Abstract
Lysobacter enzymogenes strain C3, a biological control agent for plant diseases, produces multiple extracellular hydrolytic enzymes and displays antimicrobial activity against various fungal and oomycetous species. However, little is known about the regulation of these enzymes or their roles in antimicrobial activity and biocontrol. A study was undertaken to identify mutants of strain C3 affected in extracellular enzyme production and to evaluate their biocontrol efficacy. A single mini-Tn5-lacZ1-cat transposon mutant of L. enzymogenes strain C3 that was globally affected in a variety of phenotypes was isolated. In this mutant, 5E4, the activities of several extracellular lytic enzymes, gliding motility, and in vitro antimicrobial activity were reduced. Characterization of 5E4 indicated that the transposon inserted in a clp gene homologue belonging to the Crp gene family of regulators. Immediately downstream was a second open reading frame similar to that encoding acetyltransferases belonging to the Gcn5-related N-acetyltransferase superfamily, which reverse transcription-PCR confirmed was cotranscribed with clp. Chromosomal deletion mutants with mutations in clp and between clp and the acetyltransferase gene verified the 5E4 mutant phenotype. The clp gene was chromosomally inserted in mutant 5E4, resulting in complemented strain P1. All mutant phenotypes were restored in P1, although the gliding motility was observed to be excessive compared with that of the wild-type strain. clp mutant strains were significantly affected in biological control of pythium damping-off of sugar beet and bipolaris leaf spot of tall fescue, which was partially or fully restored in the complemented strain P1. These results indicate that clp is a global regulatory gene that controls biocontrol traits expressed by L. enzymogenes C3.
The genus Lysobacter belongs to the family Xanthomonadaceae in the γ-proteobacteria and is characterized by a high G+C content, gliding motility, and a propensity to lyse other microbes, including other bacteria, fungi, and nematodes (5). Lysobacter enzymogenes is a species that is characterized by prolific production of extracellular lytic enzymes, including chitinases, β-1,3-glucanases, and proteases. Previously, Lysobacter spp. were placed with other bacteria in the myxobacter group. Evidence that extracellular enzymes produced by soil bacteria in this group contribute to lytic activity was reported as early as the 1960s (14, 21). Despite the obvious antagonistic relationships with other microbes, however, few studies have described Lysobacter spp. as potential biological control agents for plant diseases.
Three different bacterial strains characterized as biological control agents for plant diseases were recently classified as L. enzymogenes (11, 36). Strain 3.1T8 was reported to express antimicrobial activity against fungal and oomycetous plant pathogens, including various Pythium spp. (11). Strain N4-7 was described as a biocontrol agent for summer patch disease of turfgrass caused by Magnaporthe poae (22). Strain C3, formerly classified as Stenotrophomonas maltophilia, was described as an effective biological control agent for a number of fungal plant diseases, including brown patch of turfgrass caused by Rhizoctonia solani (13, 41), leaf spot of tall fescue caused by Bipolaris sorokiniana (42), and bean rust caused by Uromyces appendiculatus (40). It also displayed in vitro activity against the oomycete Pythium ultimum (Yuen, unpublished data). Consistent with the previously described characteristics of L. enzymogenes, the three strains express a number of traits thought to contribute to biocontrol activity, including a variety of lytic enzyme activities (11, 12, 36, 43). Indeed, studies with strain C3 have provided strong supporting evidence that activity resulting from multiple isoforms of chitinases has an important functional role in biocontrol (43, 44, 45). Despite the collective evidence, exact roles for various enzyme types in biocontrol activity have been difficult to establish; this has been complicated in part by the existence of different enzymes in multiple isoforms that precludes a mutagenesis approach. In addition, essentially nothing is known about the regulatory mechanisms controlling expression of enzymes and other biocontrol traits in L. enzymogenes.
Our primary goals for studying L. enzymogenes were to gain a better understanding of the roles of the various extracellular enzymes in biological control and to begin to understand the regulatory factors that control expression of these traits. In an effort to achieve these goals, we initiated a transposon mutagenesis project with strain C3 in an attempt to identify mutants that lack extracellular enzyme production. Here we report characterization of a clp gene homologue in L. enzymogenes strain C3 that belongs to the Crp gene family of global regulators. In plant-associated bacteria, Crp regulators are best known for their involvement in global control of pathogenesis-related traits in Xanthomonas campestris pv. campestris and Erwinia chrysanthemi (9, 31). To date, however, Crp family regulators in biocontrol bacteria have not been well described. In this study, we demonstrated that mutations in the clp gene in L. enzymogenes C3 affect lytic enzyme production, gliding motility, and in vitro antimicrobial activity. Mutation of the clp gene also affects biocontrol activity against pythium damping-off of sugar beet, as well as bipolaris leaf spot of tall fescue.
MATERIALS AND METHODS
Bacterial strains and media.
A rifampin-resistant variant of L. enzymogenes strain C3 (42) was used for all experiments. C3 and mutant strains were routinely grown at 30°C with shaking in medium 813 (28) containing succinic acid (0.5%, wt/vol) as a carbon source or in Luria-Bertani (LB) broth (Difco) supplemented with 50 μg of chloramphenicol per ml when appropriate. The strains also were cultured on 10% tryptic soy agar (TSA) (Sigma-Aldrich) to produce inocula for biological control experiments. Escherichia coli strains HB101 (Gibco-BRL), DH5α (Gibco-BRL), and S17-1(λpir) (34) were grown at 37°C in LB broth supplemented with appropriate antibiotics at the following concentrations unless otherwise noted: chloramphenicol, 25 μg/ml; ampicillin, 100 μg/ml; and tetracycline, 25 μg/ml.
Transposon mutagenesis and screening for loss of extracellular enzyme activities.
The transposon mini-Tn5-lacZ1-cat was constructed by replacing the kanamycin resistance (Kmr) gene from mini-Tn5-lacZ1 with the cat gene encoding chloramphenicol resistance from mini-Tn5-Cm (10). The cat gene was excised as a 3.6-kb HindIII fragment from mini-Tn5-Cm and cloned into the HindIII site of p18Not (18). The cat gene was reexcised as a NotI fragment and used to replace the 1.6-kb fragment encoding Kmr in mini-Tn5-lacZ1 to obtain mini-Tn5-lacZ1-cat.
Mutants of strain C3 were generated by conjugation of the pUT plasmid containing mini-Tn5-lacZ1-cat from E. coli S17-1(λpir) into strain C3 by using a procedure similar to the procedure described previously (10). Transposon mutants were selected and maintained on LB agar containing 100 μg of rifampin per ml and 50 μg of chloramphenicol per ml. Mutants were initially screened for the loss of lytic enzyme activity on agar media. Chitinase and protease activities were visualized as clearing zones surrounding colonies grown on medium 813 agar containing 1% (wt/vol) colloidal chitin and on medium 813 agar containing skim milk agar, respectively (22). Tween 80 hydrolysis was visualized as a zone of precipitation surrounding colonies grown on LB agar containing 1% Tween 80 (19). Hydrolysis of 1% carboxymethyl cellulose and 0.5% laminarin (soluble β-1,3-glucans) was detected by the appearance of zones of color differentiation surrounding colonies grown on medium 813 agar after staining with 1% Congo red as described previously (8, 15).
Quantification of extracellular enzyme production.
Parental and mutant strains were grown for 36 h in medium 813 supplemented with 1% (wt/vol) autoclaved yeast cell walls prepared as described previously (28). Cells were pelleted by centrifugation, and the culture supernatants were retained for enzyme assays. Chitinase activity was assayed by adding 100 μl of culture supernatant to 25 μl of 0.2 M sodium phosphate (pH 7.0), 100 μl of H2O, and 25 μl of 4 mM p-nitrophenyl-N,N′-diacetylchitobiose. The reaction mixtures were incubated for 1 h at 37°C, and the reactions were stopped by addition of 0.75 ml of 0.4 M Na2CO3. The release of p-nitrophenol was calculated by using the molar extinction coefficient of p-nitrophenol at A405 (18,000 M−1 cm−1). Protease activity was assayed by adding 10 μl of culture supernatant to 100 μl of 1% (wt/vol) azoalbumin in 0.1 M sodium acetate (pH 5.4). The reaction mixtures were incubated for 1 h at 30°C, and the reactions were stopped by addition of 100 μl of cold 10% trichloroacetic acid. The reaction mixtures were stored at 4°C for 1 h, and precipitated azoalbumin was removed by centrifugation. The absorbance at 366 nm of each reaction supernatant was measured relative to that of an enzyme-free control reaction mixture, and protease activity was expressed as the increase in A366 per minute. β-1,3-Glucanase activity was measured by determining the release of reducing sugars from hydrolysis of laminarin as described previously (28). All enzyme activities were assayed in triplicate, and the results were normalized to cell number.
Molecular characterizations and sequence analyses.
Restriction digestion, electrophoresis, ligation, and Southern hybridization were performed by using standard procedures (32). A genomic library of mutant strain 5E4 was constructed in cosmid vector pLAFR3 (35) and packaged into lambda phage with Gigapak Gold extracts (Stratagene) by a procedure described previously (23). After phage infection of E. coli HB101, cosmid clones containing the mini-Tn5-lacZ1-cat insertion were directly selected by plating cells on LB agar containing chloramphenicol and tetracycline. Three overlapping cosmid clones were recovered by this procedure, and restriction digest analysis combined with Southern hybridization analysis indicated that the mini-Tn5-lacZ1-cat element was inserted into a 4-kb SalI fragment. One cosmid clone, pFT311, was selected for further characterization. A 6-kb PstI fragment consisting of a portion of the mini-Tn5-lacZ1-cat insertion and approximately 3 kb of flanking DNA originating from C3 was subcloned from cosmid pLEC1311 into pUC119, resulting in pLEC4325. This fragment was used as a DNA probe specific for the mutated locus in strain 5E4 by random primer labeling with digoxigenin (Genius nonradioactive labeling and detection kit; Roche Diagnostics). The probe was used in colony hybridization experiments to identify corresponding clones in a cosmid genomic library of strain C3 that were constructed in pLAFR3 as described above for strain 5E4. Four overlapping clones were identified by colony hybridization. A 4-kb SalI fragment from cosmid clone pCAP130 that hybridized to the 5E4 probe was subcloned into pBluescript SK(+) (Stratagene) in both orientations, resulting in plasmids pCAP53 and pCAP55. The nucleotide sequences of both strands of the 4-kb SalI fragment were determined by fluorescence labeling by using a Big Dye version 2 labeling kit with an ABI 3100 DNA automated sequencer (Applied Biosystems Inc.). The sequence of a region consisting of approximately 50 bp and having an unusually high G+C content was not resolved despite repeated attempts with various methods to resolve secondary structure compression.
Sequences were assembled and analyzed by using the DNAStar sequence analysis software (Lasergene). Database searches with identified open reading frames (ORFs) were conducted by using BLAST (Basic Local Alignment Search Tool) (3) at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST).
Reverse transcription (RT)-PCR of clp and acetyltransferase transcript.
Two primer sets, shown in Table 1, were designed to evaluate transcriptional expression of the clp and acetyltransferase (AT) genes. The first primer set, primers CAF1 and CAR1, was designed to include 45 bases upstream of the start codon of clp and 136 bases of the coding region of the acetyltransferase gene and amplified an 878-bp fragment. This primer set spanned the region that the transposon inserted into 5E4. The second primer set, primers CAF2 and CAR2, was designed to amplify a 616-bp product downstream of the transposon insertion in 5E4, which was located 385 bases downstream from the start codon of the clp gene and 184 bases downstream from the start codon of the acetyltransferase gene.
TABLE 1.
Oligonucleotide primers used for RT-PCR and construction of various mutant strains
| Primer | Sequence (5′-3′)a | Use |
|---|---|---|
| CAF1 | CGTATTGCCCTAAGCTCGGTTGAA | RT-PCR |
| CAR1 | GCGATGCGCTCGATGGCTTT | RT-PCR |
| CAF2 | GCGCCCAAGCTGCTGTATGC | RT-PCR |
| CAR2 | CGATCTCGGCCAGCAAAAGGTAG | RT-PCR |
| clpF1 | GAGCTCGATCTCCGCCACATCCGTCA | Upstream flank for construction of mutant strains DC211 and DCA2422; SstI |
| clpR1 | GAATTCGAGCCGGAGACGACATAATAGAGG | Upstream flank for construction of mutant strains DC211 and DCA2422; EcoRI |
| clpF2 | GAATTCCGGCCGCGTGCTCAAGAAG | Downstream flank for construction of mutant strain DC211; EcoRI |
| clpR2 | CTCGAGGCCCAGCGTCTTGACGAACTTG | Downstream flank for construction of mutant strain DC211; SstI |
| clpF3 | GAATTCGCCATACCCGCTACTCGCTGAC | Upstream flank for construction of mutant strain DC2422; EcoRI |
| clpR3 | CTCGAGCGCGGCGATCGGACATAAG | Downstream flank for construction of mutant strain DC2422; SstI |
| PIF1 | GACGGGCGAGGGCCGCGA | Amplification of clp for construction of P1, complementation strain of mutant 5E4 |
| PIR1 | GGCGCGTCCCATCGGGGC | Amplification of clp for construction of P1, complementation strain of mutant 5E4 |
The underlined sequences are added restriction enzyme sites for the enzymes indicated in the column on the right.
Total RNA was isolated by using TRI REAGENT (Sigma) according to the manufacturer's instructions and was subsequently treated with RQ1 RNase-free DNase (Promega). Fifty nanograms of RNA was used as a template for RT-PCRs, which were performed by using a SuperScript one-step RT-PCR kit (Invitrogen). The initial cDNA synthesis was conducted at 48°C for 50 min and was followed by a predenaturation step of 96°C for 3 min, a 95°C melting step for 40 s, a 57°C annealing step for 40 s, and extension at 72°C for 2.75 min, which was repeated for 35 cycles before final extension for 15 min. Control experiments that were used to test for DNA in the RNA isolation procedure were performed simultaneously by substituting Taq polymerase for the SuperScript reverse transcriptase-Platinum Taq mixture. A positive control for all RT-PCRs was included for all RNA samples by using primers designed for an sctV gene homologue known to be expressed independent of clp (30).
Construction of deletion mutants and P1, a clp-complemented strain of mutant 5E4.
The gene replacement system with vector pJQ200sk (29) was used for construction of all gene replacement mutants. The primers used for all constructs are listed in Table 1. Briefly, ca. 500-bp flanking regions spanning the regions of targeted deletions were amplified by PCR by using primers that incorporated unique restriction sites at the ends. Amplified products were cloned into pGEMTeasy (Promega), recovered by digestion with the appropriate restriction enzymes, and cloned into pJQ200sk. The resulting constructs were mated into strain C3 by using E. coli S17-1. Colonies that grew on LB agar supplemented with rifampin, kanamycin, and gentamicin (100 μg/ml) represented single-event recombinant strains that contained vector constructs incorporated into the chromosome. Individual colonies were selected and grown overnight at 30°C with shaking in LB broth and then diluted 1:1,000 into fresh LB broth supplemented with 5% sucrose. After 6 h of incubation, cultures were plated onto LB agar supplemented with 5% sucrose and incubated at 30°C. Single colonies were selected and restreaked to obtain single colonies several times before replication onto LB agar containing gentamicin to confirm gentamicin sensitivity and loss of the pJQ200sk vector. Double recombination events in putative mutants were verified by Southern blot hybridization. For construction of a deletion mutation in the clp gene, the clpF1-clpR1 and clpF2-clpR2 primer pairs were designed to delete a 445-bp region beginning 182 bases from the predicted ATG start and 60 bases from the stop codon. For construction of a deletion mutation that included a portion of the clp gene and the AT gene, the clpF1-clpR1 primer pair was used for one flank and a second primer pair, primers clpF3 and clpR3, was designed to delete a 725-bp region between base 182 of the clp gene and base 211 of the AT gene (Table 1; see Fig. 3A).
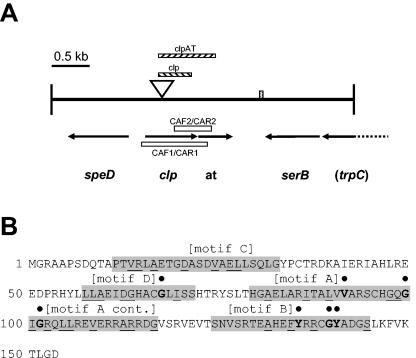
FIG. 3.
(A) Map of the 4-kb SalI fragment containing mini-Tn5-lacZ1-cat in mutant 5E4. The speckled box indicates a region where the sequence is unresolved. The arrows indicate the directions of ORFs; the corresponding gene designations are indicated below the arrows (at, acetyltransferase gene). The dashed arrow indicates the partial ORF for trpC. The open triangle indicates the position of the transposon insertion in 5E4. The striped boxes above the open triangle indicate deleted regions in the clp and clp-AT loci in DC211 and DCA2422, respectively. The open boxes indicate the positions of RT-PCR regions when the CAF1-CAR1 and CAF2-CAR2 primer sets were used. (B) Deduced amino acid sequence of the acetyltransferase, with the signature motifs (motifs A to D) of GNAT superfamily members highlighted. The residues in boldface type with solid circles above them are highly conserved, and the underlined residues are moderately conserved.
Due to the lack of a well-developed plasmid vector system for transcomplementation in L. enzymogenes strain C3, a clp-complementing strain was constructed by chromosomal insertion of the gene into the sctV gene. The sctV gene is part of a type III secretion system in L. enzymogenes strain C3, which to our knowledge is not associated with the clp gene or traits regulated by the gene (30). The complementing strain P1 was constructed by using a procedure similar to the procedure used for mutant construction. The clp gene was amplified by PCR by using primers PF1, beginning 200 bases upstream of the start codon of clp, and PR1, beginning 18 bases downstream of the stop codon of clp (Table 1), and was cloned into pGEMTeasy. The clp gene was then excised with NotI and cloned into a NotI site located between ca. 500-bp regions flanking the sctV gene cloned in pJQ200sk. The resulting construct was transformed into E. coli S17-1 and chromosomally introduced into strain C3 as described above.
Gliding motility, glass adherence, and antimicrobial activity assays.
Gliding motility was assayed by streaking bacteria onto 10% TSA and incubating the cultures at 30°C as previously described (36). Bacterial adherence to glass was observed by growing cultures of L. enzymogenes strains in glass test tubes containing 3 ml of LB at 30°C with shaking for 24 to 72 h.
The in vitro antimicrobial activities of L. enzymogenes strains were determined by coculturing bacterial strains with the fungus B. sorokiniana isolate Bs1 (42) and the oomycete P. ultimum isolate P201 (39). The bacterial strains were spot inoculated on the periphery of 10% TSA plates, and we included a mock-inoculated plate as a control. The center of each plate was inoculated with a mycelial plug containing either R. solani or P. ultimum (three plates per pathogen). After the plates were incubated for 2 to 5 days at 25°C, the radii of growth inhibition zones around bacterial colonies were measured.
Biological control assays.
C3 and mutant strains were compared for biocontrol efficacy in two pathosystems. The biocontrol assay for leaf spot of tall fescue, caused by B. sorokiniana, was conducted as described by Zhang and Yuen (42). Bacterial suspensions (108 CFU/ml) were sprayed onto foliage in pots of tall fescue turf 1 day prior to inoculation with conidia of B. sorokiniana. Pots sprayed with sterile distilled water served as controls. Inoculated pots (four pots per treatment) were placed in a moisture chamber for 24 h and then moved to a growth chamber and incubated for an additional 7 days, when the disease severity was visually assessed on the basis of the percentage of the leaf area exhibiting lesions. The experiment was conducted two times. The population levels of strain C3 and 5E4 were monitored during these experiments by collecting leaves from treated turf at various times after bacterial application and plating dilutions of leaf washes on 10% TSA amended with rifampin. The biocontrol assay for pythium damping-off of sugar beet, caused by P. ultimum, was similar to the assay described by Osburn et al. (27). The strains were coated onto sugar beet seeds (5 × 108 CFU) as cell suspensions in 1% methylcellulose. Control seeds were coated with 1% methylcellulose alone. Twenty coated seeds were planted into each pot of soil infested with P. ultimum at a concentration of approximately 100 propagules/g (16) and premoistened to field capacity. There were five replicate pots per treatment. After the pots were maintained in a growth chamber at 20°C for 5 days, control of P. ultimum was quantified by measuring seedling emergence, and the results were expressed as percentages. The experiment was conducted twice.
In both sets of experiments, the results of each experiment were analyzed separately by analysis of variance. Duncan's multiple range test was used for separation of means (α = 0.05).
Nucleotide sequence accession numbers.
The nucleotide sequence of the 2.8-kb DNA fragment containing the speD, clp, and acetyltransferase gene homologues from L. enzymogenes has been deposited in the GenBank database under accession no. AY316743. The nucleotide sequence of the 1.2-kb fragment containing the serB gene homologue and a portion of the trpC gene homologue has been deposited under accession no. AY316744.
RESULTS
Mutant strain 5E4 is affected in several phenotypes.
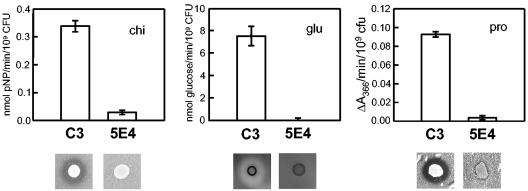
More than 2,000 Cmr isolates recovered from mini-Tn5-lacZ1-cat mutagenesis of strain C3 were screened for loss of chitinase, β-1,3-glucanase (laminarinase), protease, carboxymethyl cellulase, and Tween 80 hydrolysis activities by agar plate assays. A single mutant, designated strain 5E4, that was significantly reduced or devoid of all activities in agar plate assays was found (Table 2). Reduction of enzyme functions in 5E4 was confirmed by quantification of chitinase, β-1,3-glucanase, and protease activities relative to the activities of wild-type strain C3 (Fig. 1). Low levels of all three enzyme activities were detected with mutant 5E4 compared with the no-enzyme control; however, expression of all three enzyme activities was reduced in the mutant 10- to 20-fold compared to strain C3.
TABLE 2.
Phenotypes of L. enzymogenes wild-type strain C3 and mutant strains
| Phenotype | Strain
|
||||
|---|---|---|---|---|---|
| C3 | 5E4 | DC211 | DCA2422 | P1 | |
| Enzyme activitiesa | |||||
| Chitinase | + | − | − | − | + |
| β-1,3-Glucanase | + | − | − | − | + |
| Protease | + | +/− | +/− | +/− | + |
| Carboxymethyl cellulase | + | +/− | +/− | +/− | + |
| Tween 80 hydrolysis | + | − | − | − | + |
| Gliding motility | + | − | − | − | ++ |
| Glass adherence | − | + | + | + | − |
| Antimicrobial activitiesb | |||||
| Bipolaris sorokiniana | 9 ± 1 | 0 | 0 | 0 | 11 ± 0 |
| Pythium ultimum | 8 ± 0 | 0 | 0 | 0 | 10 ± 1 |
In vitro activities observed on agar medium after 1 to 3 days of incubation as described in Materials and Methods. +, zone of activity >2 mm from the colony edge; +/−, zone of activity >0 and <2 mm from the colony edge; −, no detectable zone of activity.
Activity expressed as the radius (in millimeters; mean ± standard deviation) of the hyphal growth inhibition zone surrounding colonies of L. enzymogenes on 10% TSA.
FIG. 1.
Specific chitinase (chi), β-1,3-glucanase (glu), and protease (pro) activities in L. enzymogenes culture filtrates from strain C3 or 5E4 cultures grown in minimal salts medium 813 supplemented with yeast cell walls. The activity values were standardized to 109 CFU, and the data are the means and standard deviations for three replicates. Below the graphs enzyme activity is shown as zones around colonies grown on medium 813 agar supplemented with 2% colloidal chitin (chi), 0.5% laminarin (glu), or 1% skim milk (pro).
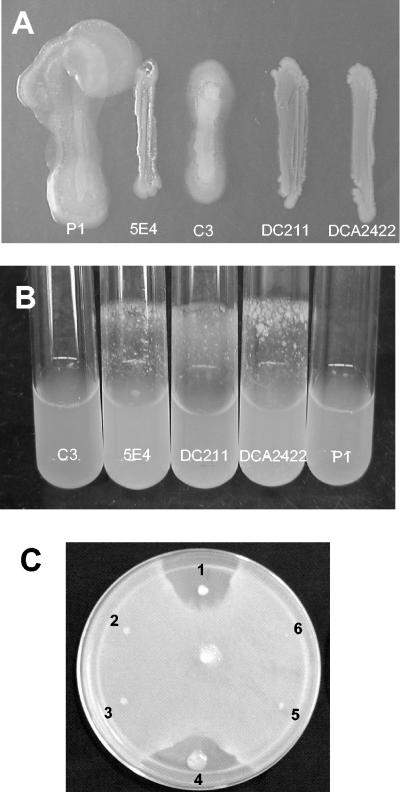
In addition to enzyme activities, a number of phenotypes, including colony morphology on agar media, were altered in mutant 5E4 compared with strain C3. Depending on the medium used for growth, colonies of 5E4 often had a rougher, drier appearance and a deeper yellow pigmentation than C3 (data not shown). Mutant strain 5E4 was also devoid of gliding motility on 10% TSA and lacked the spreading colony phenotype displayed by strain C3 (Fig. 2A and Table 2). In contrast to C3 grown in broth cultures, 5E4 typically formed aggregates and adhered to the glass surface of culture shake tubes and flasks (Fig. 2B and Table 2).
FIG. 2.
Phenotypes of L. enzymogenes wild-type and mutant strains. (A) Gliding motility assay conducted with 10% TSA. (B) Growth in LB broth, showing the adherence of mutant strains to glass. (C) In vitro antimicrobial activity assay performed with wild-type and mutant strains challenged with P. ultimum grown on 10% TSA. Spot 1, C3; spot 2, 5E4; spot 3, DC211; spot 4, P1; spot 5, DCA2422; spot 6, mock inoculation control.
Mutant 5E4 contains a transposon insertion in a clp gene homologue belonging to the Crp gene family of regulators.
The transposon insertion in 5E4 mapped in a 4-kb SalI fragment, and a sequence analysis of this fragment resulted in identification of four ORFs and a portion of a fifth ORF (Fig. 3A). The transposon insertion in 5E4 mapped between bases 222 and 223 from the start codon of the second ORF, which consisted of 690 bases and was located 1,239 bp from a SalI site. A GenBank database search and pairwise comparisons revealed that the ORF exhibited more than 75% amino acid identity to the catabolite activator protein-like protein (clp) gene from Xanthomonas campestris pv. campestris (9) and homologues originating from related members of the Xanthomonadaceae. Lower levels of similarity were detected with other members of the cyclic AMP receptor protein (crp) gene family (40 to 50% identity) from E. coli and other distantly related taxa.
Seven bases immediately downstream of the clp gene was a second ORF consisting of 462 bases. An RPS-BLAST search of the conserved domain database indicated that the predicted protein product was similar to acetyltransferases belonging to the Gcn5-related N-acetyltransferase (GNAT) superfamily (pfam00583; E value, 5e-11) (26). Similarity to these acetyltransferases was further supported by the presence of signature motifs associated with members of the GNAT superfamily (Fig. 3B). Surprisingly, in contrast to the clp gene homologue, BLAST searches of the acetyltransferase did not reveal similarities to sequences encoded by members of the Xanthomonadaceae at either the nucleotide or amino acid level. The clp and acetyltransferase ORFs were flanked on both sides by ORFs oriented in the opposite direction (Fig. 3A). The first ORF, located 219 bp upstream of clp and consisting of 795 bases, was similar (>88% amino acid identity) to the speD gene encoding S-adenosylmethionine decarboxylase from Xanthomonas and Xylella spp. Nothing resembling a true ORF was detected immediately downstream of the putative acetyltransferase gene in a region more than 400 bp long. However, several inverted repetitive sequences indicative of transcriptional termination signals were observed throughout the region.
A short nucleotide sequence region, spanning approximately 50 bp, could not be resolved due to GC compression between the region containing the 400 bp downstream of the putative acetyltransferase and a fourth ORF. This ORF exhibited more than 60% identity at the amino acid level with putative serB homologues encoding phosphoserine phosphatase from members of the Xanthomonadaceae. Part of a fifth ORF, located 35 bp upstream of the serB ORF, was similar (>60% identity) to trpC homologues encoding indole-3-glycerol phosphate synthase in members of the Xanthomonadaceae, suggesting that the two ORFs may be part of an operon involved in amino acid metabolism.
clp and acetyltransferase ORFs are encoded on a single RNA transcript.
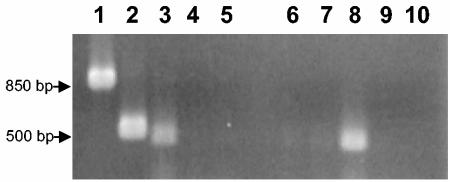
The close proximity of the clp and acetyltransferase ORFs to each other suggested that both genes were expressed on a single RNA transcript. To verify this, RT-PCR analyses were conducted by using two different primers sets. The first primer set (primers CAF1 and CAR1) was designed to amplify an 878-bp fragment and to span the entire coding region of clp and a portion of the 5′ region of the acetyltransferase gene. The second primer set (primers CAF2 and CAR2) was designed to amplify a 616-bp fragment located downstream of the transposon insertion in order to assess the polar effect of the mutation in the clp gene in 5E4. This primer set also spanned portions of both the clp and acetyltransferase ORFs (Fig. 3A). Amplified RT-PCR products of the expected sizes were obtained with both primer sets when RNA extracted from C3 was used, indicating that there was a single RNA transcript that spanned the region containing both the clp and acetyltransferase ORFs. In contrast, no RT-PCR products were observed when the same primer sets were used with RNA extracted from 5E4 (Fig. 4).
FIG. 4.
RT-PCR products from L. enzymogenes wild-type strain C3 (lanes 1 to 5) and mutant strain 5E4 (lanes 6 to 10) when constructed primers that span the clp and acetyltransferase ORFs were used. The CAF1-CAR1 primer set was used for lanes 1, 4, 6, and 9, and the CAF2-CAR2 primer set was used for lanes 2, 7, 5, and 10. Lanes 4, 5, 9, and 10 contained DNA-negative controls for the primer sets. Lanes 3 and 8 contained RT-PCR-positive controls in which primers for a gene (sctV) known to be expressed independent of the clp and acetyltransferase genes in L. enzymogenes were used.
Deletions within the clp gene and clp-AT gene loci and gene complementation verified the 5E4 mutant phenotype.
To verify that the phenotypes observed with mutant 5E4 were due to disruption of the clp-AT locus by insertion of mini-Tn5-lacZ1-cat, deletion mutations in the clp gene and a region spanning the clp and AT genes were chromosomally inserted into strain C3. Strains DC211 and DCA2242 contained deletion mutations in the clp gene and clp-AT genes, respectively (Fig. 3A). These strains were similar to 5E4 for all phenotypes tested (Table 2).
To further verify that the 5E4 mutant phenotype was due to disruption of the clp gene, the mutation in 5E4 was complemented by chromosomal insertion of the clp gene, creating strain P1. All altered phenotypes of 5E4 were restored in P1, except that it displayed excessive gliding motility compared to wild-type strain C3 (Fig. 2A and Table 2).
Antimicrobial activity and biological control of plant diseases are affected in mutants disrupted in the clp gene.
Antimicrobial activity against the plant pathogens B. sorokiniana and P. ultimum was clearly evident from the zones of growth inhibition observed around colonies of strains C3 and P1 on 10% TSA. In contrast, growth of B. sorokiniana or P. ultimum was not inhibited around colonies of mutant strains 5E4, DC211, and DCA2422 (Fig. 2C and Table 2).
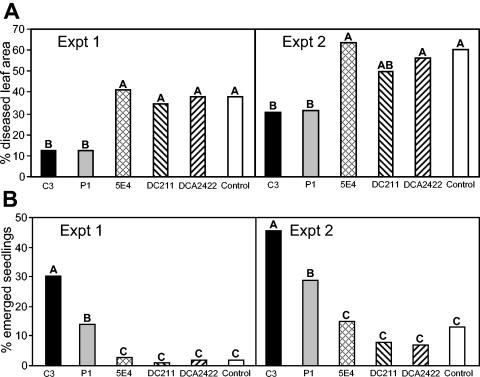
In experiments to evaluate the effects of strains on bipolaris leaf spot, treatment of tall fescue leaves with parent strain C3 or with P1 reduced the leaf spot severity by more than 50% compared with the control. In contrast, the disease severity on leaves treated with mutant strain 5E4, DC211, or DC2422 did not differ significantly from the disease severity in the control plants (Fig. 5A). Throughout these experiments, the sizes of the populations of C3 and 5E4 on leaves ranged from around 7.0 to 8.4 log CFU/g; there was no significant difference between the leaf populations of the two strains on any sampling date (data not shown).
FIG. 5.
Biocontrol effects of L. enzymogenes strains on the severity of leaf spot caused by B. sorokiniana on tall fescue (A) and on pythium damping-off of sugar beet as measured by seedling emergence (B). In each experiment, bars with same letters are not significantly different at α = 0.05.
Seed treatment with C3 improved the emergence of sugar beet seedlings in soil infested with P. ultimum, whereas treatment with mutants 5E4, DC211, and DCA2422 resulted in no significant difference compared with the methylcellulose-treated control in two separate experiments (Fig. 5B). In contrast, treatment with strain P1 resulted in an intermediate response, in which the seedling emergence with P1 treatment was significantly greater than the seedling emergence with clp-disrupted mutant strains but less than the seedling emergence with wild-type strain C3.
DISCUSSION
Members of the Lysobacter group characteristically express several traits thought to contribute to antagonism of fungi and other microbes. These traits include production of extracellular degradative enzymes, such as chitinases, glucanases, and proteases, as well as production of antibiotic compounds (5, 20, 36, 37). Mutations in the clp gene had a global effect on a variety of L. enzymogenes phenotypes, including significant reductions in extracellular enzyme production, antimicrobial activity, and biocontrol activity. These observations provide further support for the hypothesis that extracellular factors, including lytic enzymes and possibly secondary metabolites such as antibiotics, play important roles in the biocontrol activity of L. enzymogenes strain C3. Chitinases have been implicated previously in the biological control activity displayed by L. enzymogenes. Zhang and Yuen (44) demonstrated that chitinases purified from culture filtrates enhanced biocontrol of bipolaris leaf spot by strain C3. In much earlier studies, Gillespie and Cook (14) described an enzymatic lytic fraction termed lysin from L. enzymogenes strain 495 (formerly classified as a member of the myxobacter group) that displayed activity against bacteria and nematodes. Although the nature of this fraction is unclear, its presence is consistent with antagonistic effects of extracellular factors produced by the species.
Global regulation of traits associated with biocontrol has been best described for fluorescent pseudomonads, such as Pseudomonas fluorescens. The regulators involved at the transcriptional level consist of components belonging to complex signal transduction pathways, including two-component systems, alternate sigma factors, RNA binding proteins, and regulatory RNAs (1, 6, 12, 17, 24, 33). Global regulators belonging to the Crp family have not been extensively described in P. fluorescens or other plant biocontrol bacteria, but they have been characterized in the plant pathogens X. campestris and E. chrysanthemi (9, 31). In both of the latter cases, the regulators controlled numerous traits important in the pathogenetic process. The vfr gene characterized in Pseudomonas aeruginosa is also a member of the Crp gene family and regulates a variety of virulence traits, including production of exotoxin A, pyocyanin, and elastase, production of type IV pili and twitching motility, and quorum sensing (2, 4, 38). While vfr gene homologues have been identified in genomic sequences of P. fluorescens strains (Kobayashi, unpublished data), no experimental evidence linking the regulator to expression of biocontrol traits has been obtained yet.
The mutation in strain 5E4 affected not only expression of clp but also a putative acetyltransferase belonging to the GNAT superfamily. Although a role for the acetyltransferase in gene regulation was not established in this study, it is conceivable that both genes could be involved in a complex regulatory pathway. GNAT acetyltransferases are ubiquitous among organisms and have been associated with a number of cellular functions, including transcriptional regulation (26). To date, however, GNAT superfamily members that function in gene regulation have not been as extensively characterized from prokaryotic sources as they have been from eukaryotic sources.
Several similarities between the clp gene previously characterized from X. campestris pv. campestris and the clp gene from L. enzymogenes C3 are evident. The physical linkage between the speD and clp genes in C3 is identical to that described for X. campestris pv. campestris (25). In addition, the clp gene in X. campestris pv. campestris was found to globally regulate production of various extracellular enzymes, pigmentation, and the extracellular polysaccharide xanthan gum (9). Our observations of altered colony color and morphology in mutant 5E4 are consistent with clp-mediated changes in pigmentation and extracellular polysaccharide production. Furthermore, the reduction in fungal antagonism in mutant 5E4 parallels the finding that the phytopathogenicity of the X. campestris pv. campestris clp mutant strain was significantly reduced (9).
Despite the similarities noted above, one distinct difference is the absence of apparent acetyltransferase homologues in X. campestris pv. campestris and other related members of the Xanthomonadaceae, including Xanthomonas and Xylella spp. whose genomic sequences have been determined. This finding suggests that the organizational relationship and quite possibly a functional relationship between the clp gene and the acetyltransferase gene are unique to L. enzymogenes, at least among members of the Xanthomonadaceae.
Traits controlled by the clp regulator were found to be responsible for biocontrol activity against bipolaris leaf spot of tall fescue and pythium damping-off of sugar beet. The finding that wild-type strain C3 and mutant strain 5E4 were similar in terms of colonization of tall fescue leaves supports the hypothesis that the lack of bipolaris leaf spot biocontrol ability in 5E4 and other clp mutants is due to reduced production of extracellular factors rather than impaired colonization ability. For both disease assays, host infection relies on a spore-based inoculum that is activated rapidly and synchronously; disease suppression in these scenarios could be achieved largely or entirely by inhibition of spore germination by extracellular antimicrobial products of strain C3.
The nature of the affected regulatory clp gene and the control that it has on various traits has complicated genetic complementation of mutant 5E4. Unlike the crp counterparts that have been characterized in enteric bacteria, clp functions in X. campestris pv. campestris independent of cyclic AMP to control gene expression. We have obtained evidence that in L. enzymogenes strain C3 expression of chitinases is cyclic AMP dependent, while expression of β-1,3-glucanases is cyclic AMP independent (Zhou, Reedy, Yuen, and Kobayashi, Phytopathology 92:S91, 2002).
Complementation of strain 5E4 required chromosomal insertion of the clp gene, because a stable plasmid system for strain C3 was not present. All phenotypes tested were complemented in the resulting strain, P1, except that in this strain at least two phenotypes were altered compared to wild-type strain C3. P1 displayed excessive gliding motility (Fig. 2A), and biocontrol of pythium damping-off was not fully restored to the levels achieved with strain C3 (Fig. 5B). Based on the chromosomal insertion of clp in strain P1, it is possible that these altered phenotypes may have resulted from (i) altered expression of the clp gene compared to the wild-type strain, (ii) lack of complementation of the acetyltransferase, or (iii) a second mutational effect that occurred due to the position at which the clp gene was placed in P1. The latter explanation is intriguing, since identification in C3 of a type III secretion pathway (30; Reedy and Kobayashi, Phytopathology 91:S75, 2001), a central component for bacterial pathogenicity for eukaryotic hosts (7), suggests that there is a possible pathogenic interaction that also contributes to the biocontrol activity of C3 against P. ultimum. Nonetheless, observations made in this study speak to the complexity of the regulatory network governing factors contributing to the antimicrobial activity expressed by L. enzymogenes, as well as the factors involved in the interactions between the bacterium and fungal or oomycetous hosts. Efforts are in progress to further define steps in the regulatory pathway and to identify roles of specific traits associated with antimicrobial and biocontrol activities expressed by L. enzymogenes.
Acknowledgments
This work was funded in part by a grant from EcoSoils Systems, Inc. and by the New Jersey Agricultural Research Station.
We thank G. MacDonald, N. El-Barrad, and C. Jochum for technical assistance.
Footnotes
University of Nebraska, Lincoln, Agricultural Research Division Journal Series no. 14221.
REFERENCES
- 1.Aarons, S., A. Abbas, C. Adams, A. Fenton, and F. O'Gara. 2000. A regulatory RNA (PrrB RNA) modulates expression of secondary metabolite genes in Pseudomonas fluorescens F113. J. Bacteriol. 182:3913-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beatson, S. A., C. B. Whitchurch, J. L. Sargent, R. C. Levesque, and J. S. Mattick. 2002. Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J. Bacteriol. 184:3605-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen, P., and F. D. Cook. 1978. Lysobacter, a new genus of nonfruiting, gliding bacteria with high base ratio. Int. J. Syst. Bacteriol. 28:367-393. [Google Scholar]
- 6.Corbell, N., and J. E. Loper. 1995. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf-5. J. Bacteriol. 177:6230-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 8.Côté, F., S. El Ouakfaoui, and A. Asselin. 1991. Detection of β-glucanase activity on various β-1,3- and β-1,4-glucans after native and denaturing polyacrylamide gel electrophoresis. Electrophoresis 12:69-74. [DOI] [PubMed] [Google Scholar]
- 9.de Crecy-Lagard, V., P. Glaser, P. Lejeune, O. Sismeiro, C. E. Barber, M. J. Daniels, and A. Danchin. 1990. A Xanthomonas campestris pv. campestris protein similar to catabolite activation factor is involved in regulation of phytopathogenicity. J. Bacteriol. 172:5877-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Folman, L. B., J. Postma, and J. A. Van Veen. 2003. Characteisation of Lysobacter enzymogenes (Christensen and Cook 1978) strain 3.1T8, a powerful antagonist of fungal diseases of cucumber. Microbiol. Res. 158:1-9. [DOI] [PubMed] [Google Scholar]
- 12.Gaffney, T. D., S. T. Lam, J. Ligon, K. Gates, A. Frazelle, J. Di Maio, S. Hill, S. Goodwin, N. Torkewitz, A. M. Allshouse, H. J. Kempf, and J. O. Becker. 1994. Global regulation of expression of anti-fungal factors by a Pseudomonas fluorescens biological control strain. Mol. Plant-Microbe Interact. 7:455-463. [DOI] [PubMed] [Google Scholar]
- 13.Giesler, L. J., and G. Y. Yuen. 1998. Evaluation of Stenotrophomonas maltophilia strain C3 for biocontrol of brown patch disease. Crop Prot. 17:509-513. [Google Scholar]
- 14.Gillespie, D. C., and F. D. Cook. 1965. Extracellular enzymes from strains of Sorangium. Can. J. Microbiol. 11:109-118. [DOI] [PubMed] [Google Scholar]
- 15.Grenier, J., and A. Asselin. 1993. Detection of β-1,3-glucanase activity in gels containing alkali-soluble yeast glucan. Anal. Biochem. 212:301-302. [DOI] [PubMed] [Google Scholar]
- 16.Hancock, J. G. 1985. Fungal infection of feeder rootlets of alfalfa. Phytopathology 75:1112-1120. [Google Scholar]
- 17.Heeb, S., C. Blumer, and D. Haas. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 184:1046-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen, R. G. 1983. Detection and determination of lipase (acylglycerol hydrolase) activity from various sources. Lipids 18:650-657. [DOI] [PubMed] [Google Scholar]
- 20.Kato, A., S. Nakaya, Y. Ohashi, and H. Hirata. 1997. WAP-8294A2, a novel anti-MRSA antibiotic produced by Lysobacter sp. J. Am. Chem. Soc. 119:6680-6681. [Google Scholar]
- 21.Katznelson, H., D. C. Gillespi, and F. D. Cook. 1964. Studies on the relationship between nematodes and other soil microorganisms. Can. J. Microbiol. 10:699-704. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi, D. Y., and N. El-Barrad. 1996. Selection of bacterial antagonists using enrichment cultures for the control of summer patch disease of Kentucky bluegrass. Curr. Microbiol. 32:106-110. [Google Scholar]
- 23.Kobayashi, D. Y., R. M. Reedy, J. A. Bick, and P. V. Oudemans. 2002. Characterization of a chitinase gene from Stenotrophomonas maltophilia and its involvement in biological control. Appl. Environ. Microbiol. 68:1047-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laville, J., C. Voisard, C. Keel, M. Maurhofer, G. Defago, and D. Haas. 1992. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. USA 89:1562-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, T.-C., S.-T. Chen, M.-C. Lee, C.-M. Chang, C.-H. Chen, S.-F. Weng, and Y.-H. Tseng. 2001. The early stages of filamentous phage φLf infection require the host transcription factor, Clp. J. Mol. Microbiol. Biotechnol. 3:471-481. [PubMed] [Google Scholar]
- 26.Neuwald, A. F., and D. Landsman. 1997. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22:154-155. [DOI] [PubMed] [Google Scholar]
- 27.Osburn, R. M., M. N. Schroth, J. G. Hancock, and M. Hendson. 1989. Dynamics of sugarbeet colonization by Pythium ultimum and Pseudomonas spp.: effects on seed rot and damping-off. Phytopathology 79:709-716. [Google Scholar]
- 28.Palumbo, J. D., R. F. Sullivan, and D. Y. Kobayashi. 2003. Molecular characterization and expression in Escherichia coli of three β-1,3-glucanase genes from Lysobacter enzymogenes strain N4-7. J. Bacteriol. 185:4362-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quadt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 30.Reedy, R. M. 2004. Ph.D. thesis. Rutgers, The State University of New Jersey, New Brunswick.
- 31.Reverchon, S., D. Expert, J. Robert-Baudouy, and W. Nasser. 1997. The cyclic AMP receptor protein is the main activator of pectinolysis genes in Erwinia chrysanthemi. J. Bacteriol. 179:3500-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sarniguet, A., J. Kraus, M. D. Henkels, A. M. Muehlchen, and J. E. Loper. 1995. The sigma factor σs affects antibiotic production and biological control activity of Pseudomonas fluorescens Pf-5. Proc. Natl. Acad. Sci. USA 92:12255-12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 35.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan, R. F., M. A. Holtman, G. J. Zylstra, J. F. White, Jr., and D. Y. Kobayashi. 2003. Taxonomic positioning of two biological control agents for plant diseases as Lysobacter enzymogenes based on phylogenetic analysis of 16S rDNA, fatty acid composition and phenotypic characteristics. J. Appl. Microbiol. 94:1079-1086. [DOI] [PubMed] [Google Scholar]
- 37.Tymiak, A. A., T. J. McCormick, and S. E. Unger. 1989. Structure determination of lysobactin, a macrocyclic peptide lactone antibiotic. J. Org. Chem. 54:1149-1157. [Google Scholar]
- 38.West, S. E., A. K. Sample, and L. J. Runyen-Janecky. 1994. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J. Bacteriol. 176:7532-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuen, G. Y., M. L. Craig, and F. Avila. 1993. Detection of Pythium ultimum with a species-specific monoclonal antibody. Plant Dis. 77:692-698. [Google Scholar]
- 40.Yuen, G. Y., J. R. Steadman, D. T. Lindgren, D. Schaff, and C. Jochum. 2001. Bean rust biological control using bacterial agents. Crop Prot. 20:395-402. [Google Scholar]
- 41.Yuen, G. Y., and Z. Zhang. 2001. Control of brown patch using the bacterium Stenotrophomonas maltophilia C3 and culture fluid. Int. Turfgrass Soc. Res. J. 9:742-747. [Google Scholar]
- 42.Zhang, Z., and G. Y. Yuen. 1999. Biological control of Bipolaris sorokiniana on tall fescue by Stenotrophomonas maltophilia C3. Phytopathology 89:817-822. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, Z., and G. Y. Yuen. 2000. Effects of culture fluids and preinduction of chitinase production on biocontrol of bipolaris leaf spot by Stenotrophomonas maltophilia C3. Biol. Control 18:277-286. [Google Scholar]
- 44.Zhang, Z., and G. Y. Yuen. 2000. The role of chitinase production by Stenotrophomonas maltophilia strain C3 in biological control of Bipolaris sorokiniana. Phytopathology 90:384-389. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Z., G. Y. Yuen, G. Sarath, and A. R. Penheiter. 2001. Chitinases from the plant disease biocontrol agent, Stenotrophomonas maltophilia C3. Phytopathology 91:204-211. [DOI] [PubMed] [Google Scholar]