1 | AY278550 | |
| P2S/Mog66-4 | AY278551 | |
| P2S/Mog65-2 | AY278552 | |
| 102050 | AJ544513 | |
| BeniSuef/98 | AF551838 | |
| Beheira/97 | AF551837 | |
| 4623_I/ISR98 | AJ288066 | |
| Sahag/00 | AF551841 | |
| Alexandria/98 | AF551840 | |
| Giza/98 | AF551839 | |
| Aswan/95 | AF551836 | |
| 4808_1/ISR98 | AJ288075 | |
| 4774_1/ISR98 | AJ288074 | |
| 4625_1/ISR98 | AJ288067 | |
| 4588_1/ISR98 | AJ288064 | |
| CAV9 | GRIGGS | D00627 |
| 03-171FCR2 | AB167980 | |
| 03-144NPC3 | AB167979 | |
| 03-132NPR2 | AB167978 | |
| BE01-2839 | AY342746 | |
| BE99-1823 | AY342619 | |
| BE01-5294 | AY342745 | |
| BE01-4075 | AY342744 | |
| BE99-5750 | AY342623 | |
| BE99-8726 | AY342628 | |
| CBV1 | Japan | M16560 |
| P-2264/CB1/Kanagawa/2003 | AB162736 | |
| P-2199/CB1/Kanagawa/2003 | AB162733 | |
| P-2240/CB1/Kanagawa/2003 | AB162735 | |
| 04-29NGC1 | AB167989 | |
| P-2200/CB1/Kanagawa/2003 | AB162734 | |
| P-2346/CB1/Kanagawa/2003 | AB162737 | |
| 10159 | AY373092 | |
| 10165 | AY373098 | |
| CBV2 | Nancy | AF081485 |
| 10180 | AY373113 | |
| 10179 | AY373112 | |
| 10178 | AY373111 | |
| 10177 | AY373110 | |
| 10176 | AY373109 | |
| 10175 | AY373108 | |
| 10174 | AY373107 | |
| 10173 | AY373106 | |
| 10172 | AY373105 | |
| 10171 | AY373104 | |
| 10170 | AY373103 | |
| 10169 | AY373102 | |
| 10168 | AY373101 | |
| CBV4 | JVB | X05690 |
| P234pak92 | AF160018 | |
| Pk1pak92 | AF160019 | |
| BE00-117 | AF521311 | |
| 48112fin98 | AF160025 | |
| 9128net93 | AF160021 | |
| 10199 | AY373132 | |
| 10198 | AY373131 | |
| 10197 | AY373130 | |
| 10196 | AY373129 | |
| 10195 | AY373128 | |
| 10194 | AY373127 | |
| 10193 | AY373126 | |
| 10192 | AY373125 | |
| 10191 | AY373124 | |
| 10190 | AY373123 |
RESULTS
A total of 40 VIRADEN isolates were concentrated from six sampling procedures carried out at two sites, Nicosia and Limassol (Table 1). From sampling A (Nicosia, April 2003), six plaques were picked and characterized by RT-PCR. Similarly, seven plaques from sampling B (Nicosia, May 2003), six plaques from sampling C (Nicosia, September 2003), 10 plaques from sampling D (Limassol, September 2003), four plaques from sampling E (Nicosia, December 2003), and seven plaques from sampling F (Limassol, December 2003) were examined.
All the samples produced cytopathogenic effects in the BGM cell line, which indicated the presence of propagating, potentially infectious viruses. The expected amplification products, which were approximately 435 nucleotides long with primers UC53 and UG52 and approximately 350 nucleotides long with primers 292 and 222, were successfully obtained (Fig. 1a). Finally, amplification of all the isolates with the α-tubulin-specific primers yielded positive results (data not shown), which proved the effectiveness of the GuSCN-based RNA extraction procedure for both the recovery of a sufficient quantity of RNA from the environmental samples and the elimination of PCR-inhibiting substances.
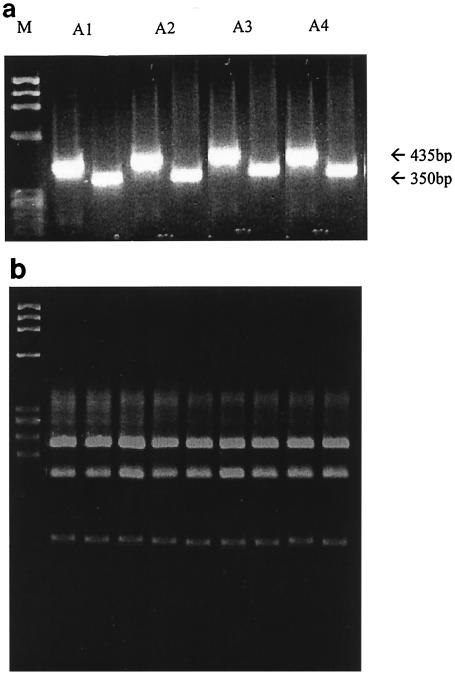
FIG. 1.
(a) Representative results for UG52-Uc53 (435-bp) amplicons and 292-222 (350-bp) amplicons for isolates A1, A2, A3, and A4. Lane M contained a molecular weight marker (φX174 HaeIII-generated restriction fragments; Gibco BRL). (b) Representative results of RFLP analysis of UC53-UG52-produced RT-PCR amplicons of the enterovirus isolates with the HpaII restriction endonuclease (for a description, see the text). All of the isolates except one (isolate A5 [data not shown]) belonged to the same genetic subcluster, and the restriction profile comprised four HpaII-generated fragments with lengths of 213, 149, 55, and 18 nucleotides. The lane on the left contained a molecular weight marker (φX174 HaeIII-generated restriction fragments; Gibco BRL).
The results of the RFLP analysis of the initial subgrouping of the enterovirus isolates with restriction endonuclease HpaII are shown in Fig. 1b. All of the isolates except one (isolate A5) belonged to the same genetic subcluster, and the restriction profile comprised four HpaII-generated fragments with lengths of 213, 149, 55, and 18 nucleotides. This genetic subcluster included 38 different serotypes of the Human enterovirus B species, which was previously described in detail (35), and therefore, an initial assumption was made about the possible serotypic group that the isolates belonged to. Isolate A5 was the only isolate which belonged to a different genetic subcluster, and the restriction profile included five HpaII-generated fragments that were 148, 121, 108, 40, and 18 nucleotides long. This genetic subcluster contained three enteroviruses of the polio-like cluster on the basis of the 5′-UTR classification, the PV2 Sabin strain, CAV11, and CAV22, which also provided an indication of the possible serotypic identity of the isolate.
The partial VP1 sequences were phylogenetically compared with enterovirus sequences available from the GenBank sequence database, and the results are shown in Table 3. The levels of partial VP1 sequence identity with previously studied enterovirus reference and wild-type strains ranged from 82 to 98%. The second-highest levels of identity were less than 70% in all cases. All but one of the isolates were nonpolio enteroviruses; in particular, 17 of 40 isolates (42.5%) were CBV4, 12 of 40 isolates (30%) were CAV9, 7 of 40 isolates (17.5%) were CBV2, and 3 of 40 isolates (7.5%) were CBV1. Also, all isolates of the same serotype had very similar VP1 sequences (98 to 100%), indicating that they may have been the same strain of the specific serotype circulating in the population. It was observed that the HpaII digests predicted the genetic subclusters of the clinical isolates of all isolates when the five corresponding HpaII-generated subclusters of most human enterovirus serotypes studied by Siafakas et al. (35) were used as references. Specifically, each the CAV9, CBV4, CBV2, and CBV1 isolates had an HpaII-generated restriction profile that was identical to the profile of a cluster of enteroviruses that included the corresponding strains of the same serotype. One poliovirus strain (2.5%) was isolated, and the VP1 sequence revealed that it was a Sabin type 2 isolate. The HpaII-produced restriction profile placed this isolate in a subcluster that contained PV2 and two other CAVs of the polio-like cluster of 5′-UTR, as mentioned above; most importantly, this subcluster contained only the PV2 Sabin strain and no other wild-type PV2 prototype strain, providing clues about the possible vaccine origin of the isolate.
TABLE 3.
Results of RFLP analysis, partial VP1 sequencing, and phylogenetic comparison of our 40 isolates with enterovirus sequences available in the GenBank sequence database.
| Isolatea | VP1 Serotype | % Nucleotide sequence identity |
|---|---|---|
| A1 | CAV9 | 90 |
| A2 | CAV9 | 91 |
| A3 | CAV9 | 90 |
| A4 | CAV9 | 91 |
| A5 | Sabin 2 | 98 |
| A6 | CAV9 | 92 |
| B1 | CAV9 | 92 |
| B4 | CAV9 | 91 |
| B5 | CAV9 | 90 |
| B6 | CAV9 | 91 |
| B8 | CAV9 | 92 |
| B9 | CAV9 | 90 |
| B10 | CAV9 | 91 |
| LK1 | CBV4 | 95 |
| LK2 | CBV4 | 94 |
| LK5 | CBV4 | 93 |
| LK8 | CBV4 | 93 |
| LK11 | CBV4 | 93 |
| LK14 | CBV4 | 93 |
| L1 | CBV2 | 87 |
| L2 | CBV2 | 87 |
| L3 | CBV1 | 83 |
| L4 | CBV2 | 87 |
| L5 | CBV2 | 87 |
| L6 | CBV2 | 86 |
| L7 | CBV4 | 93 |
| L8 | CBV4 | 94 |
| L9 | CBV2 | 87 |
| L10 | CBV2 | 87 |
| LE1 | CBV4 | 93 |
| LE2 | CBV4 | 95 |
| LE3 | CBV4 | 93 |
| LE4 | CBV4 | 93 |
| LM1 | CBV4 | 95 |
| LM2 | CBV4 | 94 |
| LM3 | CBV4 | 95 |
| LM4 | CBV4 | 96 |
| LM5 | CBV4 | 96 |
| LM6 | CBV1 | 82 |
| LM7 | CBV1 | 83 |
Most isolates belonged to HpaII genetic cluster I; the only exception was isolate A5, which belonged to cluster IV. In all cases there was agreement between HpaII classification and VP1 partial serotyping results.
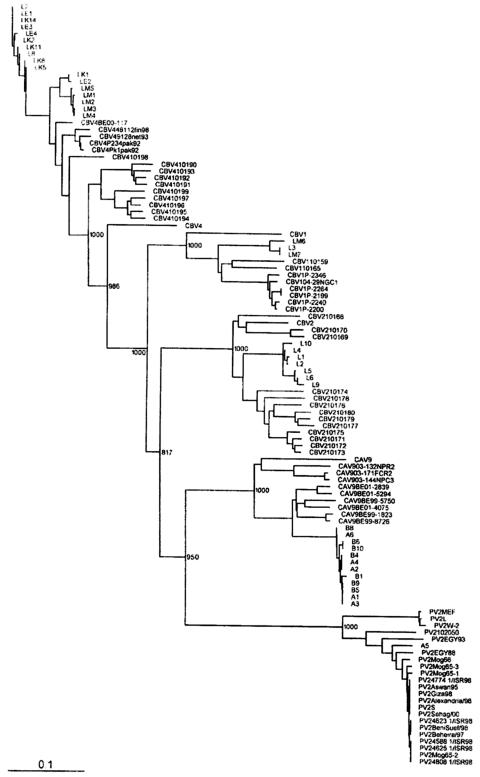
Finally, the phylogenetic comparison between these isolates and other enterovirus reference and wild-type strains showed the serotype-specific pattern of strain classification into single clusters (Fig. 2). It was also very interesting that the PV2, CBV4, and, to a lesser extent, CAV9 isolates showed the greatest levels of VP1 alignment with other enterovirus strains isolated elsewhere in Europe, in Asia, in northern Africa, and in the Middle East (Fig. 2), implying a possible epidemiological relationship of all these isolates. Specifically, the PV2 isolates were most closely related to PV2 strains of vaccine origin isolated in Israel in 2002 (unpublished data obtained from GenBank) and in Egypt from 1983 to 1993 (41), reinforcing the Sabin-like character of the isolates. CBV and CAV9 isolates were related to similar strains isolated in Belgium from 1999 to 2002 (38), and especially the CBV4 isolates also showed correlation with strains of the same serotype isolated during the last four decades in France, Finland, Romania, Denmark, North America, and Pakistan (21).
FIG. 2.
Dendrogram showing the relationships among enterovirus environmental strains isolated in the present study and reference and other clinical strains, as determined by using the partially sequenced VP1-encoding region.
DISCUSSION
Environmental surveillance has been successfully used to assess the extent or duration of epidemic enterovirus circulation in specific populations (25, 31). Furthermore, screening of wastewater has been shown to be a more sensitive tool for detection of wild-type polioviruses in communities than surveillance for acute flaccid paralysis (AFP) (12), since wild-type polioviruses have frequently been isolated from sewage in the absence of cases of AFP (5, 17, 37, 40). It may also prove to be useful for monitoring the effectiveness of vaccination against poliomyelitis and for detecting the introduction of wild-type strains into communities previously considered polio-free (7, 27). All these things indicate the importance of environmental surveillance in prevention of a possible enterovirus outbreak.
An effective surveillance scheme is a prerequisite for epidemiological investigation of disease outbreaks, providing important information about the viruses circulating in susceptible populations and, consequently, guiding a reliable system of early warning and prevention (26). In the present study we attempted to contribute to this goal by developing a simple and effective method for detection and identification of infectious enteroviruses in sewage by the VIRADEN method (cellulose nitrate membrane adsorption and direct cell culture from the membrane) and RT-PCR. In our procedure virus plaques are produced on a cell monolayer while the virus grows on the membrane filter. To our knowledge, this is the only method which produces plaques directly from the membrane filter (the virus adsorption site) without a need for elution. No special equipment or decontamination procedures are necessary, and all isolates from environmental specimens can be identified within a few days after sampling. Moreover, since cell lines other than BGM have the advantage of growing on cellulose nitrate membrane filters and since viruses other than enteroviruses can adsorb to such filters, VIRADEN has the potential of being used for detection and characterization of a wide range of viruses with different cell lines (26).
Previous studies reported detection of enteroviruses by RT-PCR from culture-negative environmental samples, showing that cytopathogenic viruses are only a minor component of the enteroviruses present in water environments (12, 28, 31). Nevertheless, a combination of cell culture amplification and detection by RT-PCR still is more sensitive for detection of enteric viruses than either method alone (28, 29, 31). Cell culture amplification also has the advantage of increasing the frequently low virus titer in environmental samples and eliminating the action of substances that inhibit RT-PCR. Moreover, it is our view that although a culture-negative, RT-PCR-positive result indicates detection of nucleic acid from an intact capsid particle, what is really important from a clinical and epidemiological point of view is detection of cytopathogenic and consequently infectious and potentially hazardous viruses.
Partial sequencing of the VP1-encoding gene produced unambiguous results regarding serotyping of the isolates, showing the effectiveness of the specific serotyping system and also showing that the cultures that were obtained were pure and that each contained only one virus clone. Moreover, the distinction between nonpolio and polio-like enteroviruses was accurate, something which is extremely important for the eradication of wild-type polioviruses from the environment and for surveillance for vaccine-associated cases of AFP (4, 8, 9, 10, 11, 13). Thoelen et al. (38) reported a subtype-specific clustering pattern for enteroviruses on the basis of the 5′-UTR and proposed that the suggestive value of the 5′-UTR for enterovirus serotype determination and further investigation should not be underestimated.
An interesting finding of the present study was that the PV2, CBV4, and, to a lesser extent, CAV9 isolates showed the highest levels of VP1 alignment with other enterovirus strains isolated elsewhere in Europe, in North America, in Asia Manor (21, 38), in northern Africa, and in the Middle East (41) (Fig. 2), implying the possible epidemiological relationships of all these isolates. Cyprus is located at a crossing point between Europe, the Middle East, and northern Africa, and the fact that the molecular analysis of the environmental isolates in the present study provided some evidence about the possible introduction of these enteroviruses from individuals traveling between these geographic regions was very interesting. It would be very interesting to carry out a more detailed and combined genotypic analysis of all these strains in an attempt to follow enterovirus circulation and evolution in the populations. The possible epidemiological association also gave us confidence in the value of the environmental surveillance for enteroviruses that was carried out in the present and previous studies.
In conclusion, the combination of virus concentration by cellulose nitrate membrane adsorption and cell culture (VIRADEN), followed by detection with RT-PCR and serotyping by partial VP1 sequencing, seems to be very promising for environmental surveillance of enterovirus circulation and epidemiology, with all the significant effects that this could have on public health. Nevertheless, the predictive value of the simple RFLP analysis of the 5′-UTR in this and previous studies can also be considered an asset for the initial subclassification of enterovirus isolates. It would also be interesting to determine the epidemiological relationship of the environmental isolates identified in the present study with viruses isolated elsewhere in Europe, in North America, in Asia, and in northern Africa in previous years. Finally, an effective means of environmental surveillance should facilitate the monitoring of potential sources for new recombinant enteroviruses and identification of newly discovered or previously unidentified strains.
Acknowledgments
This study was cofunded by the European Union (75%) and the Greek government (25%) under the framework of the Education and Initial Vocational Training Program Archimedes. It was also partially funded by a United Nations program through the United Nations Development Office of Project Services (WSE-PS-4049) in Cyprus and by the Délégation Génerale au Réseau International des Instituts Pasteur et Instituts assosiés (AC 02 Vaccin Polio Oral) in Greece.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 251:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. 2003. Guidelines for environmental surveillance of poliovirus circulation. World Health Organization Department of Vaccines and Biologicals, Geneva, Switzerland.
- 4.Blomqvist, S., A. L. Bruu, M. Stenvik, and T. Hovi. 2003. Characterization of a recombinant type 3/type 2 poliovirus isolated from a healthy vaccinee and containing a chimeric capsid protein VP1. J. Gen. Virol. 84:573-580. [DOI] [PubMed] [Google Scholar]
- 5.Bottiger, M., and E. Herrstrom. 1992. Isolation of polioviruses from sewage and their characteristics: experience over two decades in Sweden. Scand J. Infect. Dis. 24:151-155. [DOI] [PubMed] [Google Scholar]
- 6.Casas, I., L. Powell, P. E. Klapper, and G. M. Cleator. 1995. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assay. J. Virol. Methods 53:25-36. [DOI] [PubMed] [Google Scholar]
- 7.Cochi, S. L., H. F. Hull, and N. A. Ward. 1995. To conquer poliomyelitis forever. Lancet 345:1589-1590. [DOI] [PubMed] [Google Scholar]
- 8.Cuervo, N. S., S. Guillot, N. Romanenkova, M. Combiescu, A. Aubert-Combiescu, M. Seghier, V. Caro, R. Crainic, and F. Delpeyroux. 2001. Genomic features of intertypic recombinant Sabin poliovirus strains excreted by primary vaccinees. J. Virol. 75:5740-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furione, M., S. Guillot, D. Otelea, J. Balanant, A. Candrea, and R. Crainic. 1993. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology 196:199-208. [DOI] [PubMed] [Google Scholar]
- 10.Georgescu, M. M., J. Balanant, A. Macadam, D. Otelea, M. Combiescu, A. A. Combiescu, R. Crainic, and F. Delpeyroux. 1997. Evolution of the Sabin type 1 poliovirus in humans: characterization of strains isolated from patients with vaccine-associated paralytic poliomyelitis. J. Virol. 71:7758-7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georgopoulou, A., and P. Markoulatos. 2001. Sabin type 2 polioviruses with intertypic vaccine/vaccine recombinant genomes. Eur. J. Clin. Microbiol. Infect. Dis. 20:792-799. [DOI] [PubMed] [Google Scholar]
- 12.Grabow, W. O., K. L. Botma, J. C. de Villiers, C. G. Clay, and B. Erasmus. 1999. Assessment of cell culture and polymerase chain reaction procedures for the detection of polioviruses in wastewater. Bull. W. H. O. 77:973-980. [PMC free article] [PubMed] [Google Scholar]
- 13.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King, A. M. Q., F. Brown, P. Christian, T. Hovi, T. Hyypid, N. J. Knowles, S. M. Lemon, P. D. Minor, A. C. Palmenberg, T. Skern, and G. Stanway. 2000. Picornaviridae, p. 657-673. In M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, C. H. Calisher, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, New York, N.Y.
- 15.Markoulatos, P., A. Georgopoulou, N. Siafakas, E. Plakokefalos, G. Tzanakaki, and J. Kourea-Kremastinou. 2001. Laboratory diagnosis of common herpesvirus infections of the central nervous system by a multiplex PCR assay. J. Clin. Microbiol. 39:4426-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melnick, J. L. 1996. Enteroviruses: polioviruses, coxsackieviruses, echoviruses and newer enteroviruses, p. 655-712. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 17.Metcalf, T. G., J. L. Melnick, and M. K. Estes. 1995. Environmental virology: from detection of virus in sewage and water by isolation to identification by molecular biology—a trip of over 50 years. Annu. Rev. Microbiol. 49:461-487. [DOI] [PubMed] [Google Scholar]
- 18.Mocé-Llivina, L., J. Jofre, X. Méndez, D. Akkelidou, F. Lucena, and G. T. Papageorgiou. 2002. Counting cytopathogenic virus adsorbed to cellulose nitrate membrane filters as a simple method for counting viruses in raw sewage and sewage effluents. J. Virol. Methods 102:83-92. [DOI] [PubMed] [Google Scholar]
- 19.Moce-Llivina, L., J. Jofre, and M. Muniesa. 2003. Comparison of polyvinylidene fluoride and polyether sulphonate membranes in filtering viral suspensions. J. Virol. Methods 109:99-101. [DOI] [PubMed] [Google Scholar]
- 20.Muir, P., U. Kammerer, K. Korn, M. N. Mulders, T. Pöyry, B. Weissbrich, R. Kandolf, G. M. Cleator, and A. M. van Loon. 1998. Molecular typing of enteroviruses: current status and future requirements. The European Union Concerted Action on Virus Meningitis and Encephalitis. Clin. Microbiol. Rev. 11:202-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulders, M. N., M. Salminen, N. Kalkkinen, and T. Hovi. 2000. Molecular epidemiology of coxsackievirus B4 and disclosure of the correct VP1/2Apro cleavage site: evidence for high genomic diversity and long-term endemicity of distinct genotypes. J. Gen. Virol. 81:803-812. [DOI] [PubMed] [Google Scholar]
- 22.Oberste, M. S., K. Maher, D. R. Kilpatrick, M. R. Flemister, B. A. Brown, and M. A. Pallansch. 1999. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37:1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oberste, M. S., K. Maher, D. R. Kilpatrick, and M. A. Pallansch. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oberste, M. S., W. A. Nix, K. Maher, and M. A. Pallansch. 2003. Improved molecular identification of enteroviruses by RT-PCR and amplicon sequencing. J. Clin. Virol. 26:375-377. [DOI] [PubMed] [Google Scholar]
- 25.Pallin, R., A. P. Wyn-Jones, B. M. Place, and N. F. Lightfoot. 1997. The detection of enteroviruses in large volume concentrates of recreational waters by the polymerase chain reaction. J. Virol. Methods 67:57-67. [DOI] [PubMed] [Google Scholar]
- 26.Papageorgiou, G. T., L. Moce-Llivina, C. G. Christodoulou, F. Lucena, D. Akkelidou, E. Ioannou, and J. Jofre. 2000. A simple methodological approach for counting and identifying cytopathogenic viruses adsorbed to cellulose nitrate membrane filters. Appl. Environ. Microbiol. 66:194-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patriarca, P. A. 1994. Polio outbreaks: a tale of torment. Lancet 344:630-631. [DOI] [PubMed] [Google Scholar]
- 28.Puig, M., J. Jofre, F. Lucena, A. Allard, G. Wadell, and G. Girones. 1994. Detection of adenoviruses and enteroviruses in polluted waters by nested PCR amplification. Appl. Environ. Microbiol. 60:2963-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds, K. A., C. P. Gerba, and I. L. Pepper. 1996. Detection of infectious enteroviruses by an integrated cell culture-PCR procedure. Appl. Environ. Microbiol. 62:1424-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rusin, P., C. Enriquez, D. Johnson, and C. Gerba. 2000. Enteric viruses, p. 472-484. In R. Maier, I. Pepper, and C. Gerba (ed.), Environmental microbiology, 1st ed. Academic Press, London, United Kingdom.
- 31.Shieh, Y. S., R. S. Baric, and M. D. Sobsey. 1997. Detection of low levels of enteric viruses in metropolitan and airplane sewage. Appl. Environ. Microbiol. 63:4401-4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siafakas, N., A. Georgopoulou, P. Markoulatos, and N. Spyrou. 2000. Isolation of polioviruses and other enteroviruses in south Greece between 1994 and 1998. J. Clin. Lab. Anal. 14:157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siafakas, N., A. Georgopoulou, P. Markoulatos, N. Spyrou, and G. Stanway. 2001. Molecular detection and identification of an enterovirus during an outbreak of aseptic meningitis. J. Clin. Lab. Anal. 15:87-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siafakas, N., P. Markoulatos, G. Stanway, G. Tzanakaki, and J. Kourea-Kremastinou. 2001. A reliable RT-PCR/RFLP assay for the molecular classification of enterovirus reference and wild type strains to either of the two genetic clusters on the basis of the 5′-UTR. Mol. Cell. Probes 16:209-216. [DOI] [PubMed] [Google Scholar]
- 35.Siafakas, N., P. Markoulatos, C. Vlachos, G. Stanway, G. Tzanakaki, and J. Kourea-Kremastinou. 2003. Molecular sub-grouping of enterovirus reference and wild type strains into distinct genetic clusters using a simple RFLP assay. Mol. Cell. Probes. 17:113-123. [DOI] [PubMed] [Google Scholar]
- 36.Stanway, G. 1990. Structure, function and evolution of picornaviruses. J. Gen. Virol. 71:2483-2501. [DOI] [PubMed] [Google Scholar]
- 37.Tambini, G., J. K. Andrus, E. Marques, J. Boshell, M. Pallansch, C. A. deq Uadros, and O. Kew. 1993. Direct detection of wild poliovirus circulation by stool surveys of healthy children and analysis of community wastewater. J. Infect. Dis. 168:1510-1514. [DOI] [PubMed] [Google Scholar]
- 38.Thoelen, I., E. Moës, P. Lemey, S. Mostmans, E. Wolants, A. M. Lindberg, A. M. Vandamme, and M. Van Ranst. 2004. Analysis of the serotype and genotype correlation of VP1 and the 5′ noncoding region in an epidemiological survey of the human enterovirus B species. J. Clin. Microbiol. 42:963-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van der Avoort, H. G., J. H. Reimerink, A. Ras, M. N. Mulders, and A. M. Van Loon. 1995. Isolation of epidemic poliovirus from sewage during the 1992-3 type 3 outbreak in The Netherlands. Epidemiol. Infect. 114:481-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, C.-F., T. Naguib, S.-J. Yang, E. Nasr, J. Jorba, N. Ahmed, R. Campagnoli, H. van der Avoort, H. Shimizu, T. Yoneyama, T. Miyamura, M. Pallansch, and O. Kew. 2003. Circulation of endemic type 2 vaccine-derived poliovirus in Egypt from 1983 to 1993. J. Virol. 77:8366-8377. [DOI] [PMC free article] [PubMed] [Google Scholar]