Abstract
The incorporation and delivery of bifidobacterial strains as probiotic components in many food preparations expose these microorganisms to a multitude of environmental insults, including heat and osmotic stresses. We characterized the dnaK gene region of Bifidobacterium breve UCC 2003. Sequence analysis of the dnaK locus revealed four genes with the organization dnaK-grpE-dnaJ-ORF1, whose deduced protein products display significant similarity to corresponding chaperones found in other bacteria. Northern hybridization and real-time LightCycler PCR analysis revealed that the transcription of the dnaK operon was strongly induced by osmotic shock but was not induced significantly by heat stress. A 4.4-kb polycistronic mRNA, which represented the transcript of the complete dnaK gene region, was detected. Many other small transcripts, which were assumed to have resulted from intensive processing or degradation of this polycistronic mRNA, were identified. The transcription start site of the dnaK operon was determined by primer extension. Phylogenetic analysis of the available bifidobacterial grpE and dnaK genes suggested that the evolutionary development of these genes has been similar. The phylogeny derived from the various bifidobacterial grpE and dnaK sequences is consistent with that derived from 16S rRNA. The use of these genes in bifidobacterial species as an alternative or complement to the 16S rRNA gene marker provides sequence signatures that allow a high level of discrimination between closely related species of this genus.
Bifidobacteria are important components of the gastrointestinal microflora and have been the subject of growing scientific interest due to their suspected role in the maintenance of gastrointestinal health (21, 32, 36). The ability of these microorganisms to survive and adapt to changing environmental circumstances depends on their ability to resist harsh conditions encountered in their natural environment or during food manufacture in the case of probiotic strains (35). Under stressful situations, misfolded proteins may accumulate and/or aggregate, a process that may harm bacteria. Bacterial cells respond to stressful conditions by synthesizing molecular chaperones, which promote proper protein folding, thereby preventing the formation of insoluble protein aggregates. The major molecular chaperones include the DnaK complex (composed of DnaK, DnaJ, and GrpE) and the GroESL complexes (composed of GroES and GroEL) (8). Through cycles of protein binding and release, which are fuelled by ATP, the DnaK chaperone complex facilitates substrate folding to the native state (15).
In eubacteria, the dnaK operon has been investigated in detail in low-G+C-content bacteria, such as Escherichia coli and Bacillus subtilis. Cloning, sequencing, and transcriptional analyses of the dnaK operon in B. subtilis revealed four genes in the order hrcA-grpE-dnaK-dnaJ; the products of these genes constitute the dnaK chaperone machine (16). In these bacteria transcriptional regulation of the dnaK operon involves an inverted repeat element termed CIRCE (controlling inverted repeat of chaperone expression) located between the transcription initiation site and the dnaK coding region. This organization seems to be the basic and most common organization for dnaK operons in low-G+C-content gram-positive bacteria (29). A similar organization, hrcA-grpE-dnaK (without the dnaJ gene), was described for Lactococcus lactis and Chlamydia (28). A second variant of this organization, grpE-dnaK-dnaJ, was found in spirochetes, whereas in other low-G+C-content gram-negative bacteria (proteobacteria) the genes are organized into two transcriptional units; one operon contains hrcA-grpE, while the second operon contains dnaK-dnaJ. The first gene of the B. subtilis dnaK operon, hrcA, encodes a repressor that binds to the CIRCE element (30, 45, 46). Basic knowledge of the genetics of the dnaK locus in high-G+C-content gram-positive bacteria was obtained through analysis of the Streptomyces coelicolor genome sequence. In this microorganism the dnaK operon encodes four proteins, DnaK, GrpE, DnaJ, and HspR (5, 6), and the latter protein was shown to function as a repressor of the S. coelicolor dnaK operon. This gene inventory appears to be unique to Actinobacteridae (7). There is no sequence similarity between the Hsp-inverted repeat and HrcA-CIRCE systems, showing that Streptomyces has evolved a different strategy for controlling synthesis of the DnaK chaperone machine. Interestingly, a homologue of the hrcA gene, which in low-G+C-content bacteria is frequently part of the dnaK operon, is not found in this operon but was found to be part of an operon with a second copy of the dnaJ gene (12).
The genes constituting the dnaK operons are very well conserved across the archaeal, eubacterial, and eukaryal domains of life. Due to their ubiquitous distribution, functional preservation, and sequence conservation, the dnaK operon genes are considered valuable molecular markers for bacterial phylogenetic investigations. They have been successfully used for identification of Mycobacterium (31), Staphylococcus (1), Streptococcus (1), Legionella (22), and Ruminococcus (2) species. With the introduction of polyphasic taxonomy (34), many molecular markers that are alternatives to the 16S rRNA gene sequences have been described. Recently, alternative molecules, such as the gene encoding elongation factor Tu (39, 40), ATPase subunits (38), the recA gene (19, 40), and groEL (37), have been used to examine phylogenetic relationships between bifidobacterial species. The use of these molecular markers corroborates and completes the evolutionary history of bifidobacterial species. However, other molecular markers, which provide finer taxonomic detail to distinguish closely related species (e.g., Bifidobacterium lactis and Bifidobacterium animalis or Bifidobacterium catenulatum and Bifidobacterium pseudocatenulatum), are required.
To date, very little is known about the dnaK operon of bifidobacteria (26). In this report we describe the dnaK operon of Bifidobacterium breve UCC 2003. We tested the heat and osmotic inducibility of this operon by performing Northern blot hybridization analyses, primer extension experiments, and real-time PCR. Moreover, we describe below partial sequencing of the genes coding for the GrpE and DnaK chaperones from 14 bifidobacterial species in order to provide new molecular markers for investigation of the phylogeny of the genus Bifidobacterium.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All Bifidobacterium strains were grown anaerobically in MRS (Difco, Detroit, Mich.) supplemented with 0.05% l-cysteine-HCl and incubated at 37°C for 16 h. The bacterial strains used are summarized in Table 1.
TABLE 1.
Strains used in this study and grpE and dnaK sequence accession numbers
| Species or subspecies | Straina | grpE accession no.b | dnaK accession no.b |
|---|---|---|---|
| Bifidobacterium animalis | ATCC 25527 | AY642878 | AY642871 |
| Bifidobacterium animalis | LMG 18900 | AY642890 | AY642872 |
| Bifidobacterium lactis | LMG 18906 | AY642889 | AY642873 |
| Bifidobacterium lactis | LMG 11615 | AY642887 | AY642876 |
| Bifidobacterium lactis | DSM 10140 | AY667066 | AY642912 |
| Bifidobacterium breve | UCC 2003 | AY642911 | AY642911 |
| Bifidobacterium adolescentis | JCM 1275 | AY642885 | AY642864 |
| Bifidobacterium longum | NCC 2705 | NC_004307 | NC_004307 |
| Bifidobacterium infantis | JCM 1222 | AY642882 | AY642867 |
| Bifidobacterium suis | JCM 1269 | AY642883 | AY642869 |
| Bifidobacterium bifidum | JCM 1255 | AY642888 | AY642868 |
| Bifidobacterium dentium | JCM 1195 | AY642886 | AY642866 |
| Bifidobacterium catenulatum | JCM 1194 | AY642884 | AY642875 |
| Bifidobacterium pseudocatenulatum | JCM 1200 | AY642881 | AY642865 |
| Bifidobacterium angulatum | JCM 7096 | AY642880 | AY642870 |
| Bifidobacterium magnum | JCM 1218 | AY642877 | AY642863 |
| Bifidobacterium thermophilum | JCM 1207 | AY642879 | AY642874 |
| Lactobacillus plantarum | WCFSI | NC_004567 | NC_004567 |
| Lactobacillus gasseri | ATCC 33323 | NZ_AAA002000001 | NZ_AAA002000001 |
| Lactobacillus johnsonii | NCC 533 | NC_002662 | NC_002662 |
| Lactococcus lactis subsp. lactis | IL1403 | NC_002662 | NC_002662 |
| Streptococcus pyogenes | MIGAS | NC_002737 | NC_002737 |
| Enterococcus faecalis | V583 | NC_004668 | NC_004668 |
| Clostridium acetobutylicum | ATCC 824 | NC_003030 | NC_003030 |
| Mycobacterium bovis | AF 2122/97 | NC_002945 | NC_002945 |
| Streptomyces coelicolor | A3 | NC_003888 | NC_003888 |
| Corynebacterium glutamicum | ATCC 13032 | NC_003450 | NC_003450 |
| Listeria monocytogenes | EGD | NC_003210 | NC_003210 |
| Escherichia coli | K12 | NC_000913 | NC_000913 |
ATCC, American Type Culture Collection; DSM, Deutsche Sammlung von Mikroorganismen; JCM, Japanese Collection of Microorganisms; LMG, Bacteria Collection, Universiteit Gent.
For the strains whose genome sequences are available, the grpE and dnaK sequences were retrieved from the complete bacterial genome, and the accession numbers are indicated.
DNA amplification and cloning of the dnaK and grpE genes.
PCR was used to amplify the dnaK gene of all Bifidobacterium strains investigated. A DNA fragment corresponding to the dnaK gene was amplified by using oligonucleotides DNAK-1 (5′-GTCAGGCTGTGACCAAC-3′) and DNAK-2 (5′-GGTCATTTCCTCGAAGTG-3′), while the grpE-containing DNA fragment of Bifidobacterium strains was amplified by employing oligonucleotides GRE-UNI (5′-GAGTTCATCAACTACCGCA-3′) and GRE-REV (5′-CTTCCACCACGGTGTC-3′). Each PCR mixture (50 μl) contained 20 mM Tris-HCl, 50 mM KCl, each deoxynucleoside triphosphate at a concentration of 200 μM, 50 pmol of each primer, 1.5 mM MgCl2, and 1 U of Taq DNA polymerase (Gibco BRL, Paisley, United Kingdom). Each PCR cycling profile consisted of an initial denaturation step of 3 min at 95°C, followed by amplification for 30 cycles as follows: denaturation for 30 s at 95°C, annealing for 30 s at 50°C, and extension for 1 min at 72°C. The PCR was completed with an elongation phase consisting of 10 min at 72°C. The resulting amplicons were separated on a 1.5% agarose gel, and this was followed by ethidium bromide staining. PCR fragments were purified with a PCR purification spin kit (Genomed, Lohne, Germany) and were subsequently sequenced.
The upstream region of the dnaK operon of B. lactis DSM 10140 was determined by inverse PCR (24). One microgram of DSM 10140 chromosomal DNA was digested with restriction endonuclease EcoRI or ClaI, and the restriction fragments were self-ligated and amplified by using primers DNAK-1-inv (5′-GTTGGTCACAGCCTGAC-3′) and DNAK-2-inv (5′-CACTTCGAGGAAATGACC-3′) as described by Sambrook and Russell (24). The inverse PCR products obtained were used for direct sequencing with synthetic oligonucleotides.
DNA sequencing and phylogenetic study.
Nucleotide sequencing of both strands of PCR amplicons was performed by MWG-Biotech AG (Ebersberg, Germany) with primers DNAK-1, DNAK-2, GRE-UNI, and GRE-REV. The dnaK and grpE partial gene sequences of all Bifidobacterium strains examined in this study were used for comparison. Sequence data assembly and analysis were performed by using the DNASTAR software (version 5.05; DNASTAR, Madison, Wis.). Sequence alignment was performed by using the MultiAlign and Clustal W programs (33). Phylogenetic calculations, including distance calculations and generation of phylogenetic trees, were performed by using the PHYLIP package (9). Trees were calculated by the neighbor-joining method as implemented in the neighbor module of PHYLIP. Also, dendrograms of gene sequences were constructed by using the Clustal X program and were visualized with the TreeView program.
RNA isolation and Northern blot analysis.
B. breve UCC 2003 cells were grown to an optical density at 600 nm of 0.6. Heat stress was applied by transferring the culture to 43°C, while osmotic stress was applied by addition of prewarmed medium containing 5 M NaCl to obtain a final NaCl concentration of 0.3, 0.5, 0.7, or 1.5 M. At various times, 30 ml of culture was collected and briefly centrifuged to harvest cells. Total RNA was isolated by using macaloid acid (37) and was treated with DNase (Roche, Lewes, East Sussex, United Kingdom). Fifteen micrograms of RNA was subjected to electrophoresis on a 1.5% agarose-3.3% formaldehyde denaturing gel, transferred to a Zeta-Probe blotting membrane (Bio-Rad, Hemel Hempstead, Hertfordshire, United Kingdom) as described by Sambrook and Russell (24), and fixed by UV cross-linking by using a Stratalinker 1800 (Stratagene, La Jolla, Calif.). PCR amplicons obtained by using primer combinations that were designed to target each of the genes of the B. breve UCC 2003 dnaK operon were radiolabeled (24). Prehybridization and hybridization were carried out at 65°C in 0.5 M NaHPO4 (pH 7.2)-1.0 mM EDTA-7.0% sodium dodecyl sulfate (SDS). Following 18 h of hybridization, the membrane was rinsed twice for 30 min at 65°C in 0.1 M NaHPO4 (pH 7.2)-1.0 mM EDTA-1% SDS and twice for 30 min at 65°C in 0.1 mM NaHPO4 (pH 7.2)-1.0 mM EDTA-0.1% SDS and then exposed to X-OMAT autoradiography film (Eastman Kodak, Rochester, N.Y.).
cDNA synthesis and quantification of specific transcripts by LightCycler PCR.
Five micrograms of mRNA was treated with DNase (Roche) and used as a template in a 100-μl reaction mixture containing 20 ng of random primers, each deoxyribonucleoside triphosphate at a concentration of 0.125 mM, and Superscript enzyme (Invitrogen, Paisley, United Kingdom) used according the manufacturer's instructions to produce cDNA. The cDNA generated was used as a template for reverse transcription (RT)-PCRs to determine the level of transcription of grpE and ORF1 as representative genes of the dnaK operon.
In order to accurately determine and compare the transcript levels of any differentially regulated gene, it was necessary to normalize cDNA levels from the individual times. To do this, by using primers 16SRT2 (5′-GTTCCACGGGTTCCGTG-3′) and 16SRT3 (5′-GAGCTGACGACGACCATG-3′) and previously described parameters (23), the constitutively expressed 16S rRNA gene was standardized for each time by adjusting the volume of cDNA added prior to PCR until the crossing point values were identical for all samples. Subsequently, by using the standardized volumes of cDNA, the relative expression of the target genes was determined as outlined below. To prepare quantification standards for the LightCycler PCR, a 285-bp region of the grpE gene was amplified by using primers GRE-UNI and GRE-REV, and a 300-bp region of ORF1 was amplified by using primers TRA-1 (5′-GACCAAAATGCGTGCGATG-3′) and TRA-2 (5′-CAACCAGATGAAGCAGTT-3′). The PCR products were purified with a PCR purification spin kit (Genomed) and were subsequently quantified with a GenQuant spectrophotometer (Pharmacia). The copy number was calculated, and appropriate dilutions containing from 101 to 108 copies of the standards were prepared in elution buffer (QIAGEN), divided into aliquots, and stored at 4°C. A Faststart SYBR Green master mixture kit (Roche) was used for all LightCycler PCRs. By using the standards generated as described above, the conditions were optimized for the real-time PCR as recommended by the manufacturer. This included titrating the MgCl2 and primer concentrations, as well as optimizing the annealing temperatures and extension times. The final optimized conditions used in all subsequent analyses were 2 μl of either the standard dilution or standardized dilution of each RT reaction mixture as a template, 2 mM MgCl2, 2 μl of the Faststart SYBR Green master mixture (Roche), and each relevant primer at a concentration of 0.5 μM in a 20-μl (total volume) reaction mixture. The PCR parameters for amplification included denaturation for one cycle at 95°C for 8 min and 40 cycles of 95°C for 5 s, 50°C for 5 s, and 72°C for 8 s. A melting curve analysis was performed at 65 to 90°C. The melting curve for each bacterial sample was generated manually to determine the melting temperature (Tm). The Tm was the peak of the curve determined by measuring the derivative of fluorescence with respect to temperature (−dF/dT). By using the manual Tm function of the LightCycler software, the Tm was defined as the temperature at which the cursor identified the highest point on the melting curve (the −dF/dT curve). If the highest point was a plateau, the midpoint of the plateau was taken to be the Tm.
All LightCycler assay data analyses were carried out by using the fit points option of the Roche LightCycler software (version 3.53). The ability to quantify unknown targets was evaluated by using eight separate DNA samples containing various amounts of template (range, 101 to 108 copies) that were analyzed by using RT-PCR in conjunction with known standards in this range. The number of copies of each sample transcript was then determined with the LightCycler software. The standard dilutions were used to generate crossing point values, which were used to quantify the transcript levels of grpE and ORF1. By fitting the crossing point values (beginning of the exponential phase) of the known standards to those of dilutions of the unknown real-time reaction mixtures, the number of gene transcripts for each target was calculated. Each experiment was performed in triplicate, and the standard deviations were calculated to ensure statistical integrity.
Primer extension analysis.
The 5′ ends of the RNA transcripts were determined as described in a previous study (38). The synthetic oligonucleotides used were as follows: dnaK-prom (5′-GTGGTGCGCATGCCCTC-3′), grpE-prom (5′-CTCCTTCATTGGAAGGAGCCTG-3′), dnaJ-prom (5′-CTCGGCTTCCTTGGTCTTG-3′), and ORF1-prom (5′-GTACTGACGGAGGGTCTG-3′), located at positions 90 to 106, 148 to 162, 125 to 143, and 162 to 179, respectively, in the respective nucleotide sequences.
PCR identification of B. lactis.
Amplification reactions were performed in 50-μl (total volume) solutions containing 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, each deoxynucleoside triphosphate (Gibco BRL) at a concentration of 200 μM, 10 pmol of gro-1 (37), 10 pmol of gro-2 (37), 50 pmol of B. lactis-specific primer b-dnaK1 (5′-CACAGATCCTCATGAAGCTG-3′), 50 pmol of B. lactis-specific primer b-dnaK2 (5′-CGACGTAGCCTGCACCTGG-3′), 25 ng of template DNA, and 2.5 U of Taq DNA polymerase (Gibco BRL). Amplifications were performed with a DNA thermocyler (Primus) with the following temperature profile: one cycle of 95°C for 5 min; 30 cycles of 95°C for 30 s, 50°C for 1 min, and 72°C for 1 min; and one cycle of 72°C for 7 min. For routine identification, cells were lysed by using a rapid DNA extraction protocol (43) and were used as direct PCR templates. PCR amplicons were analyzed by 1.5% (wt/vol) agarose gel electrophoresis in Tris-acetate-EDTA buffer at a constant voltage of 7 V/cm, were visualized with ethidium bromide (0.5 μg/ml), and were photographed under UV light at 260 nm.
Nucleotide sequence accession numbers.
The GenBank accession numbers for partial grpE and dnaK gene sequences generated in this study are shown in Table 1. The nucleotide sequence data for the dnaK operon of B. breve UCC 2003 have been deposited in the GenBank database under accession number AY642911.
RESULTS
Genetic organization of the dnaK operon.
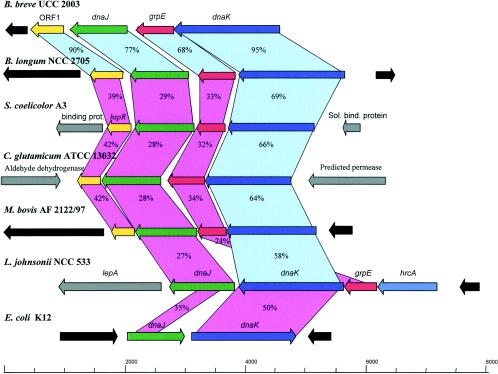
Analysis of the B. breve UCC 2003 genome sequence (S. Leahy, J. A. Moreno Munoz, M. O'Connell-Motherway, D. Higgins, G. F. Fitzgerald, and D. van Sinderen, unpublished data) allowed us to identify a 4.4-kb DNA fragment encompassing the dnaK operon, which is represented by four open reading frames. The gene order of the dnaK operon was dnaK-grpE-dnaJ-ORF1, which is identical to that observed in other high-G+C-content gram-positive bacteria (28). In fact, a comparison with other Actinobacteridae (Bifidobacterium longum, S. coelicolor, Corynebacterium glutamicum, and Mycobacterium bovis) revealed that the genetic organizations of the dnaK operon are identical (Fig. 1). The gene inventory of the dnaK operon in unrelated taxa (e.g., Lactobacillus and Escherichia) was shown to be different, which is in accordance with their distant phylogenetic positions (27).
FIG. 1.
Comparison of the dnaK operon in B. breve UCC 2003 with corresponding loci in various other bacteria. Each arrow indicates an open reading frame, and the length of the arrow is proportional to the length of the predicted open reading frame. Corresponding genes are indicated by the same color. The putative function of the protein is indicated above each arrow. Genes exhibiting ≥55% amino acid similarity are linked by light blue shading, and genes exhibiting ≤54% amino acid similarity are linked by light violet shading. Levels of amino acid identity, expressed as percentages, are indicated. Sol. bind. protein, soluble binding protein.
A protein comparison (Fig. 1) showed that the proteins most similar to the B. breve DnaK, GrpE, and DnaJ chaperones were proteins of B. longum. Notably, levels of identity of ≥24% were observed for these three proteins with the corresponding proteins of Lactobacillus johnsonii. The DnaK chaperone is the most conserved protein encoded by the operon in high- and low-G+C-content bacteria. Furthermore, a comparison of the B. breve dnaK, grpE, and dnaJ products with proteins deposited in the publicly available databases showed highly significant sequence similarities for the complete DnaK, GrpE, chaperones of other high-G+C-content gram-positive bacteria (e.g., Mycobacterium, Streptomyces, Corynebacterium, and Propionibacterium). In contrast, the DnaJ chaperone homologue of B. breve exhibited considerable similarity to the DnaJ chaperone found in low-G+C-content bacteria (Bordellia, Nostoc, and Staphylococcus).
ORF1, located downstream of dnaJ, is a gene which is predicted to encode a protein containing 195 amino acids and having a calculated mass of 21.8 kDa that displays significant similarity to other predicted heat-controlled transcriptional regulators belonging to the Mer family. These regulators include those identified for Mycobacterium leprae (level of identity, 43%), Aquifex aeolicus (level of identity, 39%), and Deinococcus radiodurans (level of identity, 32%). Interestingly, the N-terminal regions of these proteins are predicted to contain a classical helix-turn-helix motif, which is assumed to be involved in DNA recognition and DNA binding processes (4). Thus, this finding suggests that the ORF1 product is also involved in such interactions.
The predicted B. breve UCC 2003 DnaK protein contains the consensus HSP70 domains, which have been described as being involved in substrate binding and release (pfam0012). The predicted DnaJ chaperone of B. breve contains the J domain, which is highly conserved in DnaJ homologs and which is involved in protein interaction and folding (8).
The DNA regions surrounding the dnaK operon in B. breve do not display homology to the regions flanking the dnaK loci of other available bifidobacteria, indicating that the observed conservation of gene order in the two sequenced bifidobacteria appears to be restricted to this operon (data not shown).
Interestingly, the initiation methionine codon of the grpE gene overlaps the TGA termination codon of the dnaK gene, in a −1 reading frame, suggesting that the two genes are translationally coupled. The dnaJ gene and ORF1, on the other hand, are separated from the surrounding genes by 232- and 159-bp intergenic regions, respectively.
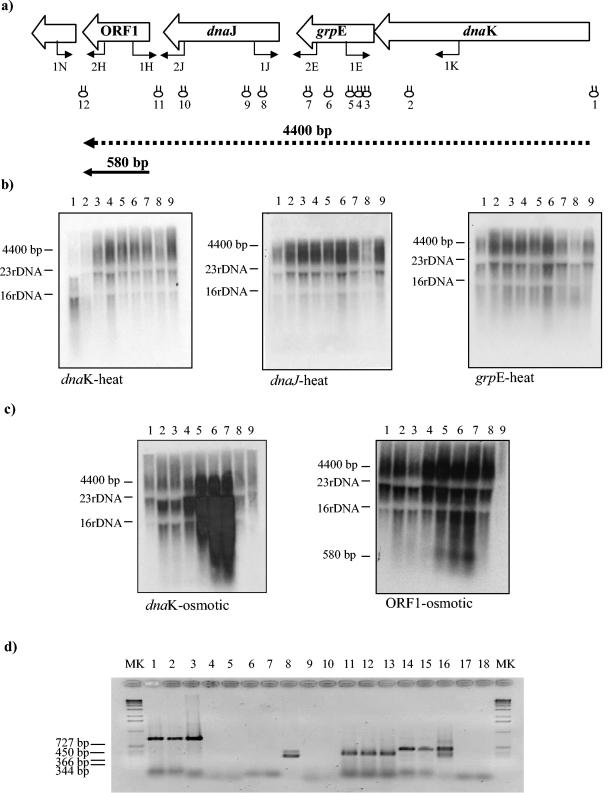
Analysis of the DNA sequences of the dnaK operon revealed several inverted repeats which could form potential stem-loop structures and thereby serve as cleavage and/or transcription terminator sites. These regions are shown in Fig. 2a as stem-loop structures above the genes and are designated 1 though 12. Stem-loop structures 1 and 12 may serve as rho-independent transcription terminators (since they occur outside the structural operon), whereas stem-loop structure 11 may represent a potential processing site while stem-loop structures 2 to 10 may be degradation sites (as they are situated within structural genes).
FIG. 2.
Transcriptional organization of the B. breve UCC 2003 dnaK operon. (a) Schematic diagram of the B. breve UCC 2003 dnaK operon. Hairpin symbols indicate predicted secondary structures. Transcripts are indicated by arrows, which point toward the 3′ end of the mRNA. (b) Northern blot analysis of B. breve UCC 2003 dnaK operon performed by using total mRNA isolated from cultures grown at 37°C (lanes 1 to 5) and 43°C (lanes 6 to 9) for the following lengths of time after a temperature shift: none (beginning of the experiments) (lanes 1), 25 min (lanes 2 and 6), 50 min (lanes 3 and 7), 100 min (lanes 4 and 8), and 150 min (lanes 5 and 9). The probes used are indicated below the panels. (c) Northern blot analysis of B. breve UCC 2003 dnaK operon performed by using total mRNA isolated from cultures grown in the absence of NaCl (lanes 1)and under hyperosmotic conditions (lanes 2 to 9) with the following NaCl concentrations and for the following times: 0.3 M NaCl for 50 min (lane 2), 0.3 M NaCl for 100 min (lane 3), 0.5 M NaCl for 50 min (lane 4), 0.5 M NaCl for 100 min (lane 5), 0.7 M NaCl for 50 min (lane 6), 0.7 M NaCl for 100 min (lane 7), 1.5 M NaCl for 50 min (lane 8), and 1.5 M NaCl for 100 min (lane 9). The probes used are indicated below the panels. (d) Products generated by RT-PCR. PCR products were obtained with primers spanning the intergenic regions between dnaK and grpE (primers 1E and 1K) (lanes 1 to 5), between ORF1 and ORFX (primers 1N and 2H) (lanes 6 to 8), between grpE and dnaJ (primers 1J and 2E) (lanes 9 to 13), and between dnaJ and ORF1 (primers 1H and 2J) (lanes 14 to 18). The positions of the primer pairs used are shown in panel a. Products of the PCR were derived under the following conditions: cDNA prepared from B. breve RNA (lanes 1, 2, 6, 7, 11, 12, 14, and 15), a negative control in which B. breve RNA was used but reverse transcriptase was omitted (lanes 4, 9, and 17), and no template (lanes 5, 10, and 18). A positive control contained B. breve DNA (lanes 3, 8, 13, and 16). Lanes MK contained DNA molecular marker X (Roche).
Transcription mapping of the dnaK operon.
In order to determine transcription patterns for the dnaK operon, total RNA extracted from B. breve UCC 2003 cells grown under unstressed and stressed conditions was hybridized with DNA probes targeting the four genes of the dnaK operon. The results indicated that a long transcript is synthesized, which is subject to extensive degradation, as indicated by the presence of a conspicuous smear in all blots (Fig. 2). A probe encompassing the dnaK gene revealed a smear of transcripts, the largest of which had an estimated size of 4.4 kb. This transcription pattern was also observed for all other genes found in the dnaK operon, suggesting that the four genes are transcribed as a single polycistronic mRNA (Fig. 2a and b).
Interestingly, when an ORF1-specific probe was used, a smaller hybridization signal corresponding to a 580-bp transcript was observed in addition to the smear of transcripts (Fig. 2b).
Northern blot analysis provided evidence that the ORF1, dnaJ, grpE, and dnaK genes constitute an operon since they appear to be transcribed as a polycistronic mRNA. Further proof for the existence of a polycistronic transcript was provided by RT-PCR experiments (Fig. 2d). cDNA templates were obtained by RT of total B. breve UCC 2003 mRNA isolated under heat stress conditions (Fig. 2d, lanes 2, 7, 12, and 15) or osmotic stress conditions (Fig. 2d, lanes 1, 6, 11, and 14). Each cDNA was used as the template in subsequent PCRs with different combinations of primer pairs spanning the dnaK, grpE, dnaJ, and ORF1 genes (Fig. 2a). RNA, which was prepared in the presence of heat and osmotic stresses, produced RT-PCR amplicons that comigrated with the products obtained when chromosomal DNA of B. breve UCC 2003 was used as a positive control (Fig. 2d, lanes 3, 8, 13, and 16). The RT-PCR products were also shown to be dependent on RNA in the cDNA preparation; in fact, no products were observed with mock cDNA prepared in the absence of reverse transcriptase (Fig. 2d, lanes 4, 9, and 17). Furthermore, no RT-PCR product was observed in the absence of any template (Fig. 2d, lanes 5, 10, and 18).
Northern blot analysis was performed by using RNA collected from cells following heat shock and osmotic stress. The hybridization signals exhibited the maximum level of intensity for RNA samples collected following exposure for 100 min to a medium containing 0.7 M NaCl (Fig. 2c, lanes 7). On the other hand, after exposure to heat stress, transcription of the dnaK operon of B. breve UCC 2003 appeared to be only weakly activated (Fig. 2b).
Induction of dnaK operon mRNA in response to environmental stresses.
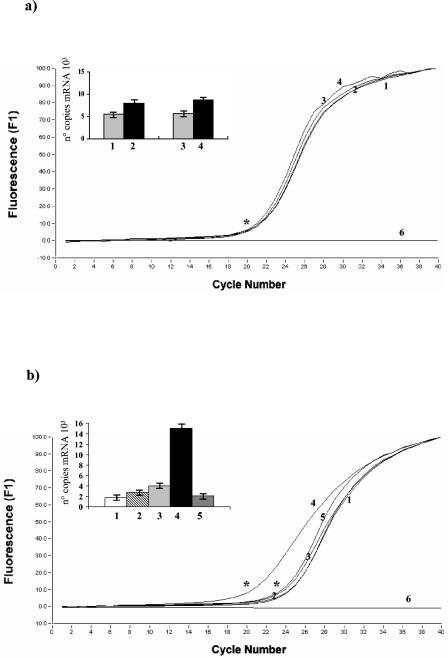
To further investigate the extent to which expression of the B. breve dnaK operon is modulated by an osmotic or temperature upshift, quantitative real-time PCR assays were performed. PCR products, obtained by using cDNA samples from different times as templates, were amplified based on the grpE and ORF1 sequences. The purified PCR products were used as standards, and dilutions of each standard were used to generate a standard curve. The sensitivity of the LightCycler PCR was estimated to be 10 copies of the grpE and ORF1 sequences, which was the cutoff point for accurate detection of DNA. Dilutions corresponding to copy numbers ranging from 101 to 108 were used in all LightCycler PCRs as standards. Using the LightCycler software, we determined the numbers of copies of the grpE and ORF1 transcripts.
In a previous study the heat shock response in B. breve UCC 2003 was evaluated (37). A slot blot hybridization procedure was used to test total RNA, which was isolated from B. breve UCC 2003 cultures grown for different lengths of time at temperatures ranging from 37 to 50°C. Based on the intensity of the hybridization signal, the most effective temperature for inducing heat shock gene expression was 43°C (37). In contrast to what was previously observed for the dnaK gene in Bifidobacterium adolescentis and B. longum (26), a relatively low level of heat induction of grpE and ORF1 gene transcription was observed. A 1.4-fold increase in the copy number of grpE transcripts was observed when cultures were shifted from 37 to 43°C for 150 min (Fig. 3a). Similarly, a 1.5-fold upshift of the ORF1 transcripts was observed upon heat stress (data not shown). Conversely, the levels of grpE and ORF1 mRNAs dramatically increased following osmotic shock. In fact, an osmotic upshift to 0.7 M NaCl in the media led to 15- and 14-fold increases in the grpE and ORF1 mRNAs, respectively (Fig. 3b and data not shown).
FIG. 3.
Osmotic and heat shock induction of the B. breve UCC 2003 grpE and ORF1 sequences. Total RNA was isolated from B. breve UCC 2003 following exposure to various extremes of osmolarity and temperature for a specific length of time and analyzed by quantitative real-time PCR assays. (a) Curves indicating the PCR amplification cycle profiles of the grpE sequences obtained by using cDNA samples from different times after heat shock as templates. The histogram indicates the numbers of copies of grpE mRNAs for the specific samples. The cDNAs used were synthesized by using RNA collected from cultures grown at 37°C (samples 1 to 2) and 43°C (lanes 3 to 4) for the following lengths of time after a temperature shift: none (beginning of the experiments) (sample 1), 150 min (samples 2 and 4), and 50 min (sample 3). Sample 6 was the negative control (no template). (b) Curves indicating the PCR amplification cycle profiles of the grpE sequences obtained by using cDNA samples from different NaCl concentrations as templates. An asterisk indicates the crossing point. The histogram indicates the numbers of copies of grpE mRNAs for the specific samples. The cDNAs used were synthesized by using RNA collected from cultures grown in media with the following NaCl concentrations: no NaCl (sample 1), 0.3 M NaCl (sample 2), 0.5 M NaCl (sample 3), 0.7 M NaCl (sample 4), and 1.5 M NaCl (sample 5). Sample 6 was the negative control (no template).
Identification of the transcription start site of the dnaK operon.
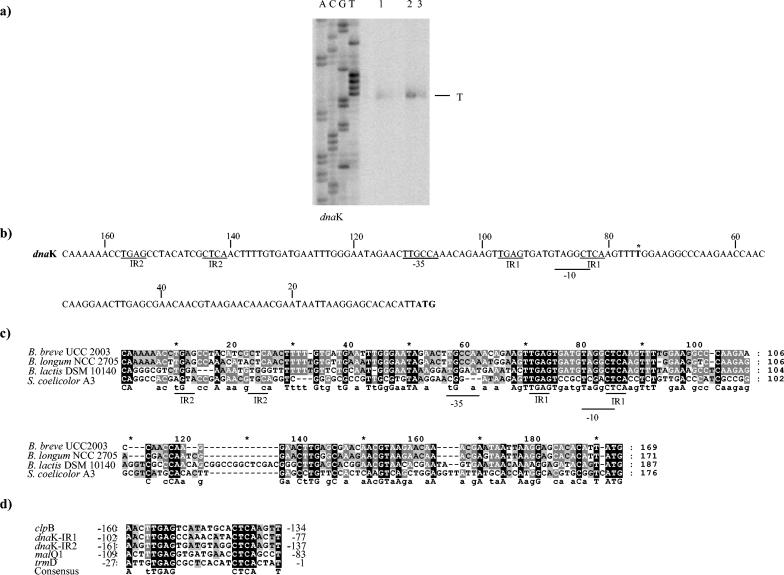
To determine the transcriptional start point of the dnaK operon, primer extension analyses were performed with RNA extracted from B. breve cells grown at 37°C or subjected to heat or osmotic shock (Fig. 4). A major extension product was identified 75 nucleotides 5′ to the translational start site of the dnaK gene (Fig. 4a). The transcription start sites were in the same positions at 37°C, at 43°C, and under osmotic shock conditions (Fig. 4a). The analysis of the putative promoter region of dnaK revealed a potential promoter-like sequence resembling the consensus −10 hexamer and −35 box (Fig. 4c) (36, 37). A specific primer extension product was also mapped within the intercistronic region between ORF1 and dnaJ (Fig. 4b). However, screening for promoter consensus sequences of ORF1 did not reveal any typical E. coli −10 and −35 sequences, and computer analysis predicted a high degree of RNA secondary structures in this region. Therefore, we cannot rule out the possibility that the mapped 5′ end actually represents a cleavage site of the polycistronic mRNA.
FIG. 4.
Determination of the dnaK transcription initiation site of B. breve UCC 2003 by primer extension analysis. (a) Primer extension results obtained by using an oligonucleotide targeting the 5′ end of dnaK. Lane 1, primer extension product obtained by using mRNA extracted from cells grown at 37°C; lane 2, primer extension product obtained by using mRNA extracted from cells grown with 0.7 M NaCl; lane 3, primer extension product obtained by using mRNA extracted from cells grown at 43°C. (b) Putative promoter sequences for the dnaK gene. The −10 and −35 putative hexamers are underlined; boldface type with an asterisk indicates the transcription start point; and boldface type without an asterisk indicates the start codon. IR1 and IR2 are putative regulator sequences. (c) Comparison of the promoter regions of four dnaK genes from different members of the Actinobacteridae. Shaded residues indicate that the level of identity for the sequences is 50% or higher. (d) Alignment of consensus IR sequences from B. longum NCC 2705. Identical nucleotides are shaded. The numbers indicate positions relative to the translational start site.
Primer extension experiments were also performed to investigate the existence of other transcription initiation sites upstream of the assumed start codons of the dnaJ and grpE genes, but all these attempts failed to generate extension products. This suggests that these genes do not possess their own promoter regions, which is consistent with the presence of a polycistronic 4.4-kb transcript.
Comparison of the upstream regulatory regions of dnaK operons from Actinobacteridae.
The upstream regions of the dnaK genes of two bifidobacterial strains and one Streptomyces strain were aligned in an attempt to identify putative regulated elements (Fig. 4c). For completeness, we determined by inverse PCR the putative promoter region of the dnaK gene for a more distantly related Bifidobacterium species (B. lactis). As Fig. 4c shows, a consensus promoter sequence could be deduced from the three sequences. The putative −10 region was conserved in all bifidobacterial sequences, while the putative −35 box was less conserved. Moreover, a clear consensus inverted repeat sequence (TGAGN9CTCA), which was designated IR, was identified. Notably, the promoter region of the dnaK gene in B. breve and B. longum contained two copies of this IR sequence (IR1 and IR2), whereas B. lactis possessed only a single copy. Interestingly, this inverted repeat resembled a regulatory structure designated HAIR (CTTGAGTN7ACTCAAG) (5), which has been demonstrated to be the operator sequence of the heat shock protein regulator (HspR) in high-G+C-content gram-positive bacteria (11). Preliminary screening of the publicly available B. longum NCC 2705 genome sequence for the consensus sequence (TGAGN9CTCA) revealed the presence of this inverted repeat in the intergenic regions of a classical stress-induced gene, such as clpB (Fig. 4d). Furthermore, this consensus sequence was identified in the promoter regions of the trmD and malQ1 genes, which are predicted to encode a methyltransferase and a glucanotransferase, respectively (25).
Phylogenetic analysis.
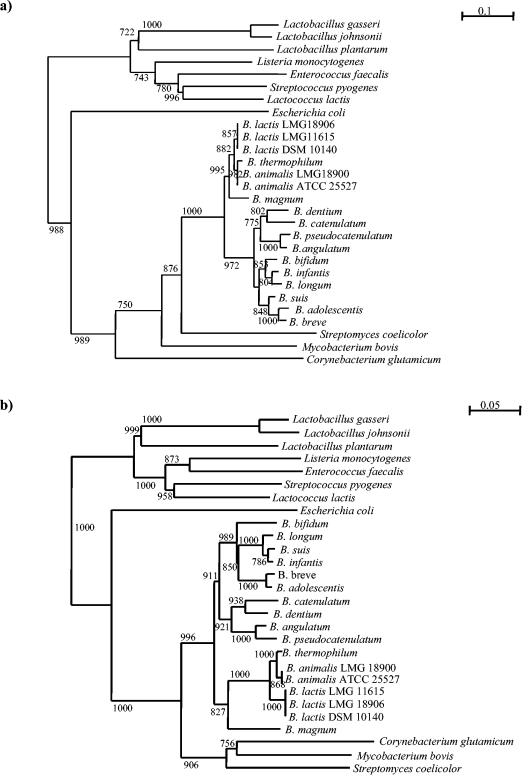
The grpE and dnaK DNA sequences of a number of selected bacterial species were aligned and compared. Two highly conserved regions for each gene were identified, and a pair of primers (GRE-UNI and GRE-REV) amplifying a 285-bp region was designed for the grpE gene. Furthermore, another primer pair (DNAK-1 and DNAK-2) was designed to amplify a 535-bp region. These primers allowed amplification of grpE and dnaK sequences from 14 Bifidobacterium species. The alignments of grpE DNA and dnaK sequences were used to generate two phylogenetic trees by the neighbor-joining method (Fig. 5). The data were supported by the bootstrap values obtained. For completeness we included in the analysis the grpE DNA and dnaK sequences of other strains belonging to different genera representing the lactic acid bacterial group (e.g., Lactococcus, Streptococcus, Lactobacillus, and Enterococcus) and other gram-positive and gram-negative bacteria with high and low G+C contents, including some Actinobacteridae (e.g., Corynebacterium, Streptomyces, and Mycobacterium). These trees showed two major clusters representing the low-G+C-content gram-positive bacteria (the genera Lactobacillus, Lactococcus, Streptococcus, and Enterococcus) and the high-G+C-content gram-positive bacteria (the genus Bifidobacterium).
FIG. 5.
Phylogenetic trees obtained by using the grpE gene sequences (a) and the dnaK gene sequences (b). The bars indicate phylogenetic distances. Bootstrap values for a total of 1,000 replicates are indicated at the nodes.
A comparison of the phylogenetic trees based on the grpE and dnaK sequences revealed very similar phylogenetic topologies (Fig. 5).
A phylogenetic tree was also constructed on the basis of the 16S rRNA gene sequences available in databases. The phylogenetic positions of bacteria based on 16S rRNA gene sequences were generally in agreement with grpE- and dnaK-based phylogenies (data not shown). Interestingly, closely related strains which had nearly identical 16S rRNA gene sequences (e.g., B. animalis and B. lactis, B. catenulatum and B. pseudocatenulatum, or B. longum and Bifidobacterium infantis) clearly branched separately in the grpE and dnaK sequence-based tree (Fig. 5).
The similarity between the partial grpE DNA sequences of bifidobacterial species was less than 96.6% in all cases except B. longum-Bifidobacterium suis and B. infantis (99.8%). The sequence divergence in grpE was 2.7 times greater than that in the 16S rRNA gene, indicating the greater variance of grpE. The dnaK sequences were less divergent than those of grpE by a factor of 1.4. The relationship between the pairwise dissimilarities of grpE and other genes was linear, which suggests that the degree of sequence divergence was proportional to the time needed for speciation. In addition, the pairwise distances between the dnaK and 16S rRNA genes exhibited a close linear correlation (data not shown). Unfortunately, the dnaK gene failed to act as a discriminating molecular marker for typing purposes. In fact, the phylogenetic distances between B. lactis strains, as well as those between B. animalis strains, are virtually zero.
Most of the base substitutions in the grpE and dnaK genes were synonymous; i.e., they did not result in amino acid changes but constituted nucleotide signatures that may be used for tracing bifidobacterial species.
Specific identification of bifidobacteria by melting point analysis and by using species-specific primers.
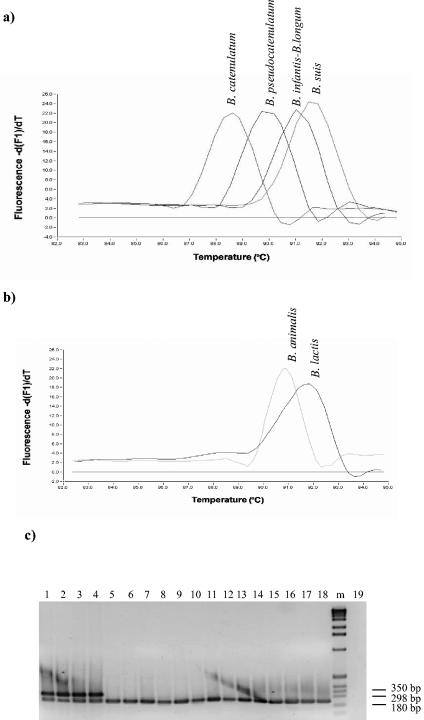
Following amplification of 285-bp grpE sequences of the individual bifidobacterial strains, a melting curve of the amplicon was generated by continuously monitoring the fluorescence while the temperature was raised from 82 to 95°C. The Tms for the partial grpE gene sequences remained within a narrow range, but the Tm was different for each of the species examined. The only exceptions were B. longum and B. infantis, which showed overlapping melting curves. Characteristic melting curves for closely related taxa, like B. catenulatum and B. pseudocatenulatum or B. longum and B. suis, as well as for B. lactis and B. animalis, are shown in Fig. 6a and b. The Tm of B. lactis DSM 10140 was 92°C, which was unequivocally higher than that of B. animalis ATCC 25527 (90.8°C) (Fig. 6b). Similar findings were obtained when other B. lactis and B. animalis strains were used (data not shown). In addition, the assay clearly differentiated B. catenulatum from B. pseudocatenulatum, which had Tms of 88.63 and 89.73°C, respectively.
FIG. 6.
Determination of the Tms for closely related bifidobacterial species in grpE real-time PCRs. (a and b) −dF/dT (change in fluorescence/change in temperature) plotted against temperature. (c) PCR products of various Bifidobacterium species obtained by using a multiplex PCR approach with B. lactis species-specific PCR primers and Bifidobacterium genus-specific primers. Lane 1, B. lactis DSM 10140; lane 2, B. lactis LMG 11615; lane 3, B. lactis LMG 18906; lane 4, B. lactis LMG 11580; lane 5, B. animalis LMG 18900; lane 6, B. animalis ATCC 25527; lane 7, B. angulatum JCM 7096; lane 8, B. pseudocatenulatum JCM 1200; lane 9, B. breve UCC 2003; lane 10, B. magnum JCM 1218; lane 11, B. thermophilum JCM 1207; lane 12, B. pullorum DSM 20433; lane 13, B. bifidum JCM 1255; lane 14, B. dentium JCM 1195; lane 15, B. suis JCM 1269; lane 16, B. infantis JCM 1210; lane 17, B. adolescentis JCM 1275; lane 18, B. catenulatum JCM 1194; lane 19, negative control. Lane m, 1-kb DNA ladder.
The analysis of the partial dnaK gene sequences of two very closely related taxa, B. lactis and B. animalis, from which the B. lactis-specific primers b-dnaK1 and b-dnaK2 were designed, revealed few nucleotide differences. Primers gro-1 and gro-2 (37), which target two conserved regions within the groES gene of the genus Bifidobacterium, were also included as a positive control in the same PCR. When a multiplex PCR was performed with this mixture of four primers, two amplicons (290 and 200 bp) were detected only in the presence of DNA isolated from B. lactis. Amplification of all other Bifidobacterium DNAs resulted in only one amplicon (200 bp). The results of these PCR assays, based on the dnaK sequences data, are shown in Fig. 6c. The 290-bp amplicon was apparent for three B. lactis strains (Fig. 6c, lanes 1, 2, 3, and 4) but not at all for any of the B. animalis strains (Fig. 6c, lanes 5 and 6). Furthermore, we confirmed the overall absence of PCR amplicons for all other bifidobacterial type strains (Fig. 6c and data not shown).
DISCUSSION
The dnaK operon of B. breve UCC 2003 was identified and shows consistent levels of similarity to previously described dnaK operons (28). Our results clearly indicate that the B. breve dnaK operon consists of four genes: dnaK, grpE, dnaJ, and ORF1. This gene organization is identical to that of presumed dnaK regions of other sequenced Bifidobacterium species and to that of other available sequences of members of the Actinobacteridae. In fact, for S. coelicolor A3 it has been demonstrated that the dnaK operon is tetracistronic and includes the dnaK, grpE, and dnaJ genes and a gene encoding a heat shock regulator (HspR) (7).
Northern blots indicated that a 4.4-kb transcript encompassing the complete operon (dnaK-grpE-dnaJ-ORF1) is produced. This primary transcript seems to be very unstable and subject to rapid processing. It is possible that the many stem-loop structures located within the coding regions, as well as between the dnaJ and grpE genes, may act as processing sites. A similar situation has previously been described in B. subtilis (16), where four hairpin-like structures were implicated in the processing of dnaKJ mRNAs. In addition, in both Clostridium acetobutylicum and Nitrosomonas europaea intensive specific mRNA degradation of the polycistronic dnaK transcript has been described (3, 17). The significance of this phenomenon is unknown, but cleavage at processing sites may lead to a product with an enhanced half-life (16). Moreover, it has been postulated that the processing of dnaK mRNAs may be an important component of regulation of expression of the genes in the dnaK operon (3, 20).
Although relatively little is known about the heat and osmotic stress responses, a previous report (26) and the present study demonstrated that the dnaK operon is part of the general stress response of bifidobacteria. We are primarily interested in the role of the DnaK chaperone machine in the heat and osmotic stress responses. In fact, identification of the genetic basis of heat and osmotic resistance for industrially applicable bifidobacteria is highly desirable for selecting strains that resist stressful conditions encountered during food manufacture.
Transcription of the dnaK operon from B. breve occurs at 37°C and in the absence of NaCl in the medium and is increased upon a shift to higher salt concentrations. The mRNA levels for the dnaK operon were shown to increase 14- to 15-fold following an osmotic challenge, while the change in transcriptional levels was shown to be not significant when cells were subjected to heat stress (approximately 1.5-fold). Interestingly, in contrast to B. breve, the dnaK operons of B. adolescentis and B. longum were shown to be strongly induced upon heat stress (25). These results indicate that in bifidobacteria the amount and type of chaperones necessary for an adaptive response to heat stress are variable. The observed overlap between the heat and osmotic stress responses have also been described for other gram-positive bacteria (e.g., L. lactis and Streptococcus mutans), in which heat-inducible proteins are also involved in the osmotic stress response, although the induction factors for DnaK chaperones in heat-shocked or osmotically shocked cells vary (4, 18, 35).
In B. breve UCC 2003 the level of temperature-induced activation of the dnaK operon appeared to be much lower than the levels of temperature-induced activation of the groEL and groES operons, which are induced approximately 8- and 12-fold (37). The groESL and dnaK operons of many bacteria, including the groES operon of B. breve UCC 2003, contain a conserved inverted repeat sequence (CIRCE), which is believed to be involved in the transcriptional regulation of these genetic units (28, 29, 37, 46). The dnaK operon of B. breve UCC 2003 does not appear to contain such a CIRCE sequence near its transcription start site or upstream of it.
Of course, we cannot rule out the possibility that when applied under different regimens (e.g., different exposure times and/or different severity of stress), some of these stimuli might result in higher expression of dnaK. We will address this question in future work and also test the influence of additional stresses.
The comparison of putative regulatory elements for the dnaK operon of three bifidobacteria allowed identification of a putative consensus promoter sequence (IR), which exhibited a consistent degree of homology with the HspR binding site (HAIR). The HAIR sequences were shown to be involved in heat shock regulation in Streptomyces, in Mycobacterium, and in Helicobacter pylori (5, 11, 29). Consequently, it is possible that ORF1 acts as a transcriptional regulator of the dnaK operon by binding to the putative operator sequence (IR). Future research will be focused on verifying this hypothesis by demonstrating direct protein-DNA interactions by means of gel retardation and DNase footprinting experiments.
Screening of the publicly available B. longum NCC 2705 genome sequence revealed that the IR putative operator sequence is located in the promoter region of the clpB gene, which has been extensively described previously as being involved in a stress response. Interestingly, the clpB gene has been shown to be part of the HspR regulon in Streptomyces albus (11). Moreover, the IR putative operator sequence is found in the promoter region of two other genes (trmD and malQ1), both of which have been described as being associated with the stress response of L. lactis (10). This suggests that all these genes may be governed by the same regulatory system.
In this study we investigated the occurrence of the genes encoding the DnaK and GrpE chaperones in different species of the genus Bifidobacterium. These genes are ideal target candidates for diagnostic purposes because they are highly conserved and ubiquitous in bacteria (13, 14). They fulfill all of the prerequisites for suitable phylogenetic markers, such as very high genetic stability and a wide distribution. These alternative molecular markers might corroborate and help to complete the evolutionary history of various bifidobacterial species. Polyphasic taxonomy (35) has been recognized by the International Committee on Systematic Bacteriology as a new tool for describing species and for revising the current nomenclature of some bacterial groups. In view of its demonstrated effectiveness, sequence analysis of protein-encoding genes (e.g., grpE and dnaK genes) and use of these genes as alternative phylogenetic markers could be added to the arsenal of rRNA sequence databases and to the relatively small groEL (37), recA (21, 40), atpD (38), and tuf (39) sequence databases. The data presented here show that the phylogenetic positions of bifidobacteria based on grpE sequences are generally in agreement with the dnaK-based phylogeny, suggesting that these genes had the same evolutionary development. Moreover, the grpE and dnaK trees had a topology similar to that obtained with the 16S rRNA genes. Nevertheless, in contrast to the 16S rRNA gene tree, the grpE and dnaK trees provide higher phylogenetic resolution for closely related taxa.
The grpE and dnaK gene analysis has many advantages compared to analyses of other genes. In fact, these genes are common in bifidobacteria, are present as single copies on the genome (25), and contain conserved nucleotide signatures that are suitable targets for PCR primers. In spite of the sequence's limited size, grpE and dnaK sequence analyses may allow resolution of phylogenetic lineages of bifidobacteria at the species and subspecies levels. In fact, they have an advantage over 16S rRNA gene-based differentiation of species in that they allow differentiation of B. lactis and B. animalis, as well as B. catenulatum and B. pseudocatenulatum.
In conclusion, the use of several markers, such as grpE, in combination with dnaK-or 16S rRNA gene-based tracing techniques (41, 42, 43, 44), may be necessary for reliable identification and phylogenetic analysis of Bifidobacterium species. Such an approach should increase the accuracy of molecular identification of bifidobacterial species in microbial ecology studies.
Although the work presented here can only be considered a first step toward an understanding of stress responses in bifidobacteria, detailed analysis of the regulatory mechanisms of the B. breve UCC 2003 dnaK gene and other stress-induced genes represents a suitable model system for the genus Bifidobacterium.
Acknowledgments
This work was financially supported by Enterprise Ireland (grant BR/1998/202), by the Higher Education Authority Programme for Research in Third Level Institutions, by the Science Foundation Ireland Alimentary Pharmabiotic Centre located at University College Cork, and by Marie Curie Development Host Fellowship HPMD-2000-00027 to M.V.
We thank J. O'Mahony and S. Cusack for excellent discussions and technical assistance with the LightCycler PCR technology. We also thank T. Cross and J. Coughlan of the Department of Zoology, Ecology and Plant Science, University College Cork for allowing us to use the LiCor sequencer.
REFERENCES
- 1.Ahmad, S., A. Selvapandiyan, and R. K. Bratnagar. 2000. Phylogenetic analysis of gram-positive bacteria based on grpE, encoded by the dnaK operon. Int. J. Syst. Bacteriol. 50:1761-1766. [DOI] [PubMed] [Google Scholar]
- 2.Antonopoulos, D. A., W. M. Russel, and B. A. White. 2003. Phylogenetic reconstruction of Gram-positive organisms based on comparative sequence analysis of molecular chaperones from the ruminal microorganism Ruminococcus flavefaciens FD-1. FEMS Microbiol. Lett. 227:1-7. [DOI] [PubMed] [Google Scholar]
- 3.Bahl, H., H. Muller, S. Behrens, H. Joseph, and F. Narberhaus. 1995. Expression of heat shock genes in Clostridium acetobutylicum. FEMS Microbiol. Rev. 17:341-348. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi, A. A., ad F. Baneyx. 1999. Hyperosmotic shock induces the σ32 and σE stress regulon of Escherichia coli. Mol. Microbiol. 34:1029-1038. [DOI] [PubMed] [Google Scholar]
- 5.Bucca, G., A. M. E. Brassington, H. J. Schonfeld, and C. P. Smith. 2000. The HspR regulon of Streptomyces coelicolor: a role for the DnaK chaperone as a transcriptional co-repressor. Mol. Microbiol. 38:1093-1103. [DOI] [PubMed] [Google Scholar]
- 6.Bucca, G., G. Ferina, A. M. Puglia, and C. P. Smith. 1995. The dnaK operon of Streptomyces coelicolor encodes a novel heat-shock protein which binds to the promoter region of the operon. Mol. Microbiol. 17:663-674. [DOI] [PubMed] [Google Scholar]
- 7.Bucca, G., Z. Hindle, and C. P. Smith. 1997. Regulation of the dnaK operon of Streptomyces coelicolor A3(2) is governed by HspR, an autoregulatory repressor. J. Bacteriol. 179:5999-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukau, B., and A. L. Horwich. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92:351-366. [DOI] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package), version 3.5c. Department of Genetics, University of Washington, Seattle.
- 10.Frees, D., P. Varmanen, and H. Igmer. 2001. Stress tolerance and proteolysis in L. lactis. Mol. Microbiol. 41:93-103. [DOI] [PubMed] [Google Scholar]
- 11.Grandvalet, C., V. Crecy-Lagard, and P. Mazodier. 1999. The ClpB ATPase of Streptomyces albus G belongs to the HspR heat shock regulon. Mol. Microbiol. 31:521-532. [DOI] [PubMed] [Google Scholar]
- 12.Grandvalet, C., G. Rapoport, and P. Mazodier. 1998. hrcA, encoding the repressor of the groEL genes in Streptomyces albus G, is associated with a second dnaJ gene. J. Bacteriol. 180:5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta, R. S. 1995. Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of proteins and origin of eukaryotic cells. Mol. Microbiol. 15:1-11. [DOI] [PubMed] [Google Scholar]
- 14.Gupta, R. S. 2002. Phylogeny of bacteria: are we now close to understanding it? ASM News 68:284-291. [Google Scholar]
- 15.Hartl, F. U. 1996. Molecular chaperones in cellular protein folding. Nature 381:571-580. [DOI] [PubMed] [Google Scholar]
- 16.Homuth, G., S. Masuda, A. Mogk, Y. Kobayashi, and W. Shumann. 1997. The dnaK operon of Bacillus subtilis is heptacistronic. J. Bacteriol. 179:1153-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iizumi, T., and K. Nakamura. 1997. Cloning, nucleotide sequence, and regulatory analysis of the Nitrosomonas europaea dnaK gene. Appl. Environ. Microbiol. 63:1777-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kilstrup, M., S. Jacobsen, K. Hammer, and F. K. Vogensen. 1997. Induction of heat shock proteins DnaK, GroEL, and GroES by salt stress in Lactococcus lactis. Appl. Environ. Microbiol. 63:1826-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kullen, M., L. J. Brady, and D. J. O'Sullivan. 1997. Evaluation of using a short region of the recA gene for rapid and sensitive speciation of dominant bifidobacteria in the human large intestine. FEMS Microbiol. Lett. 154:377-383. [DOI] [PubMed] [Google Scholar]
- 20.Lemos, J. A. C., Y. Y. M. Chien, and R. A. Burne. 2000. Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J. Bacteriol. 183:6074-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lievin, V, I. Pfeiffer, S. Hudault, F. Rochat, D. Brassart, J. R. Neeser, and A. L. Servin. 2000. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut 47:646-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, H., Y. Li, X. Huang, Y. Kawamura, and T. Ezaki. 2003. Use of the dnaJ gene for the detection and identification of all Legionella pneumophila serogroups and description of the primers used to detect 16S rDNA sequences of major members of the genus Legionella. Microbiol. Immunol. 47:859-869. [DOI] [PubMed] [Google Scholar]
- 23.O'Mahony, A., T. F. O'Sullivan, Y. Walsh, A. Vaughan, M. Maher, G. F. Fitzgerald, and D. van Sinderen. 2000. Characterization of antimicrobial producing lactic acid bacteria from malted barley. J. Int. Brew. 106:403-410. [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt, G., and R. Zink. 2000. Basic features of stress response in three species of bifidobacteria: B. longum, B. adolescentis, and B. breve. Int. J. Food Microbiol. 55:41-45. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt, G., C. Hertel, and W. P. Hammes. 1999. Molecular characterisation of the dnaK operon of Lactobacillus sakei LTH681. Syst. Appl. Microbiol. 22:321-328. [DOI] [PubMed] [Google Scholar]
- 28.Segal, G., and E. Z. Ron. 1996. Regulation and organization of the groE and dnaK operons in Eubacteria. FEMS Microbiol. Lett. 138:1-10. [DOI] [PubMed] [Google Scholar]
- 29.Servant, P., and P. Mazodier. 2001. Negative regulation of the heat shock response in Streptomyces. Arch. Microbiol. 176:237-242. [DOI] [PubMed] [Google Scholar]
- 30.Shultz, A., B. Tzschaschel, and W. Shumann. 1994. Isolation and analysis of mutants of the dnaK operon of Bacillus subtilis. Mol. Microbiol. 15:421-429. [DOI] [PubMed] [Google Scholar]
- 31.Takewaki, S. I., K. Okuzumi, H. Ishiko, K. I. Nakahara, A. Ohkubo, and R. Nagai. 1993. Genus-specific polymerase chain reaction for the mycobacterial dnaJ gene and species-specific oligonucleotide probes. J. Clin. Microbiol. 31:446-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tannock, G. W. 1994. The acquisition of the normal microflora of the gastrointestinal tract, p 1-16. In S. A. Gibson (ed.), Human health: the contribution of microorganisms. Springer, London, United Kingdom.
- 33.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van de Guchte, M., P. Serror, C. Chervaux, T. Smokvina, S. D. Ehrlich, and E. Manguin. 2002. Stress responses in lactic acid bacteria. Antonie Leeuwenhoek 82:187-216. [PubMed] [Google Scholar]
- 35.Vandamme, P., B. Pot, M. Gillis, P. de Vos, K. Kersters, and J. Swings. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60:407-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ventura, M., D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie Leeuwenhoek 86:205-223. [DOI] [PubMed] [Google Scholar]
- 37.Ventura, M., C. Canchaya, R. Zink. G. F. Fitzgerald, and D. van Sinderen. 2004. Characterization of the groEL and groES loci in Bifidobacterium breve UCC 2003: genetic, transcriptional and phylogenetic analyses. Appl. Environ. Microbiol. 70:6197-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ventura, M., C. Canchaya, D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Bifidobacterium lactis DSM 10140: identification of the atp (atpBEFHAGDC) operon and analysis of its genetic structure, characteristics, and phylogeny. Appl. Environ. Microbiol. 70:3110-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ventura, M., C. Canchaya, V. Meylan, T. R. Klaenhammer, and R. Zink. 2003. Analysis, characterization, and loci of the tuf genes in Lactobacillus and Bifidobacterium and their direct application for species identification. Appl. Environ. Microbiol. 69:6908-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ventura, M., and R. Zink. 2003. Comparative sequence analysis of the tuf and recA genes and restriction fragment length polymorphism of the internal transcribed spacer region sequences supply additional tools for discriminating Bifidobacterium lactis from Bifidobacterium animalis. Appl. Environ. Microbiol. 69:7517-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ventura, M., V. Meylan, and R. Zink. 2003. Identification and tracing of Bifidobacterium species by enterobacterial repetitive intergenic consensus sequences. Appl. Environ. Microbiol. 69:4296-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ventura, M., and R. Zink. 2002. Rapid identification, differentiation, and proposed new taxonomic classification of Bifidobacterium lactis. Appl. Environ. Microbiol. 68:6429-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ventura, M., M. Elli, R. Reniero, andR. Zink. 2001. Molecular microbial analysis of Bifidobacterium isolates from different environments by the species-specific amplified ribosomal DNA restriction analysis (ARDRA). FEMS Microbiol. Ecol. 36:113-121. [DOI] [PubMed] [Google Scholar]
- 44.Ventura, M., R. Reniero, and R. Zink. 2001. Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolates by a combined multiplex-PCR approach. Appl. Environ. Microbiol. 67:2760-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan, G., and S. Wong. 1995. Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J. Bacteriol. 177:6462-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuber, U., and W. Schumann. 1994. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J. Bacteriol. 176:1353-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]