Abstract
The development of ectomycorrhizal symbiosis leads to drastic changes in gene expression in both partners. However, little is known about the spatial regulation of symbiosis-regulated genes. Using cDNA array profiling, we compared the levels of expression of fungal genes corresponding to approximately 1,200 expressed sequenced tags in the ectomycorrhizal root tips (ECM) and the connected extraradical mycelium (EM) for the Paxillus involutus-Betula pendula ectomycorrhizal association grown on peat in a microcosm system. Sixty-five unique genes were found to be differentially expressed in these two fungal compartments. In ECM, a gene coding for a putative phosphatidylserine decarboxylase (Psd) was up-regulated by 24-fold, while genes coding for urea (Dur3) and spermine (Tpo3) transporters were up-regulated 4.1- and 6.2-fold in EM. Moreover, urea was the major nitrogen compound found in EM by gas chromatography-mass spectrometry analysis. These results suggest that (i) there is a spatial difference in the patterns of fungal gene expression between ECM and EM, (ii) urea and polyamine transporters could facilitate the translocation of nitrogen compounds within the EM network, and (iii) fungal Psd may contribute to membrane remodeling during ectomycorrhiza formation.
Soils of temperate forests show both spatial and temporal heterogeneities in nutrient availability, particularly the availability of nitrogen, which is essential for growth processes (50). To access more nutrients, trees have developed a mycorrhizal strategy, in which the expanding mycelium of ectomycorrhizal fungi is able to explore a larger soil volume than the root alone (45). The ectomycorrhizal association is therefore a great advantage for controlling plant nutrient status and growth.
The well-characterized structure of Paxillus involutus-Betula pendula ectomycorrhizae (7) is formed by three components: a sheath enclosing the root, an intraradicular network of hyphae, and an outwardly growing system of hyphae, which form essential connections with both the soil and the fruit bodies (50). The ectomycorrhizal mantle provides a structure suitable for nutrient storage and plays a key role in controlling nutrient transfer between the fungus and the plant through its intimate contact with the root surface (35). The extraradical mycelium (EM), which extends from the mantle as single hyphae or linear aggregates of such hyphae, is of additional importance, because these structural attributes form the connection between the mantle and the soil and thus provide pathways for nutrient exchange (41). The positive effect of ectomycorrhizal fungi on plant nutrition could be attributed largely to the activity of EM. The ability of P. involutus to take up and transfer nitrogen compounds to its host, B. pendula Roth, was demonstrated by 15N feeding experiments (14, 18, 23). It was shown that labeled N was incorporated into a range of amino acids in the fungal mycelium at considerable distances from the fungal sheath. The main sinks for assimilated N appeared to be Glu-Gln, Asp-Asn, and Ala (16, 32). In addition, nutrient mobilization from natural organic substrates in the fermentation horizon of forest soils may be a function of the vegetative mycelium of mycorrhizal systems. An increase in the activities of nutrient-mobilizing enzymes in P. involutus colonizing birch litter and a significant decline in the nutrient contents of the colonized litter were demonstrated (4, 37).
Moreover, mycorrhizal roots act as greater sinks for newly fixed 14C-photosynthates than do nonmycorrhizal roots (NMR) in Eucalyptus pilularis-Pisolithus sp. associations, especially in young mycorrhizae (9). These labeled compounds, which are likely to be in the form of trehalose, mannitol, and arabitol, are translocated at rates in excess of 20 cm h−1 through the mycorrhizal mycelium from the host toward the advancing hyphal front (16, 17). The exchange of nutrients and carbohydrates in ectomycorrhizal symbiosis likely follows rules of a simple “fair trade,” probably regulated by signaling substances in response to environmental changes (60). Indeed, it was shown that elevated levels of CO2 increase the trading potential of the plant, whereas elevated levels of mineral N increase the trading potential of the fungus (21).
Conventional molecular approaches and large-scale gene profiling experiments have identified several symbiosis-regulated genes in model systems such as Eucalyptus globulus-Pisolithus microcarpus (59), Tilia platyphyllos-Tuber borchii (40), Pinus sp.-Laccaria bicolor (39), and B. pendula-P. involutus (25). Nevertheless, no global molecular approach has yet been applied in investigations of spatial differences in gene expression to study gene regulation in the different compartments of the symbiosis. Instead, global transcription studies so far conducted have been designed to allow comparisons of gene expression in ectomycorrhizae, root tissues, or pure cultures of fungi in axenic systems. One therefore needs to consider an intact ectomycorrhizal system providing ideal natural simulations for determining mycorrhizosphere-driven nutrient cycling in forest soils by allowing the formation of EM. Such a system was developed by Read and coworkers with mycorrhizal pine and birch seedlings (4, 17) and was used to study nutrient translocation within a symbiotic association as described before (14, 16, 37).
Previous results suggested that the amount of NADP-Gdh in Cenococcum geophilum decreased progressively from the peripheral cells of the sheath to the most internal cells of the Hartig net (5, 11), suggesting that EM possesses primary N-assimilating functions. It was then hypothesized that functional compartmentation occurs in intact ectomycorrhizal symbiosis.
In addition, some studies have provided examples of differential gene expression in the sheath and the Hartig net. Ectomycorrhizae of Amanita muscaria-Populus tremula × tremuloides were dissected (35), and it was found by reverse transcription (RT)-PCR that hexose transporter (AmMst1) expression was enhanced about sixfold in hyphae of the Hartig net compared with those of the fungal sheath. In contrast, phenylalanine ammonium lyase (AmPal) was only barely detectable in the Hartig net but was highly expressed in the fungal sheath. Moreover, as determined by in situ hybridization, Gln synthetase of T. borchii was expressed in symbiosis-engaged hyphae with T. platyphyllos; higher hybridization signals were seen in hyphae that were penetrating root cells (34).
Gene profiling with an intact ectomycorrhizal system including EM should allow the determination of specific cellular functions operating solely at the symbiotic interface or in the external mycelial network, especially in terms of nutrient acquisition and transfer. In this study, we combined the use of a two-dimensional peat microcosm to allow the formation of an intact mycorrhizal network from the P. involutus-B. pendula association (41) and a cDNA array approach to investigate the differential expression of fungal genes in EM and in the ectomycorrhizal root tips (ECM). Amino acid pools were also analyzed by gas chromatography (GC)-mass spectrometry (MS) to assess the nutrient status of the two compartments.
MATERIALS AND METHODS
Growth conditions and ectomycorrhiza synthesis.
The ectomycorrhizal fungus P. involutus (Batsch) Fr. (ATCC 200175) was originally isolated from a fruiting body associated with 15- to 30-year-old B. pendula trees growing on coal waste in Midlothian, Scotland. The fungus was grown on cellophane-covered agar medium containing modified Melin-Norkrans medium (MMN). MMN contained (in milligrams liter−1) the following: KH2PO4, 500; (NH4)2HPO4, 250; CaCl2 · 2H2O, 50; NaCl, 25; MgSO4 · 7H2O, 15; thiamine hydrochloride, 0.1; and FeCl3 · 6H2O, 1. The glucose concentration was 10 g liter−1. For library constructions, Gln at 30 mg liter−1 or (NH4)2HPO4 at 250 mg liter−1 was used. All pHs were adjusted to 5.5.
To synthesize ectomycorrhizae, seeds of birch (B. pendula Roth) from Docksta (Sweden) were stored dried at +5°C until required. They were surface sterilized with 5% (wt/vol) calcium hypochlorite for 30 min, rinsed in several changes of sterile distilled water, and then placed on agar petri dishes to germinate. Two-week-old seedlings were transferred to petri dishes on which the fungus P. involutus had been grown on peat-vermiculite (1:4 [vol/vol]) substrate moistened with double-strength MMN solution and supplemented with malt extract at 10 g liter−1 and glucose at 1.25 g liter−1. Root systems were grown under aseptic conditions, whereas shoot parts were in contact with the atmosphere. Dishes were incubated in a growth chamber maintained with a 16-h photoperiod (150 μmol m−2 s−1); day and night temperature and relative humidity were 22 and 18°C and 85 and 65%, respectively. After 10 weeks, mycorrhizal birch seedlings were transferred to humidified peat microcosms and returned to the growth chamber (16). At 2 weeks after transfer of the seedlings to the peat microcosms, EM and ECM were harvested from four different microcosms and pooled before RNA or amino acid extractions. ECM were made up of the Hartig net, mantle hyphae, and root cells, whereas EM, which had extended from ECM and had spread out onto the peat substrate, was collected at the margin (i.e., 10 cm away from the root tips). Three independent RNA or amino acid extractions were carried out.
Axenic mycorrhizal birch seedlings were produced as described previously (7). Pure cultures of mycelium and mycorrhizal roots were harvested at 4, 8, and 12 days after contact from four petri dishes and pooled before RNA extractions.
The Escherichia coli strain used was DH5α. Classical procedures for manipulating E. coli cells and DNA were essentially based on those described by Sambrook et al. (46).
cDNA library construction.
Total RNA from fungi grown on NH4+- or Gln-containing MMN were extracted from approximately 100 mg of frozen mycelium by using an RNeasy plant minikit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. Two double-stranded cDNA populations were synthesized from total RNA by using a Smart PCR cDNA synthesis kit (Clontech Laboratories Inc., Palo Alto, Calif.). Suppression subtractive hybridization (SSH) was performed by using a PCR-Select cDNA subtraction kit (Clontech). A population enriched for differentially expressed cDNAs with Gln (Gln cDNA population) was obtained after the SSH procedure when the Gln and NH4+ cDNA populations were considered the tester and the driver, respectively. Similarly, a population enriched for differentially expressed cDNAs with NH4+ (NH4+ cDNA population) was obtained with the original tester as a driver and the driver as a tester. In further experiments, both Gln and NH4+ cDNA populations were used for the construction of a plasmid cDNA library. PCR products from the SSH procedure were ligated into plasmid vector pGEM-T (Promega, Madison, Wis.) and transferred to DH5α competent cells. Bacterial clones were stored at −70°C in 35% glycerol.
DNA sequencing.
Clones were transferred to 150 μl of 0.1 M Tris-EDTA buffer (pH 8) in 96-well microtiter plates. Cells were immediately lysed in a microwave oven and then stored frozen at −20°C until use. The bulk of cDNA sequencing was automated, with bacterial lysate as the starting material for PCR amplification of the plasmid insert and then 5′ DNA sequencing of purified PCR products. Reaction mixtures were prepared with a Biomek 2000 robot (Beckman Coulter, Roissy, France) in a 96-well microtiter format. PCR amplifications were performed with the universal T7 promoter primer (5′-TAATACGACTCACTATAGGG-3′) and the SP6 promoter primer (5′-TATTTAGGTGACACTATAG-3′) and standard PCR protocols. PCR products were purified by isopropanol precipitation (46), followed by assessment of the size and quality by gel electrophoresis. Partial nucleotide sequences of the cDNA inserts were determined by using the dideoxy chain termination method (47) with either a BigDye Terminator kit (Applied Biosystems) or a CEQ Dye Terminator cycle sequencing Quick Start kit (Beckman) and the T7 promoter primer. Sequencing products were purified by isopropanol precipitation and then loaded onto an ABI 3100 genetic analyzer (Applied Biosystems) or a CEQ 2000 XL apparatus (Beckman). Base calling of DNA sequencer traces was conducted by using the PHRED program with the quality level set to 20 (15). After quality trimming and translation into all three forward frames, sequence comparisons against the GenBank nonredundant protein database were performed by using the FASTA program (6, 36). Expressed sequence tags (ESTs) from SSH were assembled by using a contig assembly program (20). Parameters for the contig assembly program were optimized for the EST data, and the following values were set: overlen, 20; percentage, 0.85; cutoff, 40; delta, 8.5; open, 0; and pos5, 20. The entire processing of EST data was performed by using the PHOREST tool (1).
RNA isolation for target preparation.
Total RNAs from EM, ECM, and NMR of B. pendula grown in microcosms were extracted by using a hot phenol procedure. Tissues were ground (Retsch blender model MM 300; Qiagen) and homogenized in a mixture of extraction buffer (100 mM Tris-HCl [pH 8], 20 mM EDTA, 0.5 M NaCl, 0.5% sodium dodecyl sulfate [SDS], 0.1 M β-mercaptoethanol) and phenol (Aquaphenol; 5:1 [vol/vol]; Appligene Oncor, Illkirch Graffenstaden, France), followed by incubation at 65°C for 10 min. After the addition of chloroform (1:1 [vol/vol]), the mixture was maintained on ice for 15 min and then centrifuged at 9,000 × g for 10 min at 4°C. The supernatant was collected, and RNA was precipitated by the addition of 8 M LiCl to a final concentration of 2 M. The pellet was resuspended in Tris-EDTA buffer (pH 7.5) and purified by a phenol-chloroform procedure. RNA finally was precipitated overnight in 3 M sodium acetate (pH 5.2)-100% ethanol (1:6 [vol/vol]), washed twice in 70% ethanol, and resuspended in diethyl pyrocarbonate-treated water.
cDNA array construction.
One-microliter quantities of the bacterial stocks were used to PCR amplify cDNA inserts with primers present in plasmid vector pGEM-T (Promega). The purity and length of all PCR products (20 to 40 ng μl−1) were checked by agarose gel electrophoresis. The length of the PCR products varied between 300 and 1,000 bp. A total of 1,230 PCR products, which satisfied our quality control requirements (single PCR products with homogeneous band intensities), were arrayed from 384-well microtiter plates onto nitrocellulose membranes (Eurogentec, Seraing, Belgium) by using a microGrid spotting device (BioRobotics, Cambridge, United Kingdom) with a 384-pin gadget (27). The 0.4-μm pins deposited 100 nl of each PCR product in duplicate with a spacing of 500 μm between spots on filters (7 by 10 cm) saturated with NaOH (0.1 M) at a final density of approximately 55 clones per cm2. The nitrocellulose filters then were washed, blocked, and baked according to the manufacturer's protocol (Eurogentec).
cDNA array hybridization.
Complex probes were prepared by RT and PCR amplified by using the Smart PCR cDNA synthesis kit. Labeling of the cDNA probes was done in the presence of 30 μCi of [33P]dCTP, 30 μCi of [33P]dATP, and random hexamers by using a Prime-a-Gene kit (Promega) according to the manufacturer's instructions. The unincorporated labeled nucleotides were removed by using QIAquick columns (Qiagen). The nitrocellulose filters were preincubated in 30 ml of hybridization solution (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 10× Denhardt's solution, 0.5% SDS, 100 μg of shared salmon sperm DNA ml−1) for 4 h at 65°C in an HIR10M rotating hybridization incubator (Grant/Boekel, VWR International, Strasbourg, France). The filters then were incubated in 10 ml of fresh hybridization solution containing 33P-labeled probe at 65°C for 22 h. Hybridized filters were washed successively three times for 5 min each time in 2× SSC at room temperature, two times for 20 min each time in 2× SSC containing 0.5% SDS (65°C), two times for 20 min each time in 1× SSC containing 0.1% SDS (65°C), and two times for 20 min each time in 0.1× SSC containing 0.1% SDS (65°C). Air-dried filters then were wrapped in thin, flexible sheets of plastic and exposed to a phosphorimaging screen (Eastman Kodak Company, Rochester, N.Y.) for various periods (12 h to 3 days), after which the imaging plate was scanned by using Personal Molecular Imager FX (Bio-Rad Laboratories, Hercules, Calif.) at a resolution of 50 μm per pixel.
Data analysis.
To calculate the signal intensities of each spot, a grid was overlaid on phosphorimages and quantitation of signals was performed by using the volume quantitation method of XDOTREADER (Cose, Paris, France). Signal intensities lower than two times the mean of the background plus the standard deviation were eliminated, and central normalization was performed on the remaining valid data. A Bayesian statistical method (Cyber-T software; http://visitor.ics.uci.edu/genex/cybert/) based on the t test was used to test for statistically significant differences in gene expression for each pair of conditions tested. The Bayesian t test allows statistical inference to be made even when experiments are replicated only at nominal levels. It assumes that genes with similar expression levels have similar measurement errors (31).
RT-PCR analysis.
Total RNA isolation was performed as described above except that RNase-free DNase (Promega) was applied to avoid genomic DNA contamination. RT reactions were performed with total RNA and with the enzyme Omniscript (Qiagen) as recommended by the manufacturer. RT reactions were performed for 60 min at 37°C, and RT products were used for PCR. The products were amplified by PCR with a Mastercycler (Eppendorf, Le Pecq, France) under the following conditions: 95°C for 1 min, followed by 24, 30, 30, and 33 cycles for cipC (concanamycin-induced protein type C gene), dur3 (urea transporter gene), psd (phosphatidylserine decarboxylase gene), and ti (translation initiation inhibitor gene), respectively, at 95°C for 5 s, 60°C for 45 s, and 72°C for 1 min. The primers used are shown in Table 1. The suitability of the extracted RNA for RT-PCR amplification was checked by performing RT-PCR control experiments with ti and with primers ti1 and ti2 (Table 1). The control gene was one of those showing no differential expression on cDNA arrays.
TABLE 1.
Primers used for RT-PCR and RACE-PCR analyses
| Gene (function) | Primer(s) | Sequence |
|---|---|---|
| cipC (unknown) | cip1a | 5′ GGGGAAGCTTATGCCCCACCACGAT 3′ |
| cip2a | 5′ GGGGGATCCTCAGTAGCGATCTTT 3′ | |
| dur3 (urea transport) | dur1a,b | 5′ GGCCCTTGCCGGAGTCATCT 3′ |
| dur2a | 5′ ATCAGAGAAGCGGCAACG 3′ | |
| ti (translation initiation inhibitor) | ti1a | 5′ GGCAACCAACCCAAGATG 3′ |
| ti2a | 5′ GCGACGCCTTCTATCTCG 3′ | |
| psd (phosphatidylserine decarboxylase) | psd1a | 5′ TTCCAAACCTGGGACGCTTT 3′ |
| psd2a | 5′ TGAGGGTCGCCTGGGTTGAG 3′ | |
| psd3b | 5′ TCTCAAGGCTGGCTCACTGTCTCAT 3′ | |
| dal5 (allantoate transporter) | dalb | 5′ TCAAATAGTGTATCTGAAGAGAGG 3′ |
Used for RT-PCR.
Used for RACE-PCR.
RACE-PCR.
First-strand cDNA was synthesized from 1 μg of total RNA (extracted from P. involutus by using the RNeasy plant minikit) by using Superscript II reverse transcriptase (Invitrogen, Cergy Pontoise, France), an RNase H-negative derivative from Moloney murine leukemia virus, according to the manufacturer's protocol. Specific primers psd3, dur1, and dal (Table 1) were designed from the sequences obtained from the library sequencing and were used in 5′ and 3′ rapid amplification of cDNA ends (RACE) reactions performed by using a Smart RACE cDNA kit (Clontech) according to the manufacturer's instructions. 3′ RACE fragments were successfully amplified for dur3 and psd, and 5′ and 3′ RACE fragments were obtained for dal5.
Extraction of amino acids and GC-MS analysis.
Amino acids were extracted and analyzed by GC-MS as described previously (24).
RESULTS
Amino acid and urea contents.
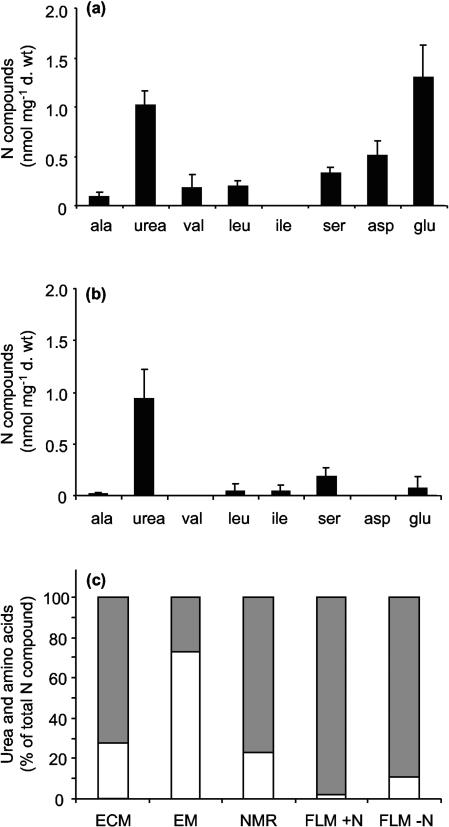
The most abundant nitrogenous compounds detected in ECM were Glu, Asp, and Urea (Fig. 1a). Including Ala, Val, Leu, and Ser, which were found at lower concentrations, the total content of N compounds was 3.6 nmol mg−1 (dry weight). In EM, urea was the major nitrogenous compound detected (Fig. 1b), representing 73.0% (0.9 nmol mg−1 [dry weight]) of the total soluble N compounds (Fig. 1c). Urea was also found in free-living mycelia (FLM) of P. involutus, although it represented only 1.9 and 11.0% of the total soluble N compounds in mycelia grown on MMN with 3.8 mM N (FLM +N) and on N-free MMN (FLM −N), respectively (Fig. 1c). Indeed, concentrations of free amino acids were much higher in free-living mycelia (110.3 and 17.2 nmol mg−1 [dry weight] on MMN with 3.8 mM N and on N-free MMN, respectively) than in the microcosm system used in the present study (Fig. 1). It must be noted that the amino acids detected in ECM may well be of plant origin, since NMR contained 28.2 nmol of free amino acids mg−1 (dry weight) (Fig. 1c).
FIG. 1.
Amino acids and urea in various components of the P. involutus-B. pendula symbiosis. (a and b) Amino acids and urea were measured in ECM (a) and in EM (b) of P. involutus-B. pendula microcosms. d. wt, dry weight. (c) Proportions of urea (white bars) and total amino acids (gray bars) were calculated and expressed as percentages of total N compounds measured; 100% represented 3.6, 1.3, 36.6, 112.5, and 19.3 nmol mg−1 (dry weight), respectively, in ECM, EM, and NMR of P. involutus-B. pendula microcosms and in free-living mycelium grown on medium with 3.8 mM N (FLM +N) or on N-free medium (FLM −N). Amino acid and urea contents were determined by GC-MS analysis as described in Materials and Methods.
EST analysis.
A set of 1,230 clones were collected after SSH performed on free-living mycelia. The SSH procedure, comparing Gln-fed mycelia (N derepression, i.e., low internal amino acid content) and ammonium-fed mycelia (N repression, i.e., high internal amino acid content), was performed to produce cDNA libraries enriched in genes involved in N nutrition and in genes that are usually underrepresented in cDNA libraries (i.e., transporter genes). A total of 892 ESTs were successfully sequenced (>99 bp) (Table 2). The ESTs were assembled into 623 tentative consensus contigs, each putatively representing one gene. Of those, 76% appeared as singletons. The redundancy seemed low compared to that seen in similar projects (25) and may indicate that the subtractive hybridization reduced a pool of more common transcripts. The assembled sequences were analyzed for homology with known sequences in databases by using the BLASTX program (http://www.ncbi.nlm.nih.gov/BLAST/). Totals of 46, 33, and 19% of the sequences, respectively, displayed low, moderate, and high degrees of similarity to protein sequences in the GenBank nonredundant protein database (1).
TABLE 2.
cDNA library characterization
| Parameter | Valuea |
|---|---|
| Total | 1,230 |
| ESTs | 892 |
| Assembled sequencesb | |
| Total | 623 |
| Singletons | 76%c |
| Sequence similarityd | |
| Orphans | 3% |
| Low (<100) | 46% |
| Moderate (100-299) | 33% |
| High (>299) | 19% |
Given as numbers, unless otherwise indicated.
Assembled sequences, putatively representing different transcripts, refer to the combined sets of contigs (assembled continuous sequences) and singletons after assembly by the contig assembly program (20).
Fraction of singletons out of the total number of assembled sequences, an indication of the level of redundancy.
ESTs are distributed according to various ranges of sequence homology scores after sequence comparison with the GenBank nonredundant protein database (1).
Among the most redundant clones, we found ESTs coding for acyl coenzyme A oxidase (nine ESTs), cytochrome P450 (eight ESTs), 40S ribosomal protein S27 (seven ESTs), 40S ribosomal protein S15 (seven ESTs), 40S ribosomal protein S12 (six ESTs), a protein involved in actin filament assembly (Arp2-3) (six ESTs), and actin (five ESTs). A large proportion of the identified sequences corresponded to proteins involved in primary metabolism (29.0%); protein synthesis (17.0%); protein fate (8.0%); transport (7.8%); control of cellular organization (4.7%); cell rescue, defense, and virulence (4.2%); regulation (3.6%); and the cell cycle (0.8%).
Differential gene expression in EM and ECM.
To assess gene expression profiles in EM and ECM, cDNAs from the subtracted libraries were arrayed at high density on nitrocellulose membranes. Probes were synthesized from EM and ECM total RNAs and used for hybridization. Twenty-eight percent of the signals showing intensities lower than two times the mean of the background plus the standard deviation were eliminated before statistical analysis. Central normalization was applied, compensating for the probable dilution of fungal transcripts in mycorrhizae due to extraction from material containing both fungal and plant RNAs. It was previously shown, by using several independent techniques, that only 30% of the transcripts extracted from P. involutus-B. pendula ectomycorrhizae are of plant origin (25). We confirmed that the higher level of transcripts found in ECM was not due to transcripts of B. pendula roots by hybridizing a membrane with a probe synthesized from RNA from NMR grown under similar conditions. No signals were detected, indicating that there was no cross hybridization between the plant target and the fungal reporters (data not shown). Principal-component analysis performed with GeneAnova software demonstrated that transcript profiles between the two compartments analyzed were statistically different (data not shown). Moreover, there were no significant variations among the three independent replicates analyzed as well as among redundantly spotted clones and duplicated spots. According to a statistical analysis based on a Bayesian t test with Cyber-T software (31), a total of 65 unique transcripts (19.5%) of P. involutus were differentially expressed (P < 5 × 10−2) between EM and ECM, with signal intensity ratios ranging from 1.9- to 24.0-fold (Tables 3 and 4). Seventeen of these 65 differentially expressed transcripts had no homology with any known sequence, and some of them may be of importance for the functioning of the symbiotic structure.
TABLE 3.
Transcripts significantly up-regulated in ECM
| Function | GenBank accession no. | Best database matcha | Expected valueb | Bayes.lnpc | PPDEd | Fold change between ECM and EM |
|---|---|---|---|---|---|---|
| Metabolism, cell rescue, and transport | CN072155 | Phosphatidylserine decarboxylase (Psd) (Arabidopsis thaliana) | 6e−04 | 4.38E−06 | 0.996 | 23.98 |
| CN072156 | CipC protein (Emericella nidulans) | 2e−12 | 9.24E−06 | 0.993 | 21.41 | |
| AF114848 | Manganese superoxide dismutase (Paxillus involutus) | 7e−43 | 5.00E−04 | 0.910 | 8.06 | |
| CN072157 | Guanine nucleotide binding protein beta subunit (Lentinula edodes) | 8e−53 | 1.37E−03 | 0.840 | 5.79 | |
| CN072158 | Polyubiquitin (Bos taurus) | 3e−48 | 8.91E−03 | 0.590 | 4.19 | |
| CN072159 | Inorganic diphosphatase (EC 3.6.1.1) (Kluyveromyces marxianus var. lactis) | 2e−33 | 2.06E−02 | 0.450 | 3.26 | |
| CN072162 | Rehydrin-like protein (Candida albicans) | 5e−41 | 1.07E−02 | 0.561 | 2.17 | |
| CN072163 | Symbiosis-related protein (Laccaria bicolor) | 2e−17 | 4.35E−02 | 0.333 | 1.97 | |
| Protein synthesis and transcription | CN072164 | Ribosomal protein L12 (Neurospora crassa) | 3e−40 | 2.61E−04 | 0.940 | 12.64 |
| CN072165 | 60S ribosomal protein 19-a (Schizosaccharomyces pombe) | 3e−35 | 5.25E−04 | 0.907 | 8.47 | |
| CN072166 | Translation initiation factor eIF1; Sui1p (Saccharomyces cerevisiae) | 8e−24 | 1.05E−03 | 0.860 | 7.92 | |
| CN072167 | 60S ribosomal protein L36 (S. pombe) | 1e−07 | 1.11E−03 | 0.854 | 6.55 | |
| CN072168 | Ribosomal protein S9 (S. pombe) | 2e−33 | 3.81E−03 | 0.718 | 5.62 | |
| CN072169 | 40S ribosomal protein S27 (S. pombe) | 7e−24 | 4.37E−03 | 0.699 | 5.51 | |
| CN072170 | Ribosomal protein S28 (N. crassa) | 1e−32 | 2.80E−03 | 0.759 | 5.33 | |
| CN072171 | Ribosomal protein L26 (Oryza sativa) | 1e−08 | 2.87E−03 | 0.755 | 5.31 | |
| CN072172 | 60S ribosomal protein L12 (Rattus norvegicus) | 4e−53 | 7.10E−03 | 0.626 | 5.30 | |
| CN072173 | Ribosomal protein L35 (Paracoccidioides brasiliensis) | 9e−10 | 4.02E−03 | 0.711 | 5.30 | |
| CN072174 | Ribosomal protein 10 (C. albicans) | 2e−27 | 3.51E−03 | 0.730 | 5.10 | |
| CN072175 | Cytoplasmic ribosomal protein S7 (Podospora anserina) | 3e−12 | 3.00E−03 | 0.750 | 4.90 | |
| CN072176 | Elongation factor 3 (Cryptococcus neoformans var. neoformans) | 1e−11 | 4.24E−03 | 0.704 | 4.51 | |
| CN072177 | Homology to rat L35a; Rpl33ap (S. cerevisiae) | 6e−33 | 1.29E−02 | 0.529 | 4.30 | |
| CN072178 | 40S ribosomal protein S12 (S. pombe) | 1e−17 | 6.85E−03 | 0.632 | 4.15 | |
| CN072179 | Ribosomal protein S3a (Cicer arietinum) | 5e−69 | 1.03E−02 | 0.567 | 4.13 | |
| CN072180 | Heat shock protein 90 (Trypanosoma cruzi) | 2e−23 | 6.10E−03 | 0.650 | 4.12 | |
| CN072181 | Elongation factor Tu; mitochondrial precursor (S. pombe) | 3e−09 | 1.04E−02 | 0.565 | 3.51 | |
| CN072197 | 40S ribosomal protein S3ae (C. albicans) | 9e−57 | 1.87E−02 | 0.466 | 3.44 | |
| CN072182 | Ribosomal protein L11 (Oryzias latipes) | 3e−18 | 1.87E−02 | 0.466 | 3.38 | |
| CN072183 | 60S ribosomal protein L27a (Mortierella alpina) | 7e−24 | 2.19E−02 | 0.441 | 3.15 | |
| CN072184 | Rat S25; Rps25ap (S. cerevisiae) | 1e−19 | 1.19E−02 | 0.543 | 3.05 | |
| CN072185 | Alpha-NACe protein (A. thaliana) | 2e−14 | 2.62E−02 | 0.411 | 3.02 | |
| CN072186 | Translation initiation factor elF4A.1 (rabbit) | 8e−93 | 4.70E−02 | 0.322 | 2.85 | |
| CN072187 | Ribosomal protein L41 (Coprinopsis cinerea) | 8e−42 | 4.38E−02 | 0.332 | 2.70 | |
| Unknown | CN072188 | No homology | ND | 8.22E−05 | 0.971 | 11.50 |
| CN072189 | No homology | ND | 2.91E−04 | 0.935 | 9.48 | |
| CN072190 | Predicted protein | 3e−12 | 1.46E−03 | 0.830 | 6.69 | |
| CN072191 | Hypothetical protein | 3e−04 | 6.23E−03 | 0.647 | 5.43 | |
| CN072192 | No homology | ND | 2.66E−03 | 0.765 | 5.26 | |
| CN072193 | No homology | ND | 5.24E−03 | 0.673 | 4.87 | |
| CN072194 | Hypothetical protein (Schizophyllum commune) | 9e−06 | 4.44E−03 | 0.697 | 4.82 | |
| CN072195 | No homology | ND | 8.00E−03 | 0.607 | 4.48 | |
| CN072196 | Hypothetical protein | 1e−05 | 1.85E−02 | 0.469 | 3.61 | |
| CN072198 | No homology | ND | 1.41E−02 | 0.514 | 3.33 |
Best database match and corresponding species obtained with BLASTX search.
Degree of similarity to known genes given by BLASTX. ND, not determined.
P value associated with the Bayesian t test of log-transformed data.
PPDE, a posteriori probability of differential gene expression between ECM and EM.
NAC, nascent.
TABLE 4.
Transcripts significantly up-regulated in EM
| Function | GenBank accession no. | Best database matcha | Expected valueb | Bayes.lnpc | PPDEd | Fold change between EM and ECM |
|---|---|---|---|---|---|---|
| Metabolism, cell rescue, and transport | CN072199 | Major facilitator superfamily multidrug efflux transporter (Tpo3) (Schizosaccharomyces pombe) | 6e−19 | 5.00E−03 | 0.680 | 6.18 |
| CN072201 | Dihydroorotase (Ustilago maydis) | 3e−14 | 5.45E−03 | 0.667 | 4.91 | |
| CN072202 | Golgi membrane sorting protein (S. pombe) | 3e−16 | 5.47E−03 | 0.667 | 4.21 | |
| CN072203 | Urea active transport protein (Dur3) (Oryza sativa) | 6e−13 | 4.30E−03 | 0.702 | 4.10 | |
| CN072204 | Alpha-mannosidase (S. pombe) | 2e−13 | 2.62E−02 | 0.411 | 3.99 | |
| CN072205 | Ubiquitin-like protein (Cicer arietinum) | 6e−09 | 7.70E−03 | 0.614 | 3.84 | |
| CN072206 | Arsenite-translocating ATPase (S. pombe) | 5e−35 | 3.13E−02 | 0.383 | 3.55 | |
| CN072207 | Subtilisin-like serine protease (Coprinopsis cinerea) | 2e−42 | 2.54E−02 | 0.416 | 3.17 | |
| CN072209 | Aldose reductase (Magnaporthe grisea) | 6e−14 | 4.74E−02 | 0.321 | 2.62 | |
| Protein synthesis and transcription | CN072210 | 40S ribosomal protein S10 (Lumbricus rubellus) | 1e−14 | 9.27E−04 | 0.869 | 8.83 |
| CN072211 | Probable translation release factor Erf3 (Neurospora crassa) | 1e−07 | 2.90E−02 | 0.395 | 3.65 | |
| Unknown | CN072212 | No homology | ND | 1.66E−03 | 0.817 | 8.98 |
| CN072213 | No homology | ND | 5.83E−04 | 0.901 | 8.86 | |
| CN072214 | No homology | ND | 9.66E−04 | 0.866 | 7.39 | |
| CN072215 | No homology | ND | 2.60E−03 | 0.768 | 7.11 | |
| CN072216 | No homology | ND | 2.31E−03 | 0.782 | 5.31 | |
| CN072200 | No homology | ND | 2.58E−03 | 0.768 | 5.22 | |
| CN072217 | No homology | ND | 1.04E−02 | 0.564 | 4.15 | |
| CN072218 | No homology | ND | 1.82E−02 | 0.471 | 3.95 | |
| CN072219 | No homology | ND | 1.02E−02 | 0.568 | 3.70 | |
| CN072220 | No homology | ND | 3.47E−02 | 0.367 | 3.29 | |
| CN072221 | No homology | ND | 3.48E−02 | 0.366 | 2.91 |
Best database match and corresponding species obtained with BLASTX search.
Degree of similarity to known genes given by BLASTX. ND, not determined.
P value associated with the Bayesian t test of log-transformed data.
PPDE, a posteriori probability of differential gene expression between EM and ECM.
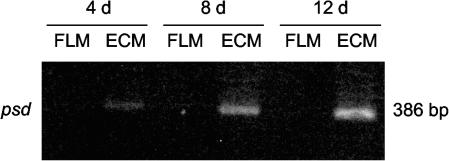
Forty-three transcripts showed up-regulated expression in ECM, with a P value of <5 × 10−2 (Table 3). Among the most abundant transcripts, a large proportion represented genes for homologues of ribosomal proteins (19 ESTs), 5 coded for translation and elongation factors or heat shock proteins, and 10 coded for proteins with unknown functions. A gene encoding a putative phosphatidylserine decarboxylase (Psd) related to Arabidopsis thaliana Psd showed 24-fold up-regulation in ECM. From the psd EST (Table 3), a forward primer was designed (psd3) (Table 1) to allow the amplification of an 866-bp 3′ fragment by RACE-PCR; this analysis revealed a 1.10−44 degree of similarity (E value) to Burkholderia fungorum Psd. This enzyme catalyzes the formation of phosphatidylethanolamine (PE) from phosphatidylserine (PS) (58). To confirm the up-regulation of psd in ECM, we measured its expression in a P. involutus-B. pendula axenic symbiotic system, which allowed us to monitor the development of the symbiotic structure in a time course series as described previously (7). Interestingly, psd transcripts were not detected in the free-living mycelium, whereas their expression increased markedly after 4, 8, and 12 days of contact between fungal and root cells (Fig. 2).
FIG. 2.
Levels of psd transcripts after RT-PCR in free-living mycelium (FLM) and ECM of P. involutus-B. pendula axenic cultures after 4, 8, and 12 days (d) of contact. Mycorrhizae were synthesized as described previously (7). The primers used are listed in Table 1. The cycle number used is given in Materials and Methods.
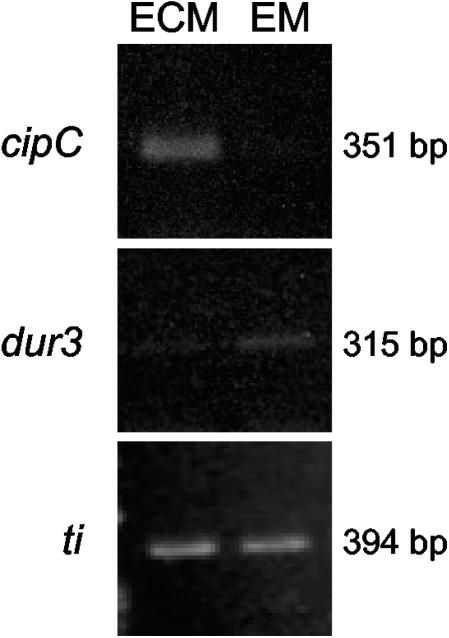
Another gene that was highly up-regulated in ECM (21.4-fold) was homologous to a gene encoding CipC protein (concanamycin-induced protein type C) from Emericella nidulans. This EST corresponds to the full-length cDNA (351 bp). The up-regulation of cipC in ECM was confirmed by RT-PCR (Fig. 3). Although its function remains unknown, its abundance was strikingly increased when E. nidulans was grown in concanamycin-containing medium, leading to drastic changes in hyphal morphology (33).
FIG. 3.
Levels of cipC, dur3, and control gene ti transcripts after RT-PCR in ECM and in EM of P. involutus-B. pendula microcosms. The primers used are listed in Table 1. The cycle number used for each gene is given in Materials and Methods.
A putative manganese superoxide dismutase homologue was up-regulated in ECM (8.1-fold), in agreement with the up-regulation of manganese superoxide dismutase in ectomycorrhizal tissue compared with the free-living fungus (25). This enzyme was previously characterized in P. involutus (22) and might function as a defense mechanism against high concentrations of reactive oxygen species. A gene coding for a putative rehydrin-like protein, a peroxiredoxin, was up-regulated 2.2-fold in ECM. This enzyme is also able to detoxify various hydroperoxides and peroxynitrites (44). A homologue for a gene coding for polyubiquitin, a highly conserved protein, implicated in the function of vital cellular processes by protein degradation, was up-regulated 4.2-fold in ECM. A homologue for a gene coding for a putative symbiosis-related protein of L. bicolor (Aut7), which has been shown to be involved in vesicular transport and autophagocytosis (26), was up-regulated 2.0-fold. The aut7 transcripts were not detected in the free-living fungus and seemed to be dependent on a symbiotic interaction with the host plant (26). The protein encoded by this gene may thus play a key role during the colonization of the root by the fungus by providing necessary components for hyphal biogenesis and differentiation. Genes coding for a putative guanine nucleotide binding protein and inorganic diphosphatase were also up-regulated in ECM (5.8- and 3.3-fold, respectively) (Table 3).
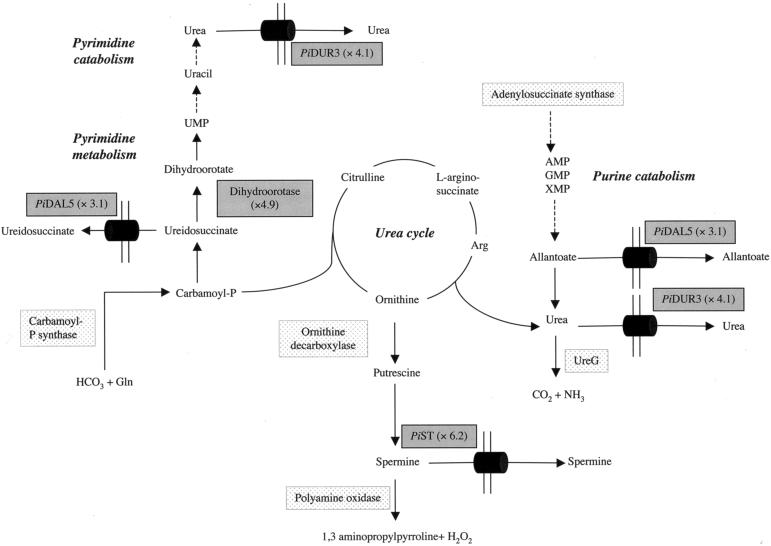
In EM, 22 genes were significantly up-regulated, with a P value of <5 × 10−2 (Table 4), although the functions of 11 of these were unknown. A gene coding for a multidrug efflux protein from the major facilitator superfamily was found to be up-regulated (6.2-fold) in EM compared to ECM. It was a homologue (E value, 5.10−11) of the Saccharomyces cerevisiae tpo3 vacuolar spermine transporter gene (56). The first step of polyamine biosynthesis is the formation of putrescine, which is synthesized from ornithine by ornithine decarboxylase. Spermine, which is derived from putrescine by the activity of spermine synthase, then is oxidized by a polyamine oxidase, leading to the formation of 1,3-aminopropylpyrroline and H2O2 (Fig. 4). Both ornithine decarboxylase- and polyamine oxidase-encoding genes were slightly up-regulated in EM (1.3- and 1.7-fold, respectively) (data not shown). A gene coding for a putative dihydroorotase, which produces dihydroorotate from ureidosuccinate during the synthesis of pyrimidines, was 4.9-fold up-regulated in EM (Fig. 4). In agreement with these data, transcripts of a putative allantoate-ureidosuccinate transporter (Dal5) were up-regulated in EM (3.1-fold). This gene was not included in Table 4 because it did not meet the condition of a P value of <5 × 10−2 (P value, 8.4 × 10−2). A 1,428 bp full-length cDNA obtained for dal5 by RACE-PCR showed a 4.10−59 degree of similarity (E value) to the S. pombe allantoate transporter gene. Allantoate can be produced by purine catabolism and degraded into urea by the activity of allantoicase. Furthermore, an EST (dur3) that was homologous to a gene coding for a putative urea transporter was found to be up-regulated 4.1-fold in EM compared to ECM. From the dur3 EST (Table 4), a forward primer was designed (Table 1) to allow the amplification of a 749-bp 3′ fragment by RACE-PCR; this analysis revealed a 2.10−18 degree of similarity (E value) to the Oryza sativa urea transporter. The up-regulation of dur3 in EM was confirmed by RT-PCR (Fig. 3). Moreover, a homologue for the gene (ureG) coding for the urease accessory protein, a component of urease, was expressed (data not shown). This result may indicate that P. involutus possesses the enzymatic capacity to produce ammonia from urea.
FIG.4.
Proposed model for urea, allantoate, and spermine synthesis and translocation in EM. Gray boxes represent proteins for which the corresponding genes were found to be up-regulated in EM. The expression ratios are given in parentheses. Stippled boxes represent expressed genes encoding proteins involved in the pathways described but not found to be up-regulated in EM. Urea could originate from the urea cycle and both pyrimidine and purine catabolism, with allantoate as an intermediate. Spermine could originate from ornithine, a urea cycle intermediate. These compounds could be translocated through EM by urea (Dur3), allantoate (Dal5), and spermine (Tpo3) transporters, which were found to be up-regulated in EM.
Genes coding for a putative Golgi membrane sorting protein, alpha-mannosidase, arsenite-translocating ATPase, serine proteinase, and aldose reductase were also up-regulated in EM (4.2-, 4-, 3.5-, 3.2-, and 2.6-fold, respectively) (Table 4).
DISCUSSION
We have shown a spatial pattern of fungal gene expression in EM and ECM that suggests differences in metabolism in the two compartments. Among the fungal genes being up-regulated the most in ECM compared to EM is a homologue for a phosphatidylserine decarboxylase (Psd) gene (Table 3). We confirmed, in a time course experiment with an axenic system described previously (7), that psd was indeed up-regulated in the mycelium attached to the roots, whereas it remained almost undetectable in the free-living mycelium (Fig. 2). In all organisms, Psd plays a key role in phospholipid metabolism by catalyzing the formation of PE from PS (58). In S. cerevisiae, two different enzymes exist; one (Psd1) is associated with the inner mitochondrial membrane (61), and the other (Psd2) is associated with the Golgi and vacuolar compartments (57). In A. thaliana, mRNA of a mitochondrial Psd was found to be expressed throughout the plant (43). In agreement with a previous report suggesting that symbiosis development leads to increased membrane formation (28), these results suggest that the up-regulation of the psd gene in ECM could allow the synthesis of newly formed membranes at the symbiotic interface. This process could allow for the incorporation into the membrane of new permeases required for the transfer of N compounds at the symbiotic interface. The up-regulation of psd expression in P. involutus-B. pendula may be also linked to changes in the phospholipid fractions observed in Pinus sylvestris-Pisolithus tinctorius ectomycorrhizal roots (29). Additionally, PE, the product of PS decarboxylation, may also be involved in the integration and stabilization of proteins in membranes and in membrane-membrane contact, as demonstrated for S. cerevisiae (13, 42).
Supported by the up-regulation of a gene coding for a vacuolar spermine transporter in EM (Tpo3), we hypothesize that the long-distance translocation of N compounds along the mycelial network, such as spermine, which has a low C/N ratio (2.5), occurs through vacuole exchange. It has repeatedly been demonstrated that vacuoles are motile structures that can be translocated from cell to cell across the dolipore septum for relatively long distances (2, 12, 49). Polyamines, whose biosynthesis, release, and uptake have been demonstrated in P. involutus (19), could be produced and stored in the vacuole, as suggested for S. cerevisiae (56). Thus, in ectomycorrhizal associations, the vacuole system may act as a conduit to move nitrogen between hyphal tips and the plant-fungus interface. This view is consistent with the observed translocation patterns of phosphorus through ectomycorrhizal mycelia in association with pine seedlings (53). Polyphosphates were detected in fungal vacuoles of ectomycorrhizae (8, 51), and Arg and Gln were proposed to be the associated cations to neutralize them (51). Through vacuole exchange, these N compounds, in particular, Arg, could be translocated from extraradical to intraradical mycelium, as already described for endomycorrhizal and ectomycorrhizal systems (3).
However, Arg and other urea cycle intermediates (ornithine and citrulline) were not detected in EM but were readily detected in P. involutus mycelia grown on standard MMN (10). It was shown previously that, after ammonium supply, Ala, Glu, or Gln and Asp or Asn were the most abundant amino acids and the major sinks for labeled N incorporation, both in EM and in mycorrhizal tips in a P. involutus-Fagus sylvatica microcosm (18). In peat medium of the microcosm, where no nutrients were applied and no amino acids were detected, the mycelium did not accumulate free amino acids (Fig. 1); the latter were probably directly incorporated into proteins. Conversely, a high proportion of urea was detected in our mycorrhizal system, indicating a rapid turnover of urea cycle intermediates, which could occur in an N-starved system (Fig. 1c). This “futile” cycle probably takes place to feed the margin of EM with carbon compounds, such as ornithine, to allow the survival of these active hyphae. Urea in EM may also originate from purine catabolism through allantoate degradation (Fig. 4). P. involutus was able to grow on 1 mM allantoate or urea, producing 29.6 and 20.9 mg of fresh weight biomass, respectively (compared with 67.2 and 2.5 mg for NH4+-grown and N-starved mycelia, respectively); these results demonstrated that it is able to use allantoate and urea as N sources for growth, as demonstrated by Schultz et al. (48). Moreover, urea has been shown to be an obligate intermediate in and the penultimate product of the catabolism of pyrimidine ring nitrogen (52). The synthesis of pyrimidines involves dihydroorotase, which is encoded by a gene observed here to be up-regulated in EM (Fig. 4). These data suggest that pyrimidines are probably synthesized at the active margin of the mycelium, while older hyphae possibly degrade them for the release of urea.
Thus, degradation products (allantoate and/or urea) of purine and pyrimidine catabolism may serve as nitrogen sources when the preferred nitrogen sources, e.g., Glu, ammonia, or Gln, are exhausted. In agreement with these conclusions, we observed the up-regulation in EM of genes homologous to the genes for a urea transporter (Dur3) and an allantoate-ureidosuccinate transporter (Dal5) (Table 4), possibly ensuring the transfer of these N sources between hyphae. Observations of 14C-α-aminoisobutyric acid transport in colonies of Phanerochaete velutina are consistent with the idea that nutrients effectively cycle through the mycelium from a loading site and are drawn off as required at different sinks (54, 55). Moreover, such circulation of nutrients in fungal mycelia should equalize differences in nutrient concentrations in various parts of the mycelia, enabling the net translocation of nutrients from areas of nutrient availability to those of nutrient demand (30).
For the growth conditions used, we could not find any statistical difference between EM and ECM in transcriptional levels for Gln synthetase, Glu synthase, Ala aminotransferase, or Asp aminotransferase, which are involved in the assimilation of ammonia. Instead, the genes for these enzymes were expressed in both compartments, a finding which may suggest that the corresponding enzymes are constitutively expressed in our system. Thus, EM does not favor primary amino acid anabolism under our starvation conditions; rather, the results indicate that the activation of urea-producing pathways may occur in EM.
The up-regulation in ECM of 25 ESTs coding for proteins involved in protein synthesis, including ribosomal proteins and translation factors, supports the fact that ECM cells need to rearrange their structure and metabolism to form an efficient symbiotic organ. It was previously shown that plant transcripts corresponding to ribosomal proteins were also up-regulated during ectomycorrhiza formation (25). Indeed, the intercellular penetration and formation of the Hartig net induce a profound change in the morphology and metabolism of fungal hyphae (7, 50). Hyphal proliferation and prolific branching, decreased interhyphal spacing, and increased hyphal density around roots are primary events that are often observed during ectomycorrhiza formation (50). We may therefore speculate that the up-regulation of cipC may be linked to changes in the growth morphology of the fungus. Interestingly, cipC homologues have also been found in several EST studies of ECM fungi, including Laccaria laccata and P. tinctorius (38).
Our results allow us to hypothesize that the spatial regulation of transport functions occurs in the intact mycorrhizal system used. It appears that the active hyphae of ECM could represent an important sink for phospholipids from the plant under the nutrient starvation conditions used, considering the up-regulation of psd.
Genes up-regulated in EM code for proteins (Dur3 and Tpo3) involved in the transport of N compounds. Long-distance translocation of specific N compounds (such as spermine) or hypha-to-hypha transfer of urea seems to occur in EM but would not be needed in ECM, where other amino acids (Glu and Asp) may serve as primary N sources.
Further investigations are needed to understand how the mycelium may adjust its metabolism under changing nitrogen conditions. A combination of transcriptional and biochemical studies should allow determination of the form in which nitrogen is transferred to the host plant, depending on the soil nitrogen status.
Acknowledgments
We thank Martina Peter and Sébastien Duplessis for helpful assistance and advice on cDNA array analysis and Nicolas Rouhier for helpful discussions. We also acknowledge Antoine Le Quéré and Eva Friman at Lund University, Pierre-Emmanuel Courty, and Christine Delaruelle for technical assistance and sequencing.
The research used in part the DNA sequencing and functional genomics facilities at INRA Nancy financed by INRA and Région de Lorraine through Institut Fédérateur de Recherche No. 110. DNA sequencing was also performed at the SWEGENE Center of Genomic Ecology at the Ecology Building, Lund University, supported by the Knut and Alice Wallenberg Foundation through the SWEGENE Consortium.
REFERENCES
- 1.Ahren, D., C. Troein, T. Johansson, and A. Tunlid. 2004. PHORESTt: a web-based tool for comparative analyses of expressed sequence tag data. Mol. Ecol. Notes 4:311-314. [Google Scholar]
- 2.Ashford, A. E. 1998. Dynamic pleiomorphic vacuoles: are they endosomes and transport compartments in fungal hyphae? Adv. Bot. Res. 28:119-159. [Google Scholar]
- 3.Bago, B., P. Pfeffer, and Y. Shachar-Hill. 2001. Could the urea cycle be translocating nitrogen in the arbuscular mycorrhizal symbiosis. New Phytol. 149:4-8. [DOI] [PubMed] [Google Scholar]
- 4.Bending, G., and D. Read. 1995. The structure and function of the vegetative mycelium of ectomycorrhizal plants. New Phytol. 130:411-417. [DOI] [PubMed] [Google Scholar]
- 5.Botton, B., and B. Dell. 1994. Expression of glutamate dehydrogenase and aspartate aminotransferase in eucalypt ectomycorrhizas. New Phytol. 126:249-257. [Google Scholar]
- 6.Brenner, S. E., C. Chothia, and T. J. Hubbard. 1998. Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proc. Natl. Acad. Sci. USA 95:6073-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brun, A., M. Chalot, R. Finlay, and B. Söderström. 1995. Structure and function of the ectomycorrhizal association between Paxillus involutus (Batsch) Fr. and Betula pendula Roth. New Phytol. 129:487-493. [Google Scholar]
- 8.Bücking, H., and W. Heyser. 1999. Elemental composition and function of polyphosphates in ectomycorrhizal fungi—an X-ray microanalytical study. Mycol. Res. 103:31-39. [Google Scholar]
- 9.Cairney, J. W. G., A. E. Ashford, and W. G. Allaway. 1989. Distribution of photosynthetically fixed carbon within root systems of Eucalyptus pilularis plants ectomycorrhizal with Pisolithus tinctorius. New Phytol. 112:495-500. [DOI] [PubMed] [Google Scholar]
- 10.Chalot, M., A. Brun, R. D. Finlay, and B. Söderström. 1994. Respiration of [14C]alanine by the ectomycorrhizal fungus Paxillus involutus. FEMS Microbiol. Lett. 121:87-91. [DOI] [PubMed] [Google Scholar]
- 11.Chalot, M., A. Brun, A. Khalid, B. Dell, R. Rohr, and B. Botton. 1990. Occurrence and distribution of aspartate aminotransferases in spruce and beech ectomycorrhizas. Can. J. Bot. 68:1756-1762. [Google Scholar]
- 12.Cole, L., D. A. Orlovich, and A. E. Ashford. 1998. Structure, function, and motility of vacuoles in filamentous fungi. Fungal Genet. Biol. 24:86-100. [DOI] [PubMed] [Google Scholar]
- 13.Dowhan, W. 1997. Phosphatidylserine decarboxylases: pyruvoyl-dependent enzymes from bacteria to mammals. Methods Enzymol. 280:81-88. [DOI] [PubMed] [Google Scholar]
- 14.Ek, H., S. Andersson, K. Arnebrant, and B. Söderström. 1994. Growth and assimilation of NH4+ and NO3− by Paxillus involutus in association with Betula pendula and Picea abies as affected by substrate pH. New Phytol. 128:629-637. [Google Scholar]
- 15.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 16.Finlay, R., H. Ek, G. Odham, and B. Söderström. 1988. Mycelial uptake, translocation and assimilation of nitrogen from 15N-labelled ammonium by Pinus sylvestris plants infected with four different ectomycorrhizal fungi. New Phytol. 110:59-66. [Google Scholar]
- 17.Finlay, R. D., and D. J. Read. 1986. The structure and function of the vegetative mycelium of ectomycorrhizal plants. I. Translocation of 14C-labelled carbon between plants interconnected by a common mycelium. New Phytol. 103:143-156. [Google Scholar]
- 18.Finlay, R. D. E., H. Odham, and B. Söderström. 1989. Uptake, translocation and assimilation of nitrogen from 15N-labelled ammonium and nitrate sources by intact ectomycorrhizal systems of Fagus sylvatica infected with Paxillus involutus. New Phytol. 113:47-55. [Google Scholar]
- 19.Fornalé, S., T. Sarjala, and N. Bagni. 1999. Endogenous polyamine content and metabolism in the ectomycorrhizal fungus Paxillus involutus. New Phytol. 143:581-587. [DOI] [PubMed] [Google Scholar]
- 20.Huang, X. 1992. A contig assembly program based on sensitive detection of fragment overlaps. Genomics 14:18-25. [DOI] [PubMed] [Google Scholar]
- 21.Ineichen, K., V. Wiemken, and A. Wiemken. 1995. Shoots, roots and ectomycorrhizal formation of pine seedlings at elevated atmospheric carbon dioxide. Plant Cell Environ. 18:703-707. [Google Scholar]
- 22.Jacob, C., M. Courbot, A. Brun, H. M. Steinman, J. P. Jacquot, B. Botton, and M. Chalot. 2001. Molecular cloning, characterization and regulation by cadmium of a superoxide dismutase from the ectomycorrhizal fungus Paxillus involutus. Eur. J. Biochem. 268:3223-3232. [DOI] [PubMed] [Google Scholar]
- 23.Javelle, A., M. Chalot, B. Söderström, and B. Botton. 1999. Ammonium and methylamine transport by the ectomycorrhizal fungus Paxillus involutus and ectomycorrhizas. FEMS Microbiol. Ecol. 30:355-366. [DOI] [PubMed] [Google Scholar]
- 24.Javelle, A., M. Morel, B. R. Rodriguez-Pastrana, B. Botton, B. Andre, A. M. Marini, A. Brun, and M. Chalot. 2003. Molecular characterization, function and regulation of ammonium transporters (Amt) and ammonium-metabolizing enzymes (GS, NADP-GDH) in the ectomycorrhizal fungus Hebeloma cylindrosporum. Mol. Microbiol. 47:411-430. [DOI] [PubMed] [Google Scholar]
- 25.Johansson, T., A. Le Quéré, B. Söderström, R. Erlandsson, J. Lundeberg, M. Uhlén, and A. Tunlid. 2004. Transcriptionnal responses of Paxillus involutus and Betula pendula during formation of ectomycorrhizal root tissue. Mol. Plant-Microbe Interact. 17:202-215. [DOI] [PubMed] [Google Scholar]
- 26.Kim, S. J. , D. Bernreuther, M. Thumm, and G. K. Podila. 1999. LB-AUT7, a novel symbiosis-regulated gene from an ectomycorrhizal fungus, Laccaria bicolor, is functionally related to vesicular transport and autophagocytosis. J. Bacteriol. 181:1963-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohler, A., C. Delaruelle, D. Martin, N. Encelot, and F. Martin. 2003. The poplar root transcriptome: analysis of 7,000 expressed sequence tags. FEBS Lett. 542:37-41. [DOI] [PubMed] [Google Scholar]
- 28.Kottke, I., and F. Oberwinkler. 1989. Amplification of root-fungus interface in ectomycorrhizae by Hartig net architecture. Ann. Sci. For. 46:737s-740s. [Google Scholar]
- 29.Laczko, E., T. Boller, and V. Wiemken. 2004. Lipids in roots of Pinus sylvestris seedlings and in mycelia of Pisolithus tinctorius during ectomycorrhiza formation: changes in fatty acid and sterol composition. Plant Cell Environ. 27:27-40. [Google Scholar]
- 30.Lindahl, B., R. Finlay, and S. Olsson. 2001. Simultaneous, bidirectional translocation of 32P and 33P between wood blocks connected by mycelial cords of Hypholoma fasciculare. New Phytol. 150:189-194. [Google Scholar]
- 31.Long, A., H. Mangalam, B. Chan, L. Tolleri, G. Hatfield, and P. Baldi. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. J. Biol. Chem. 276:19937-19944. [DOI] [PubMed] [Google Scholar]
- 32.Martin, F., and D. Canet. 1986. Biosynthesis of amino acids during 13C-glucose utilization by the ectomycorrhizal ascomycete Cenococcum geophilum monitored by 13C nuclear resonance. Physiol. Veg. 24:209-218. [Google Scholar]
- 33.Melin, P., J. Schnürer, and E. G. H. Wagner. 2002. Proteome analysis of Aspergillus nidulans reveals proteins associated with the response to the antibiotic concanamycin A, produced by Streptomyces species. Mol. Genet. Genomics 267:695-702. [DOI] [PubMed] [Google Scholar]
- 34.Montanini, B., M. Betti, A. J. Marquez, R. Balestrini, P. Bonfante, and S. Ottonello. 2003. Distinctive properties and expression profiles of glutamine synthetase from a plant symbiotic fungus. Biochem. J. 373:357-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nehls, U., A. Bock, M. Ecke, and R. Hampp. 2001. Differential expression of the hexose-regulated fungal genes AmPAL and AmMst1 within Amanita/Populus ectomycorrhizas. New Phytol. 150:583-589. [Google Scholar]
- 36.Pearson, W. R. 1994. Using the FASTA program to search protein and DNA sequence databases. Methods Mol. Biol. 24:307-331. [DOI] [PubMed] [Google Scholar]
- 37.Perez-Moreno, J. , and D. Read. 2000. Mobilization and transfer of nutrients from litter to tree seedlings via the vegetative mycelium of ectomycorrhizal plants. New Phytol. 145:301-309. [Google Scholar]
- 38.Peter, M., P. E. Courty, A. Kohler, C. Delaruelle, D. Martin, D. Tagu, P. Frey-Klett, S. Duplessis, M. Chalot, G. Podila, and F. Martin. 2003. Analysis of expressed sequence tags from the ectomycorrhizal basidiomycetes Laccaria bicolor and Pisolithus microcarpus. New Phytol. 159:117-129. [DOI] [PubMed] [Google Scholar]
- 39.Podila, G. K., J. Zheng, S. Balasubramanian, S. Sundaram, S. Hiremath, J. H. Brand, and M. J. Hymes. 2002. Fungal gene expression in early symbiotic interactions between Laccaria bicolor and red pine. Plant Soil 244:117-128. [Google Scholar]
- 40.Polidori, E., D. Agostini, S. Zeppa, L. Potenza, F. Palma, D. Sisti, and V. Stocchi. 2002. Identification of differentially expressed cDNA clones in Tilia platyphyllos-Tuber borchii ectomycorrhizae using a differential screening approach. Mol. Genet. Genomics 266:858-864. [DOI] [PubMed] [Google Scholar]
- 41.Read, D. 1984. The structure and function of the vegetative mycelium of mycorrhizal roots, p. 215-240. In D. H. Jennings and A. D. M. Rayner (ed.), The ecology and physiology of the fungal mycelium. Cambridge University Press, Cambridge, United Kingdom.
- 42.Robl, I., R. Grassl, W. Tanner, and M. Opekarova. 2001. Construction of phosphatidylethanolamine-less strain of Saccharomyces cerevisiae. Effect on amino acid transport. Yeast 18:251-260. [DOI] [PubMed] [Google Scholar]
- 43.Rontein, D., W. I. Wu, D. R. Voelker, and A. D. Hanson. 2003. Mitochondrial phosphatidylserine decarboxylase from higher plants. Functional complementation in yeast, localization in plants, and overexpression in Arabidopsis. Plant Physiol. 132:1678-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rouhier, N., E. Gelhaye, and J. P. Jacquot. 2002. Redox control by dithiol-disulfide exchange in plants. II. The cytosolic and mitochondrial systems. Ann. N. Y. Acad. Sci. 973:520-528. [DOI] [PubMed] [Google Scholar]
- 45.Rousseau, J. , D. Sylvia, and A. Fox. 1994. Contribution of ectomycorrhiza to the potentiel nutriment-absorbing surface of pine. New Phytol. 128:639-644. [Google Scholar]
- 46.Sambrook, J. , E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 47.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schultz, A. C., P. Nygaard, and H. H. Saxild. 2001. Functional analysis of 14 genes that constitute the purine catabolic pathway in Bacillus subtilis and evidence for a novel regulon controlled by the PucR transcription activator. J. Bacteriol. 183:3293-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shepherd, V. A., D. A. Orlovich, and A. E. Ashford. 1993. Cell-to-cell transport via motile tubules in growing hyphae of a fungus. J. Cell Sci. 105:1173-1178. [DOI] [PubMed] [Google Scholar]
- 50.Smith, S., and D. Read. 1997. Mycorrhizal symbiosis, 2nd ed. Academic Press, Inc., San Diego, Calif.
- 51.Strullu, D. G., and J. P. Gourret. 1981. Ultrastructure and electron-probe microanalysis of the metachromatic vacuolar granules occurring in Taxus mycorrhizas. New Phytol. 87:537-545. [Google Scholar]
- 52.Thwaites, W. M., C. H. Davis, N. Wallis-Biggart, L. M. Wondrack, and M. T. Abbott. 1979. Urea: obligate intermediate of pyrimidine-ring catabolism in Rhodosporidium toruloides. J. Bacteriol. 137:1145-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Timonen, S., R. D. Finlay, S. Olsson, and B. Söderström. 1996. Dynamics of phosphorus translocation in intact ectomycorrhizal systems: non-destructive monitoring using a β-scanner. FEMS Microbiol. Ecol. 19:171-180. [Google Scholar]
- 54.Tlalka, M., D. Hensman, P. R. Darrah, S. C. Watkinson, and M. D. Fricker. 2003. Noncircadian oscillations in amino acid transport have complementary profiles in assimilatory and foraging hyphae of Phanerchaete velutina. New Phytol. 158:325-335. [Google Scholar]
- 55.Tlalka, M., S. C. Watkinson, P. R. Darrah, and M. D. Fricker. 2002. Continuous imaging of amino-acid transport in intact mycelia of Phanerochaete velutina reveals rapid, pulsatile fluxes. New Phytol. 153:173-184. [Google Scholar]
- 56.Tomitori, H., K. Kashiwagi, T. Asakawa, Y. Kakinuma, A. J. Michael, and K. Igarashi. 2001. Multiple polyamine transport systems on the vacuolar membrane in yeast. Biochem. J. 353:681-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trotter, P. J. , and D. R. Voelker. 1995. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270:6062-6070. [DOI] [PubMed] [Google Scholar]
- 58.Voelker, D. R. 1997. Phosphatidylserine decarboxylase. Biochim. Biophys. Acta 1348:236-244. [DOI] [PubMed] [Google Scholar]
- 59.Voiblet, C., S. Duplessis, N. Encelot, and F. Martin. 2001. Identification of symbiosis-regulated genes in Eucalyptus globulus-Pisolithus tinctorius ectomycorrhiza by differential hybridization of arrayed cDNAs. Plant J. 25:181-191. [DOI] [PubMed] [Google Scholar]
- 60.Wiemken, V., and T. Boller. 2002. Ectomycorrhiza: gene expression, metabolism and the wood-wide web. Curr. Opin. Plant Biol. 5:355-361. [DOI] [PubMed] [Google Scholar]
- 61.Zinser, E., C. D. Sperka-Gottlieb, E. V. Fasch, S. D. Kohlwein, F. Paltauf, and G. Daum. 1991. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 173:2026-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]