Abstract
Background
Klebsiella pneumoniae is a frequent nosocomial pathogen causing difficult-to-treat infections worldwide. The prevalence of Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae (KPC-KP) is increasing in China. The aim of this study was to investigate the molecular epidemiology of KPC-KP in a nosocomial outbreak.
Methods
Fifty-four KPC-KP isolates were consecutively collected between November 2013 and August 2014 during a KPC-KP outbreak in a tertiary care hospital in Beijing, China. Antimicrobial susceptibility was determined by agar dilution. Carbapenemase, extended-spectrum β–lactamase, 16S rRNA methylase, AmpC β-lactamase, and plasmid-mediated quinolone resistance determinants were detected by PCR amplification. The genetic relatedness of isolates was analyzed by pulsed-field gel electrophoresis and multi-locus sequence typing.
Results
All isolates belonged to ST11 except one isolate which was identified as a new sequence type (ST2040). PFGE profile of genomic DNA revealed seven clusters, of which cluster A and C dominated the KPC-KP outbreak and cluster A was replaced by cluster C during the outbreak. PFGE of genomic DNA, S1-PFGE of plasmids, replicon typing, and drug resistant characteristics showed that clonal spread occurred during the outbreak. When compared with isolates within cluster A, all isolates in cluster C harbored rmtB and showed higher level of resistance to cefepime, amikacin, tobramycin, and tigecycline.
Conclusion
We reported a nosocomial outbreak of KPC-KP with clonal replacement and a new sequence type (ST2040) of KP. High degree of awareness and surveillance of KPC-KP should be given to avoid potential outbreaks, especially in ICU wards.
Keywords: Klebsiella pneumoniae, Carbapenemase, Clonal replacement, Epidemiology, Antimicrobial resistance
Background
Carbapenem-resistant Klebsiella pneumoniae (KP) has spread worldwide and become a major public health threat in health care facilities [1], and the mortality could reach up to 40–50% [2]. According to an antimicrobial resistance surveillance networks in China (CHINET), the rate of carbapenem-resistant KP escalated from 0.7% in 2006 to 10% in 2013 [3], which is mainly due to the rapid dissemination of Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae (KPC-KP) [4]. The bla KPC gene can be disseminated by both clonal spread and horizontal plasmid transfer [5]. In China, KPC-KP was firstly identified in 2007 [6]. Since then, this pathogen has been identified in several provinces and ST11 has been demonstrated to be the dominant clone [4, 7–16]. Nosocomial outbreaks of KPC-KP were also described previously [14, 17, 18]. However, clonal replacement of epidemic KPC-KP has not been reported in China or abroad.
Here, we reported a nosocomial outbreak of KPC-KP with clonal replacement in our hospital, which involved 54 consecutive patients and mainly occurred in ICU wards. The drug resistance and epidemiologic features of KPC-KP were also described.
Methods
Patients and bacterial isolates
All Carbapenem-resistant KP strains, which were collected between November 2013 and August 2014 in this study, were isolated from different clinical wards of our tertiary care hospital with 1500 beds. Species were identified by the Vitek 2 system, followed by 16S rDNA sequencing. Crude mortality was defined as death during hospitalization. Infection-related mortality was defined as death only as a direct consequence of KPC-KP infection during hospitalization. A KPC-KP outbreak was defined as two or more laboratory-confirmed patients that were temporally related, epidemiologically linked, and infected by the same KP variant.
Detection of antibiotic resistance genes
Isolates exhibiting resistance to at least one of the carbapenems (imipenem or ertapenem) were evaluated for the presence of bla KPC by PCR and sequencing as described previously [19, 20]. All of the KPC-positive isolates were subjected to PCR amplification and sequencing for the presence of carbapenemase genes (bla GES, bla SME, bla IMI, bla BIC, bla NDM, bla IMP, bla VIM, bla SIM, and bla SPM) [19, 20], extended-spectrum β–lactamase genes (bla TEM, bla SHV, bla CTX-M, and bla OXA-48-like) [21–23], 16S rRNA methylase genes (armA, rmtA, rmtB, rmtC, rmtD, rmtE, and npmA) [24, 25], AmpC β-lactamase genes (bla ACC, bla FOX, bla MOX, bla DHA, bla CIT /bla SPM, and bla EBC) [26], and plasmid-mediated quinolone resistance (PMQR) genes (qnrA, qnrB, qnrC, qnrS, qepA, acc(6′)-Ib-cr, oqxA, and oqxB) [27].
Antimicrobial susceptibility testing
Susceptibility tests were performed using the Vitek 2 system and the AST-GN card. MICs of various antimicrobials were determined by agar dilution method and results were interpreted according to the criteria recommended by CLSI (2014) [28].
Pulsed-field gel electrophoresis (PFGE) analysis
The clonal relatedness of KP isolates were analyzed by PFGE as described previously [29]. Prepared genomic DNA was digested using XbaI restriction enzyme on all clinical isolates. The banding patterns were analyzed by the BioNumerics software. The genetic similarity was calculated by dice coefficients and dendrograms were constructed by the unweighted pair group of arithmetic average. The analysis parameters were based on 1.5% tolerance values. Clusters were defined as DNA patterns sharing ≥85% similarity. Plasmid profiling was performed by PFGE of total bacteria genome DNA cut with nuclease S1.
Multi-locus sequence typing (MLST) analysis
Genotyping was further determined by MLST analysis. Standard DNA amplification and sequencing of seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) were performed as described previously [30]. The allele sequences and sequence types (ST) were identified at http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html.
PCR-based replicon typing (PBRT)
To determine the plasmid incompatibility groups (F, FIA, FIB, FIC, HI1, HI2, I1-Ic, L/M, N, P, W, T, A/C, K, B/O, X, Y, and FII), PBRT were carried out in all isolates as described previously [31].
Conjugation experiments
The conjugation experiments were performed by using azide-resistant E.coli J53 as the recipient strain. Cultures of donor and recipient cells in logarithmic phase (0.5 ml of each) were added to 4 ml of fresh Luria-Bertani broth and were incubated overnight without shaking. The transconjugants were selected on Mueller-Hinton agar plates containing 100 μg/ml ampicillin and 300 μg/ml sodium-azide. Antibiotic resistance genes and replicon typing were performed as described previously in this study.
Statistical analysis
All statistical analysis were performed using the SPSS software package version 20.0. Continuous variables (age and Charlson score) were summarized as medians and were compared using non-parametric Mann-Whitney U test. Categorical variables were summarized as percentages and were compared using the pearson chi-square test. A p-value of less than 0.05 was considered statistically significant.
Results
Description of isolates
A total of 54 non-duplicate KPC-KP isolates were included in this study. All patients, where the 54 isolates were isolated from, were identified as clinical infections based on the isolation of KPC-KP from clinical samples and medical diagnosis established by physicians according to the clinical manifestations and the antibacterial effects. The predominant type of samples from which KPC isolates were isolated was sputum (n = 41, 75.9%), followed by bronchoalveolar lavage fluid (n = 5, 9.3%), blood (n = 4, 7.4%), secretions (n = 2, 3.7%), urine (n = 1, 1.9%), and peritoneal fluid (n = 1, 1.9%). Of note, 77.8% (42/54) of isolates were isolated from patients in two ICU wards (ICU-1, respiratory medicine department; ICU-2, department of critical care medicine), and remaining 22.2% (12/54) of isolates were isolated from other seven medical wards. Specifically, the ICU-1 ward (with 14 beds) and ICU-2 ward (with 12 beds) belonged to two departments and were located in two different buildings with a distance of about 100 m. There were no intercommunications of patients, staffs, and medical instruments between the two ICU wards. Patients in the two ICU wards were scattered in patient rooms or beds.
Genotypic investigation of resistance
Among the 54 KPC-KP (bla KPC-2), 1.9% (1/54) carried bla SHV-11, 48.1% (26/54) carried bla SHV-11 and bla TEM-1, and the remaining 50% (27/54) carried bla TEM-1 , bla SHV (bla SHV-1, bla SHV-2, bla SHV-11 and bla SHV-12), and bla CTX (bla CTX-M-3 and bla CTX-M-14). Among rmtB-positive KPC-KP (RP-KP) isolates (n = 20), 45.0% (9/20) of isolates carried qnrS and 50.0% (10/20) of isolates carried qnrS and acc(6′)-Ib-cr. However, among rmtB-negative KPC-KP (RN-KP) isolates (n = 34), only 5.9% (2/34) of isolates carried acc(6′)-Ib-cr and 8.8% (3/34) of isolates carried qnrS and acc(6′)-Ib-cr. Resistance genes encoding class B carbapenemase (bla NDM, bla SIM, bla IMP, bla VIM, and bla SPM) and AmpC β–lactamase (bla ACC, bla FOX, bla MOX, bla DHA, bla CIT /bla SPM, and bla EBC) were not detected in any of the isolates.
Molecular typing
Through KP MLST analysis, 98.1% (53/54) of isolates belonged to ST11 (allelic profile 3–3–1-1-1-1-4). Only one isolate was identified as a new ST (ST2040, allelic profile 3–3–1-1-1-1-326), which only differed from ST11 with a substitution in the tonB allele. PFGE identified two major clusters (A, C) among 54 isolates. The cluster A and C consisted of 27 RN-KP isolates and 20 RP-KP isolates, respectively. The other seven RN-KP isolates were classified into the cluster B, D, E, F, and G (Fig. 1).
Fig. 1.
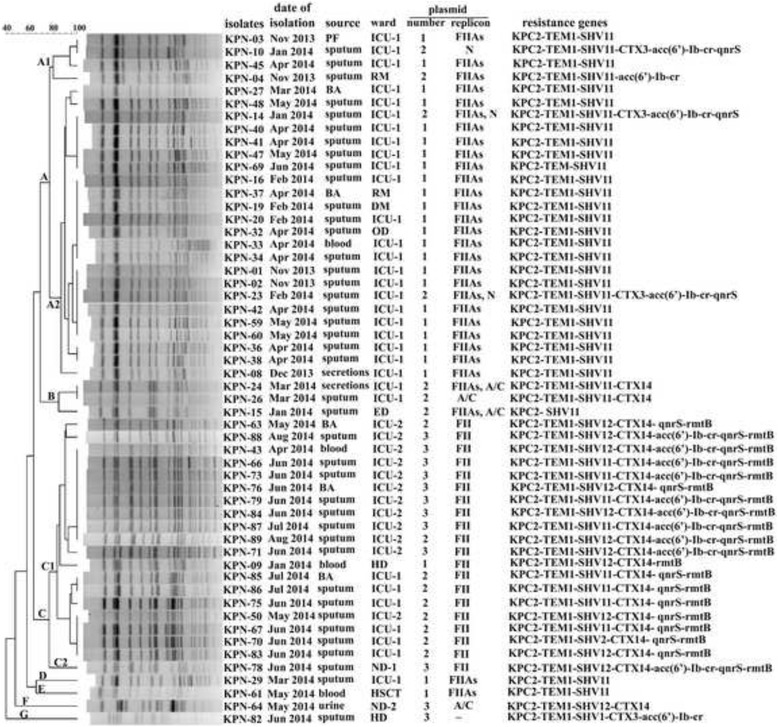
Dendrograms showing the PFGE and epidemiology profiles of 54 KPC-KP isolates. BA, bronchial aspirate; PF, peritoneal fluid; RM, respiratory medicine department; DM, digestive medicine department; OD, oncology department; ED, emergency department; HD, hematology department; ND-1, neurology department; ND-2, nephrology department; HSCT, department of hematopoietic stem cell transplantation; ICU-1, ICU ward of RM; ICU-2, ICU ward of department of critical care medicine; KPC2, KPC-2; TEM1, TEM-1; CTX, CTX-M-
Distribution of plasmid replicons
PBRT revealed that 55.6% (30/54) and 37.0% (20/54) of KP isolates contained IncFIIAs and IncFII, respectively. Specifically, 96.3% (26/27) of isolates in cluster A contained IncFIIAs, and 100% (20/20) of isolates in cluster C contained IncFII. After sequencing, the IncFIIAs confirmed in this study was identified as a new subtype of IncFII, which was characterized with low nucleotide identity (90%) when compared with the IncFIIAs replicon of Salmonella virulence plasmids.
Conjugation experiments
Five predominant isolates (KPN-01, KPN-16, KPN-34, KPN-60, and KPN-69) in cluster A and 11 predominant isolates (KPN-50, KPN-66, KPN-70, KPN-71, KPN-73, KPN-75, KPN-79, KPN-84, KPN-87, KPN-88, and KPN-89) in cluster C were used in conjugation experiments. Of the 16 tested isolates, only one isolate (KPN-69) in cluster A and four isolates (KPN-50, KPN-66, KPN-70, and KPN-88) in cluster C yielded transconjugants. As for replicon type, IncFIIAs and IncFII were confirmed in transconjugants (IncFIIAs: KPN-69; IncFII: KPN-50 and KPN-70). The transconjugant of KPN-69 harbored bla KPC-2 and bla TEM-1. The transconjugant of KPN-50 and KPN-70 harbored bla KPC-2, bla TEM-1, bla CTX-M, and rmtB. The transconjugant of KPN-66 and KPN-88 harbored bla SHV-11 (or bla SHV-12), bla CTX-M, and qnrS.
Outbreak
The epidemic curve revealed five phases (Fig. 2): phase 1 (Nov 2013 to Mar 2014), fewer cases (< 5 cases per month) mainly caused by cluster A; phase 2 (Apr 2014), a small outbreak (≥ 5 cases per month) caused by cluster A; phase 3 (May 2014), a small outbreak caused by four different clusters; phase 4 (Jun 2014), a small outbreak caused by cluster C; phase 5 (Jul 1014 to Aug 2014), fewer cases mainly caused by cluster C. Of note, cluster A was replaced by cluster C during this KP outbreak. 85.2% (23/27) of isolates in cluster A contained one plasmid with IncFIIs replicon which harbored resistance gene bla KPC, bla TEM, and bla SHV. According to PFGE of genomic DNA, S1-PFGE of plasmids, replicon typing, and drug resistant characteristics (Fig. 1), two main groups in cluster C were suggested to be derived from different origins, of which one group consisted of isolates from the ICU ward of respiratory department (ICU-1) and the other group consisted of strains from the ICU ward of department of critical care medicine (ICU-2). 75% (9/12) of isolates in the former group mainly contained three plasmids with IncFII replicon which harbored bla KPC-2 , bla TEM-1 , bla SHV-11 (or bla SHV-12), bla CTX-M-14 , acc(6′)-Ib-cr, qnrS, and rmtB. However, all strains (n = 7) in the latter group contained two plasmids with FII replicon which harbored bla KPC-2 , bla TEM-1 , bla SHV-11 (or bla SHV-12), bla CTX-M-14, qnrS, and rmtB. Specifically, clonal replacement (from cluster A to cluster C) occurred in ICU wards, especially in ICU-1 (Fig. 1).
Fig. 2.
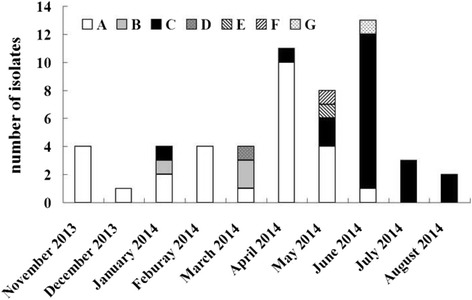
Distribution of KPC-KP isolates by month and PFGE clusters. A to G were designated as PFGE clusters as described in Fig. 1.
Antibiotic susceptibility
Resistance rates in cluster C (RP-KP, n = 20) were 100% to ampicillin, piperacillin/tazobactam, cefoxitin, cefepime, aztreonam, imipenem, amikacin, gentamycin, tobramycin, ciprofloxacin, levofloxacin, furantoin, sulfamethoxazole, and 40.0% (8/20) to tigecycline. However, resistance rates in cluster A (RN-KP, n = 27) against aforementioned antibiotics were 100% (27/27), 96.3% (26/27), 92.6% (25/27), 22.2% (6/27), 100% (27/27), 100% (27/27), 0 (0/27), 88.9% (24/27), 7.4% (2/27), 100% (27/27), 100% (27/27), 100% (27/27), and 100% (27/27), respectively, as well as 0 (0/27) to tigecycline, which was characterized with significantly lower resistance rates against cefepime (22.2% vs 100%), amikacin (0 vs 100%), tobramycin (7.4% vs 100%), and tigecycline (0 vs 40.0%) when compared with cluster C (p < 0.05).
Mortality discrepancy between clones
Medical records revealed that there were no significant differences in gender (male percentage, 81.5% vs 70.0%; median age, 75 vs 77; median Charlson scores, 6.0 vs 6.0; patient distributions (from ICU wards or transferred from ICU wards), 92.6% vs 95%) in patients between cluster A (n = 27) and cluster C (n = 20). As for the crude mortality, it was significantly higher in cluster C than those in cluster A (80.0% vs 51.9%, p < 0.05). The infection-related mortality in cluster C was higher than those in cluster A although the difference was statistically nonsignificant (25.0% vs 14.8%, p > 0.05).
Discussion
KPC-KP outbreaks have been previously reported in several provinces of China [4, 7–16]. Compared with Western countries, the prevalence of KPC-KP in China is relatively low (2.9%), as described in a 4000-bed tertiary care hospital [32]. However, KPC-KP is an increasing cause for great clinical concern due to its high antibiotic resistance and pathogenicity. In this study, a nosocomial outbreak caused by KPC-KP is described. As shown in Fig. 1, the first clonal replacement of epidemic KPC-KP occurred in ICU wards is confirmed by PFGE of genomic DNA, S1-PFGE of plasmids, replicon typing, and drug resistant characteristics. RP-KP also seems to have a higher dissemination advantage over RN-KP. Although this outbreak was severe in the ICU wards, it was terminated by effective measures, such as hand hygiene intervention, patient isolation, and environmental cleaning. This study suggests that continuous surveillance system and strict infection control measures are necessary and urgent to prevent the spread or outbreak of KPC-KP in health care facilities. It is noteworthy that bacterial culture and susceptibility test require 3–5 days and outbreak might occur during this period of time. Thus, rapid identification of the pathogens is crucial in the active surveillance culture program. On the other hand, considering that colonization frequently precedes infection, early identification of colonized cases might also reduce infections and potential outbreaks.
Although KPC-KP isolates have been identified worldwide, the spread has been proved to be caused by the dominant KP clones (ST258 and ST11). In China, ST11 is the dominant clone of KPC-KP [4]. Some new sequence types of KP have also been frequently described [17, 33]. Here, we reported a new sequence type (ST2040). Compared with the majority of isolates in cluster C (ICU-2), this clone only contains two plasmids (Fig. 1). During the outbreak period, the new ST2040 was only isolated once and didn’t become the epidemic strain. Although resistances are proved to be associated with reduced fitness and virulence of pathogens [34, 35], the clinical resistant KP isolates could survive in those high-density antibiotic environments (health care facilities and day care centers). Thus, as a multi-drug resistant clone, the fitness and pathogenicity of ST2040 might need to be investigated further.
The most frequently encountered ESBLs are TEM-, SHV-, and CTX-M-type β–lactamases. The bla CTX-M, of which prevalence is increased in KP, is carried mainly by IncFII-type plasmids [36]. In this study, the bla CTX-M genes in isolates are also carried by IncFII-type plasmids and could be transferred by conjugation. On the contrary, IncFIIAs plasmids (a new replicon subtype), which harbor bla KPC-2 , bla TEM-1, and bla SHV-11 in isolates of cluster A, seems to be hard to be conjugative.
In this study, 100% (54/54) of isolates are positive for bla SHV, consistent with the hypothesis that chromosomal bla SHV is ubiquitous in KP [37]. The majority of chromosomal SHV, including SHV-1, SHV-11, and its close relatives, are non-ESBLs enzymes. However, most of ESBLs-type SHV enzymes are plasmid-borne. In this study, the prevalence of ESBLs-type SHV enzymes (SHV-2 or SHV-12) and CTX-M enzymes in cluster C is significantly higher than those in cluster A (60% (12/20) vs 0 (0/27, 100% (20/20) vs 11.1% (3/27)). Thus, higher prevalence of cefepime-resistance KP in cluster C might be attributed to the fact that more isolates carry ESBLs enzymes.
PMQR determinants are increasingly reported in KP [14]. In this study, most (86.7%, 13/15) of the acc(6′)-Ib-cr-positive isolates also carry qnrS (Fig. 1). However, two transconjugants from donor isolates (KPN-66 and KPN-88) co-harbored qnrS and acc(6′)-Ib-cr only carry qnrS, but no acc(6′)-Ib-cr. These results suggest that qnrS and acc(6′)-Ib-cr are located in distinct plasmids, which is different from previous studies that qnr alleles are frequently co-expressed with acc(6′)-Ib-cr on the same plasmid [27, 38]. Interestingly, all 19 PQMR-determinants-producers in cluster C are also bla CTX-M -positive and two transconjugants (KPN-66 and KPN-88) co-carry qnrS and bla CTX-M, indicating a significant correlation between the two determinants among these KP isolates.
Isolates producing 16S rRNA methylase frequently exhibit high level of resistance to almost all clinically important aminoglycosides through methylation of the aminoglycoside-binding site [39]. The resistance mechanism caused by 16S rRNA methylase has been reported in KP, and the prevalence of 16S rRNA methylase genes among clinical KP isolates in China is increasing [40, 41]. KPC-KP isolates are multi-drug resistant, but are usually susceptible to aminoglycosides [42–44]. Aminoglycosides also have a significantly higher microbiologic clearance rate than polymyxin B or tigecycline [45]. Here, 37.0% (20/54) of RP-KP showed strong resistance to amikacin (100%) and tobramycin (100%). Co-production of 16S rRNA methylases in KPC-KP could leave limited therapeutic choices for antibacterial treatment and might be associated with higher mortality in this study.
Conclusions
In summary, we reported a nosocomial outbreak of KPC-KP with clonal replacement and a new sequence type (ST2040) of KP in our hospital. ST11 is the dominant clone. The outbreak mainly occurred in ICU wards. The clonal spread was responsible for this outbreak. Our study also suggested that a high degree of awareness and surveillance of KPC-KP should be given to avoid potential outbreaks, especially in ICU wards.
Acknowledgements
Not applicable.
Availability of data and materials
The data and materials can be obtained on request from the authors.
Authors’ contributions
SC designed the study and drafted the manuscript. YL and XY performed the experiments and analyzed the data. LZ collected the data. All authors read and approved the final version of this manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not required for this study.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- CLSI
Clinical and laboratory standards institute
- ICU
Intensive care unit
- KPC
Klebsiella pneumoniae carbapenemase
- KPC-KP
KPC-producing Klebsiella pneumoniae
- MIC
Minimum inhibitory concentration
- MLST
Multi-locus sequence typing
- PBRT
PCR-based replicon typing
- PFGE
Pulse field gel electrophoresis
- PMQR
Plasmid-mediated quinolone resistance
- rDNA
ribosomal deoxyribonucleic acid
- rRNA
ribosomal ribonucleic acid
Contributor Information
Yuying Liang, Email: 786847290@qq.com.
Xiuyun Yin, Email: yinxiuyun1965@163.com.
Lijun Zeng, Email: zengzeng616@gmail.com.
Shuiping Chen, Email: shpchen@hotmail.com.
References
- 1.Munoz-price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borer A, Saidel-Odes L, Riesenberg K, Eskira S, Peled N, Nativ R, Schlaeffer F, Sherf M. Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteremia. Infect Control Hosp Epidemiol. 2009;30:972–976. doi: 10.1086/605922. [DOI] [PubMed] [Google Scholar]
- 3.Hu F, Zhu D, Wang F, Jiang X, Sun Z, Chen Z, Hu Z, Li J, Xie Y, Kang M, Xu Y, Zhang X, Zhang C, Ji P, Wang C, Wang A, Ni Y, Sun J, Yu Y, Lin J, Chu Y, Tian S, Xu Y, Shen J, Shan B, Du Y, Zhuo C, Su D, Zhang H, Kong J, Wei L, Wu L, Hu Y, Ai X. CHINET 2013 surveillance of bacterial resistance in China. Chin J Infect Chemother. 2014;14:365–374. [Google Scholar]
- 4.Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66:307–312. doi: 10.1093/jac/dkq431. [DOI] [PubMed] [Google Scholar]
- 5.Adler A, Khabra E, Paikin S, Carmeli Y. Dissemination of the blaKPC gene by clonal spread and horizontal gene transfer: comparative study of incidence and molecular mechanisms. J Antimicrob Chemother. 2016;71:2143–2146. doi: 10.1093/jac/dkw106. [DOI] [PubMed] [Google Scholar]
- 6.Wei ZQ, Du XX, Yu YS, Shen P, Chen YG, LJ LI. Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob Agents Chemother. 2007;51:763–765. doi: 10.1128/AAC.01053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai JC, Zhou HW, Zhang R, Chen GX. Emergence of Serratia Marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing β-Lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob Agents Chemother. 2008;52:2014–2018. doi: 10.1128/AAC.01539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, Li L. Novel genetic environment of the carbapenem-hydrolyzing β-Lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob Agents Chemother. 2009;53:4333–4338. doi: 10.1128/AAC.00260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi Y, Wei Z, Li L, Ji S, Du X, Shen P, Yu Y. Detection of a common plasmid carrying blaKPC-2 in Enterobacteriaceae isolates from distinct cities in China. Microb Drug Resist. 2010;16:297–301. doi: 10.1089/mdr.2010.0023. [DOI] [PubMed] [Google Scholar]
- 10.Yang Q, Wang H, Sun H, Chen H, Xu Y, Chen M. Phenotypic and genotypic characterization of Enterobacteriaceae with decreased susceptibility to carbapenems: results from large hospital-based surveillance studies in China. Antimicrob Agents Chemother. 2010;54:573–577. doi: 10.1128/AAC.01099-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S, Hu F, Xu X, Liu Y, Wu W, Zhu D, Wang H. High prevalence of KPC-2-type carbapenemase coupled with CTX-M-type extended-spectrum β-lactamases in carbapenem-resistant Klebsiella pneumoniae in a teaching hospital in China. Antimicrob Agents Chemother. 2011;55:2493–2494. doi: 10.1128/AAC.00047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Cao W, Zhu X, Chen Z, Li L, Zhang B, Wang B, Tian L, Wang F, Liu C, Sun Z. Characterization of a novel Klebsiella pneumoniae sequence type 476 carrying both blaKPC-2 and blaIMP-4. Eur J Clin Microbiol Infect Dis. 2012;31:1867–1872. doi: 10.1007/s10096-011-1512-7. [DOI] [PubMed] [Google Scholar]
- 13.Sheng JF, Li JJ, Tu S, Sheng ZK, Bi S, Zhu MH, Shen XM, Li LJ. blaKPC and rmtB on a single plasmid in Enterobacter amnigenus and Klebsiella pneumoniae isolates from the same patient. Eur J Clin Microbiol Infect Dis. 2012;31:1585–1591. doi: 10.1007/s10096-011-1481-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang R, Wang XD, Cai JC, Zhou HW, Lv HX, Hu QF, Chen GX. Outbreak of Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae with high qnr prevalence in a Chinese hospital. J Med Microbiol. 2011;60:977–982. doi: 10.1099/jmm.0.015826-0. [DOI] [PubMed] [Google Scholar]
- 15.Tang HJ, Chen YT, Chiang T, Fung CP, Chuang YC, Kristopher SL. Identification of the first imported KPC-3 Klebsiella pneumoniae form the USA to Taiwan. Int J Antimicrob Agents. 2014;44:431–435. doi: 10.1016/j.ijantimicag.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 16.Xing C, Ge B, Yu K, Gao S, Liang B, Ye H. Bloodstream infections caused by Klebsiella pneumoniae carbapenemase 2-producing K.pneumoniae at a hematology Department in Wenzhou, China. Intern Med. 2016;55:2087–2091. doi: 10.2169/internalmedicine.55.6369. [DOI] [PubMed] [Google Scholar]
- 17.Zhou T, Zhang Y, Li M, Yu X, Sun Y, Xu J. An outbreak of infections caused by extensively drug-resistant Klebsiella pneumoniae strains during a short period of time in a Chinese teaching hospital: epidemiology study and molecular characteristics. Diagn Microbiol Infect Dis. 2015;82:240–244. doi: 10.1016/j.diagmicrobio.2015.03.017. [DOI] [PubMed] [Google Scholar]
- 18.Jiang Y, Wei Z, Wang Y, Hua X, Feng Y, Yu Y. Tracking a hospital outbreak of KPC-producing ST11 Klebsiella pneumoniae with whole genome sequencing. Clin Mirobiol Infect. 2015;21:1001–1007. doi: 10.1016/j.cmi.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Hong SS, Kim K, Huh JY, Jung B, Kang MS, Hong SG. Multiplex PCR for rapid detection of genes encoding class a carbapenemases. Ann Lab Med. 2012;32:359–361. doi: 10.3343/alm.2012.32.5.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 22.Edelsin M, Pimkin M, Palagin I, Edelstein I, Stratchounski L. Prevalence and molecular epidemiology of CTX-M extended-spectrum ß-Lactamase-producing Esherichia coli and Klebsiella pneumonia in Russian hospitals. Antimcrob Agents Chenother. 2003;47:3724–3732. doi: 10.1128/AAC.47.12.3724-3732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012;67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 24.Yamane K, Wachino J, Suzuki S, Shibata N, Kato H, Shibayama K, Kimura K, Kai K, Ishikawa S, Ozawa Y, Konda T, Arakawa Y. 16S rRNA Methylase-producing, gram-negative pathogens Japan. Emerg Infect Dis. 2007;13:642–646. doi: 10.3201/eid1304.060501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y, Yu H, Guo Q, Xu X, Ye X, Wu S, Guo Y, Wang M. Distribution of 16S rRNA methylases among different species of gram-negative bacilli with high-level resistance to aminoglycosides. Eur J Clin Microbiol Infect Dis. 2010;29:1349–1353. doi: 10.1007/s10096-010-1004-1. [DOI] [PubMed] [Google Scholar]
- 26.Perez FJ, Hanson ND. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo Y, Yang J, Zhang Y, Ye L, Wang L, Guo L. Prevalence of ß-lactamases and 16S rRNA methylase genes amongst clinical Klebsiella pneumoniae isolates carrying plasmid-mediated quinolone resistance determinants. Int J Antimicrob Agents. 2011;37:352–355. doi: 10.1016/j.ijantimicag.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 28.Clinical Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 24th Informational Supplement M100–S24. Wayne: CLSI; 2014.
- 29.Pereira PS, Borghi M, de Araújo CF, Aires CA, Oliveira JC, Asensi MD, Carvalho-Assef AP. Clonal dissemination of OXA-370-producing Klebsiella pneumoniae in Rio de Janeiro, Brazil. Antimicrob Agents Chemother. 2015;59:4453–4456. doi: 10.1128/AAC.04243-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 32.Yang J, Ye L, Guo L, Zhao Q, Chen R, Luo Y, Chen Y, Tian S, Zhao J, Shen D, Han L. A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin Microbiol Infect. 2013;19:E509–E515. doi: 10.1111/1469-0691.12275. [DOI] [PubMed] [Google Scholar]
- 33.Li JJ, Sheng ZK, Deng M, Bi S, Hu FS, Miao HF, Ji ZK, Sheng JF, Li LJ. Epidemic of Klebsiella pneumoniae ST11 clone coproducing KPC-2 and 16S rRNA methylase RmtB in a Chinese university hospital. BMC Infect Dis. 2012;12:373. doi: 10.1186/1471-2334-12-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giamarellos-Bourboulis EJ, Plachouras D, Tzivra A, Kousoulas V, Bolanos N, Raftogiannis M, Galani I, Dontas I, Dionyssiou-Asteriou A, Giamarellou H. Stimulation of innate immunity by susceptible and multidrug-resistant Pseudomonas aeruginosa an in vitro and in vivo study. Clin Exp. 2004;135:240–246. doi: 10.1111/j.1365-2249.2003.02365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin J, Ko KS. Comparative study of genotype and virulence in CTX-M-producing and non-extended-spectrum-β-lactamase-producing Klebsiella pneumoniae isolates. Antimicrob Agents Chemother. 2014;58:2463–2467. doi: 10.1128/AAC.02499-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin J, Choi MJ, Ko KS. Replicon sequence typing of IncF plasmids and the genetic environments of blaCTX-M-15 indicate multiple acquisitions of blaCTX-M-15 in Escherichia coli and Klebsiella pneumoniae isolates from South Korea. J Antimicrob Chemother. 2012;67:1853–1857. doi: 10.1093/jac/dks143. [DOI] [PubMed] [Google Scholar]
- 37.Hammond DS, Harris T, Bell J, Turnidge J, Giffard PM. Selection of SHV extended-spectrum-β-lactamase-dependent cefotaxime and ceftazidime resistance in Klebsiella pneumoniae requires a plasmid-borne blaSHV gene. Antimicrob Agents Chemother. 2008;52:441–445. doi: 10.1128/AAC.00359-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Y, Zhou Z, Qian Y, Wei Z, Yu Y, Hu S, Li L. Plasmid-mediated quinolone resistance determinants qnr and acc(6′)-Ib-cr in extended-spectrum β–lactamase-producing Escherichia coli and Klebsiella pneumoniae in China. J Antimicrobial Chemoth. 2008;61:1003–1006. doi: 10.1093/jac/dkn063. [DOI] [PubMed] [Google Scholar]
- 39.Doi Y, Arakawa Y. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin Infect Dis. 2007;45:88–94. doi: 10.1086/518605. [DOI] [PubMed] [Google Scholar]
- 40.Wu Q, Zhang Y, Han L, Sun J, Ni Y. Plasmid-mediated 16S rRNA methylases in aminoglycoside-resistant Enterobacteriaceae isolates in shanghai, China. Antimicrob Agents Chemother. 2009;53:271–272. doi: 10.1128/AAC.00748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Ye L, Wang W, Luo Y, Zhang Y, Han L. Diverse prevalence of 16S rRNA methylase genes armA and rmtB amonst clinical multidrug-resistant Escherichia coli and Klebsiella pneumoniae isolates. Int J Antimicrob Agents. 2011;38:348–351. doi: 10.1016/j.ijantimicag.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 42.Samuelsen O, Naseer U, Tofteland S, Skutlaberg DH, Onken A, Hjetland R, Sundsfjord A, Giske CG. Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. J Antimicrob Chemother. 2009;63:654–658. doi: 10.1093/jac/dkp018. [DOI] [PubMed] [Google Scholar]
- 43.Bogdanovich T, Adams-Haduch JM, Tian GB, Nguyen MH, Kwak EJ, Muto CA, Doi Y. Colistin-resistant, Klebsiella pneumoniae Carbapenemase (KPC)-producing Klebsiella pneumoniae belonging to the international epidemic clone ST258. Clin Infect Dis. 2011;53:373–376. doi: 10.1093/cid/cir401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daly MW, Riddle DJ, Ledeboer NA, Dunne WM, Ritchie DJ. Tigecycline for treatment of pneumonia and empyema caused by carbapenemase producing Klebsiella pneumoniae. Pharmacotherapy. 2007;27:1052–1057. doi: 10.1592/phco.27.7.1052. [DOI] [PubMed] [Google Scholar]
- 45.Satlin MJ, Kubin CJ, Blumenthal JS, Cohen AB, Furuya EY, Wilson SJ, Jenkins SG, Calfee DP. Comparative effectiveness of aminoglycosides, polymyxin B, and tigecycline for clearance of carbapenem-resistant Klebsiella pneumoniae from urine. Antimicrob Agents Chemother. 2011;55:5893–5899. doi: 10.1128/AAC.00387-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data and materials can be obtained on request from the authors.


