Abstract
A new starch-binding domain (SBD) was recently described in α-amylases from three lactobacilli (Lactobacillus amylovorus, Lactobacillus plantarum, and Lactobacillus manihotivorans). Usually, the SBD is formed by 100 amino acids, but the SBD sequences of the mentioned lactobacillus α-amylases consist of almost 500 amino acids that are organized in tandem repeats. The three lactobacillus amylase genes share more than 98% sequence identity. In spite of this identity, the SBD structures seem to be quite different. To investigate whether the observed differences in the SBDs have an effect on the hydrolytic capability of the enzymes, a kinetic study of L. amylovorus and L. plantarum amylases was developed, with both enzymes acting on several starch sources in granular and gelatinized forms. Results showed that the amylolytic capacities of these enzymes are quite different; the L. amylovorus α-amylase is, on average, 10 times more efficient than the L. plantarum enzyme in hydrolyzing all the tested polymeric starches, with only a minor difference in the adsorption capacities.
α-Amylases (1,4-α-d-glucan-4 glucanohydrolase; EC 3.2.1.1) are a widespread group of enzymes that catalyze the hydrolysis of the α-1,4 glycosidic linkages of raw and soluble starch, thereby generating smaller dextrins and oligosaccharides. They are classified into family 13 in the sequence-based classification of glycoside hydrolases (GH-13) (16, 17). They are multidomain proteins that contain, in addition to the catalytic (β/α)8 domain (domain A), domains B and C. Domain B is inserted between the third β-strand and the third α-helix of the barrel and varies greatly in length and structure (21). Domain C follows the catalytic barrel; this domain is made of β-strands and is thought to stabilize the catalytic domain by shielding hydrophobic residues of domain A from the solvent (29). Some of these enzymes contain one noncatalytic domain whose function is generally described as that of a starch-binding domain (SBD).
The SBD is a functional domain that can bind granular starch, increasing the local concentration of substrate at the active site of the enzyme, and that may also disrupt the structure of the starch surface, thereby enhancing the amylolytic rate (39, 40). In the primary structure classification of glycoside hydrolases (16, 17; http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html), the carbohydrate-binding modules (CBM) are organized into 39 families, which include several specificities such as cellulose, xylan, chitin, and starch binding. The most generalized and studied family of starch-binding modules is CBM-20. These modules are present in approximately 10% of amylolytic enzymes from GH-13 (almost all cyclomaltodextrin glucanotransferase, in a few α-amylases, and in maltotetraohydrolases, maltopentaohydrolases, maltogenic α-amylases, and acarviose transferases), GH-14 (some β-amylases), and GH-15 (most fungal glucoamylases). This domain is positioned at the C-terminal end of proteins except for the glucoamylase from Rhizopus oryzae and the Thermoactinomyces vulgaricus α-amylase, which contains the SBD at its N terminus (1, 2). The SBD is usually formed by 100 amino acids, producing several β-strand segments that form an open-sided, distorted β-barrel structure (30, 33).
Sequenced lactobacillus amyA genes (Lactobacillus amylovorus, Lactobacillus plantarum, and Lactobacillus manihotivorans) share an identity of 98% (32). The three enzymes are organized in two functional domains: the catalytic domain (amino acids 1 to 474) and the SBD (amino acids 475 to 953).
The catalytic domain belongs to GH-13; it contains the conserved regions described by Vihinen and Mäntsälä (43), Rumbak et al. (36), and Janec̆ek et al. (20). The three lactobacillus catalytic domains share 99.2% identity; they have 65.5 and 61.5% identity with Bacillus subtilis and Streptococcus bovis α-amylases, respectively (32).
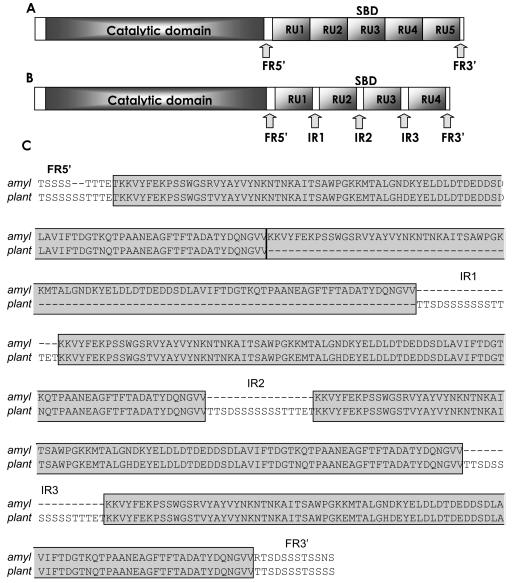
In contrast, the lactobacillus SBD (included in CBM-26; http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html) has a completely different structure from the common SBD (35). The lactobacillus α-amylases present an SBD formed by almost 500 amino acids organized in tandem repeat units (RUs) of 91 amino acids each (Fig. 1), with four repeats in L. manihotivorans (32) and L. plantarum and five repeats in L. amylovorus (11). A similar organization is found in the Bacillus sp. no. 195 α-amylase, with two repeats forming the SBD (41), and in the maltopentaose-producing amylase from an alkaliphilic gram-positive bacteria with three C-terminal repeats of unknown function (5).
FIG. 1.
Arrangement and localization of the consensus RUs with their flanking regions (FRs) and IRs in the amyA gene of L. amylovorus (U62096) (A) and the amyA gene of L. plantarum U62095 (B). (C) Alignment of the SBDs from L. amylovorus and L. plantarum. Boxed regions are the repeated units.
Flanking the RUs are two regions, one of 35 nucleotides (at the 5′ end of the RU) and the other of 21 nucleotides (at the 3′ end). Between the RUs in the L. plantarum and L. manihotivorans α-amylases are intermediary regions (IRs) consisting of 48 nucleotides (Fig. 1). These regions are rich in serine and threonine; consequently, they may increase the random coil regions and perhaps the mobility of the RUs in the L. plantarum and L. manihotivorans α-amylases in contrast with the SBD from L. amylovorus. The flanking regions and intermediary regions have a consensus sequence (TTSDSSSSSSSTTTET) that resembles the serine-threonine rich O-glycosylated Gp-I domain of glucoamylase I from Aspergillus niger involved in maintenance of protein structure against stress, adsorption onto raw starch granules, and secretion (7, 14, 15, 23, 27, 38).
In cellulolytic systems, the two functional domains are typically separated by relatively long linkers, peptides generally rich in serine, threonine, proline, and glycine, which are often glycosylated (28). These linkers favor correct conformations and independent actions of joined functional domains (10, 34).
Here we report a kinetic study of L. plantarum (four RUs with three IRs) and L. amylovorus α-amylases (five RUs without IRs) acting on starch in both granular and soluble forms. We also studied their adsorption capacities to consider the implications of a soluble enzyme acting upon a solid substrate.
MATERIALS AND METHODS
Materials.
Soluble potato starch was from Prolabo, Fontenay-sous-Bois, France. Raw cornstarch, amylopectin, amylose, dinitrosalicylic acid (DNS), and β-cyclodextrin were from Sigma Chemical Co. (St. Louis, Mo.). Sepharose was from Pharmacia Biotech (Uppsala, Sweden).
Bacterial strains.
L. amylovorus NRRL B-4540 (kindly provided by the Agriculture Research Service culture collection, U.S. Department of Agriculture, Peoria, Ill.) and L. plantarum A6 (13) were grown in MRS-medium (8). For enzyme preparation, cultures were grown in 2 liters of MRS-starch (2%) at 30°C.
Enzyme purification.
Following an 18-h batch culture, the fermentation broth was collected and centrifuged at 9,000 × g for 15 min at 4°C. Amylases were purified from the supernatant by affinity chromatography as described previously (35) by using a β-cyclodextrin epoxy-activated Sepharose 6B column.
Protein concentration was estimated by the Bradford method (4) by using bovine serum albumin as a standard (Bio-Rad protein assay). Once purified, the protein was determined at 280 nm (L. plantarum α-amylase theoretical ɛ280 = 188,800 and L. amylovorus α-amylase theoretical ɛ280 = 207,680).
Electrophoresis analysis.
Sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis (SDS-7.5% PAGE) was performed according to the Laemmli method (25). Proteins were visualized by Coomassie blue staining as described by Blakesly and Boezi (3). Activity staining was performed in the gel after renaturation of enzymes, by using the method described by Lacks and Springhorn (24). Glycosylation was determined in the SDS-PAGE gel with the Sigma glycoprotein detection kit.
Preparation of starch suspensions.
Starch granules were washed twice in ice water and then suspended in 0.1 M citrate-phosphate buffer, pH 5, and gently agitated by swirling the mixture. Mixing was carried out either at room temperature and designated native or in a boiling water bath for experiments where dissolved starch was to be used. The flasks were sealed to restrict water loss by evaporation during heating, and the flask and its contents were weighed both prior to and after 30 min in order to avoid any loss of volume. Suspensions were freshly prepared and used immediately for each experiment.
Enzyme activity assay. (i) Starch.
Amylase activity was determined by measuring the reducing sugars released in 10 min by enzymatic hydrolysis of 1% soluble potato starch, amylose, amylopectin, or corn starch in 0.1 M citrate-phosphate buffer, pH 5, at 63°C (35). Reducing sugars were quantified by the DNS method by using glucose as a standard (31). One unit of amylase activity was defined as the amount of enzyme that liberated 1 mol of glucose per s. In addition, α-amylase activity on soluble potato starch was determined by measuring its iodine-complexing ability according to the protocol described by Giraud et al. (12).
(ii) pNPG7.
Hydrolytic activity of α-amylases over benzylidene-blocked p-nitrophenyl maltoheptaoside (pNPG7) was determined by using a Randox assay amylase kit (Antrim, United Kingdom) according to the manufacturer's instructions.
Kinetics of reactions involving soluble starch.
The Michaelis constant (Km) was determined at 10 different starch concentrations (from 0 to 40 g/liter) at optimal activity temperature and pH (63°C and pH 5, respectively) (35). For pNPG7, 10 different concentrations ranging from 0.008 to 8 mmol/liter were used. Kinetic parameters were calculated by fitting initial velocities and substrate concentrations to the Michaelis-Menten equation by using the quasi-Newton minimization method (Microsoft Excel, version 5).
Hydrolysis of insoluble starch.
Amylase activity on insoluble substrates was determined by measuring the increase of reducing sugars formed by enzymatic hydrolysis of 1% soluble potato starch, amylose, amylopectin, or raw cornstarch at different times under the established conditions of pH and temperature. Reducing sugars were quantified by the DNS method by using glucose as a standard (31). One unit of amylase activity was defined as the amount of enzyme that liberated 1 mol of glucose per s.
Adsorption of α-amylases on raw starch.
Adsorption was measured at 4°C in 1.5-ml Eppendorf tubes containing L. amylovorus or L. plantarum α-amylase to a final concentration of 0 to 100 μM and a 10% raw cornstarch suspension in 100 mM citrate-phosphate buffer, pH 5, to a final volume of 60 μl. Control tubes contained protein without starch. Each mixture was incubated for 30 min with gentle shaking (6 rpm) and centrifuged for 5 min at 4°C and 15,000 × g to remove the starch. Free protein left in the clarified supernatants was measured spectrophotometrically (A280) and used to calculate the amount of the α-amylase adsorbed to starch (45). The adsorption constant (Kad in milliliters per milligram of starch) was calculated from the slope obtained from the initial linear adsorption of the purified enzymes (6).
RESULTS
Amylase production and purification.
Growth and amylase production of L. amylovorus and L. plantarum were compared. The two lactobacilli displayed comparable growth rates. In both cultures, α-amylase activity was evident from the early stages of fermentation, reaching a maximal hydrolytic rate during the late logarithmic growth phase (data not shown). Cultures were harvested at this phase (optical density at 600 nm, 3), and amylolytic activities on soluble potato starch were measured. The amylases produced by both lactobacilli were purified from the supernatant by affinity chromatography on β-cyclodextrin-Sepharose. The elution pattern showed, in both cases, a unique protein peak which was superimposable on the amylase activity of the enzyme. Purified enzymes migrated as a single band with the same mobility on SDS-PAGE and were active on a zymogram (data not shown). In order to determine whether the IRs (rich in serine and threonine residues) were glycosylated as in other hydrolases, the enzymes were treated with the Sigma glycoprotein detection kit. Neither L. plantarum α-amylase nor L. amylovorus α-amylase contains carbohydrates.
Starch hydrolysis.
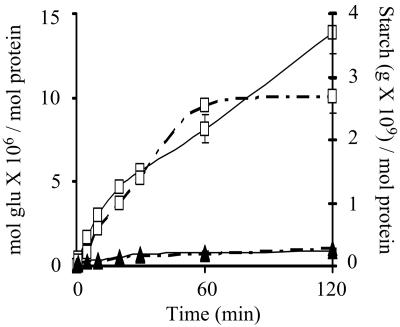
The hydrolytic capacity of both amylases was first examined on soluble potato starch by simultaneously measuring the decrease in iodine-staining power and the production of reducing sugars from starch. For both enzymes, starch hydrolysis was accompanied by a rapid reduction in the iodine-staining capability of the substrate with a correspondingly slow release of reducing sugars. As shown in Fig. 2, L. amylovorus α-amylase hydrolyzes 10 times more starch than L. plantarum α-amylase; similarly, L. amylovorus α-amylase releases 10 times more reducing sugars.
FIG. 2.
Starch degradation (grams per mole of protein) (broken lines) and release of reducing sugars (moles of glucose per mole of protein) (solid lines) from soluble potato starch by L. amylovorus (□) and L. plantarum (▴) purified α-amylases at pH 5 and 63°C.
Kinetics on soluble starch.
This study was performed with the same level of catalytic activity (10 U, based on soluble potato starch hydrolysis) in order to remain inside the detection limits; initial rates were determined at 63°C and pH 5. At the tested enzymatic concentration (10 U), the amylases followed Michaelis-type kinetics. The kinetic constants for these enzymes on different substrates are shown in Table 1. As shown, L. amylovorus α-amylase is from 3.4 to 14.6 times more efficient in hydrolyzing all tested starches. The smallest activity differences between the amylases were found when they acted on amylose, their natural substrate.
TABLE 1.
Kinetic parameters on gelatinized starch and enzymatic activity on raw starch of lactobacilli α-amylasesa
| Strain and substrate | Kinetic parameter on gelatinized starch
|
Initial-activity rate (U mol−1) on starch granules | ||
|---|---|---|---|---|
| Km (g liter−1) | kcat (s−1) | kcat/Km (s−1 g−1 liter−1) | ||
| L. amylovorus | ||||
| Soluble potato starch | 1.97 ± 0.11 | 3.1 × 104 | 1.6 × 104 | 1,578 ± 86 |
| Corn starch | 5.24 ± 0.31 | 1.4 × 103 | 2.6 × 102 | 133 ± 13 |
| Amylopectin | 2.87 ± 0.29 | 1.3 × 103 | 4.5 × 102 | 607 ± 18 |
| Amylose | 8.00 ± 0.50 | 1.8 × 103 | 2.2 × 102 | 3,020 ± 259 |
| Maltoheptaoside | 0.19 ± 0.02b | 1.4 × 107 | 7.7 × 107 | ND |
| L. plantarum | ||||
| Soluble potato starch | 1.92 ± 0.09 | 3.8 × 103 | 2.0 × 104 | 98 ± 5 |
| Corn starch | 13.00 ± 1.31 | 2.4 × 102 | 1.8 × 101 | 5 ± 0.3 |
| Amylopectin | 5.10 ± 0.40 | 1.9 × 102 | 3.8 × 101 | 16 ± 0.9 |
| Amylose | 5.00 ± 0.59 | 3.2 × 102 | 6.3 × 101 | 127 ± 10 |
| Maltoheptaoside | 0.22 ± 0.02b | 1.2 × 107 | 5.4 × 107 | ND |
Error in the model adjustment to the Michaelis-Menten equation is as follows (L. plantarum and L. amylovorus, respectively): on soluble potato starch, 8 × 10−8 and 10−7; on corn starch, 4 × 10−9 and 2 × 10−8; on amylopectin, 4 × 10−9 and 2 × 10−8; on amylose, 2 × 10−8 and 10−7; on maltoheptaoside, 3 × 10−2 and 2 × 10−1. ND, not determined.
Km in millimoles per liter.
Kinetics on pNPG7 were measured as a control of the catalytic domain activity. In contrast to the results obtained for the polymeric substrates, there are no differences in the capacities of the two enzymes to hydrolyze this small substrate.
Hydrolysis of insoluble starch.
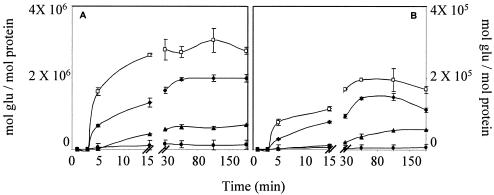
The hydrolytic rate was estimated following the release of reducing sugars. Since the catalytic activity of the two amylases against soluble starch differed, raw starch digestion was carried out under conditions in which the same catalytic activity level was present in each reaction mixture (10 U/ml). As shown in Fig. 3, after the first 5 min of incubation, amylolysis began with a linear period at the highest velocity. The rate then diminished until the system reached stability.
FIG. 3.
Digestion of several raw starches by L. amylovorus (A) and L. plantarum (B) α-amylases as measured by release of reducing sugars. Incubations were carried out at pH 5 and 63°C. □, amylose; ♦, soluble potato starch; ▴, amylopectin; •, corn starch.
As expected, both amylases hydrolyzed amylose and nongelatinized soluble potato starch better than insoluble cornstarch, but L. amylovorus α-amylase released between 11 and 23 times more reducing sugars than L. plantarum α-amylase, regardless of starch origin. In all experiments assayed, with raw or gelatinized starch, L. amylovorus α-amylase produced, on average, 10 times more reducing ends than L. plantarum α-amylase.
To compare the hydrolytic capacity on raw starch, we considered the slope of product formed in time in the linear period observed between 5 to 15 min (Fig. 3). Table 1 also shows that the L. amylovorus amylase hydrolyzes raw starch faster than L. plantarum amylase.
Raw starch binding.
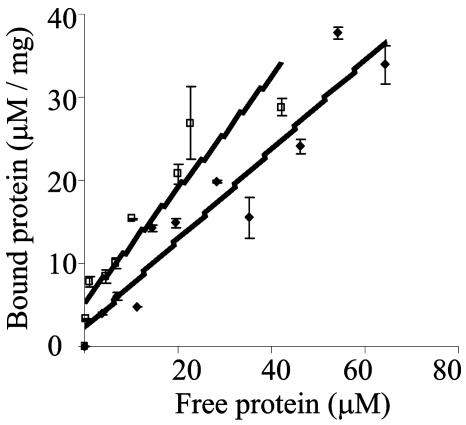
Enzyme adsorption to raw starch granules was assayed at various protein concentrations. As shown in Fig. 4, only small differences in the extent of adsorption of lactobacillus enzymes was observed; the Kad was 0.53 for L. amylovorus amylase and 0.67 for L. plantarum amylase. At the tested concentrations the curves did not reach a plateau; this maximum is reached when the protein forms a monolayer on the starch surface. This would suggest that the number of IRs does not affect the binding affinity or that not all the IRs are interacting with the starch granule (under investigation).
FIG. 4.
Adsorption of purified enzymes to native cornstarch. Linear adsorption isotherms indicate the apparent equilibrium distribution of enzymes between the solid phase (bound) and the liquid phase (free) at various protein concentrations. □, L. plantarum; ♦, L. amylovorus.
DISCUSSION
Studies of the catalytic properties of α-amylase are sometimes performed by using low-molecular-weight artificial substrates such as p-nitrophenyl α-d-maltoside (44) or different methyl-isomaltosides (9). In the present study pNPG7 was utilized only as a control to make sure that there were no differences in the action of the two catalytic domains (which was expected, given their high homology), so that the comparison of lactobacillus enzymes was made over their natural substrate, polymeric starch.
It has been shown that the α-amylase of Bacillus subtilis adsorbs to crystalline starchy materials and that this binding is a prerequisite for catalysis (26). In the L. amylovorus α-amylase, previous studies demonstrated that the tandem repeats found at the carboxyl-terminal end of the protein are responsible for raw starch adsorption (35). Our results suggest a kinetically important adsorption step, since they show that raw starch is not hydrolyzed at the early stages of the reaction. However, when the enzymes act on soluble starch, the reaction can be described by conventional Michaelis-Menten kinetics.
Kinetic constants and insoluble starch hydrolysis are greatly influenced by both the starch source and the enzyme origin. Many reasons have been proposed for the differences in the susceptibilities of different starches to amylolysis. Of definite importance, for instance, are restrictions in the substrate and product diffusion rates because of variations in viscosity that are associated with different starch materials and with the limitation of amylase accessibility to the starch itself due to particular structural features. There are very few systematic enzyme studies that test these suggestions, probably due to the inherent difficulties presented by the complexity of the system.
The sequenced lactobacillus amylases (11, 32) show a different structure from the SBDs of other origins; however, identity among them is almost 98% (Fig. 1). In spite of this identity, the catalytic properties of the enzymes are quite different. As reported for the Bacillus sp. no. 195 α-amylase (41), we thought that the presence of one more RU in the L. amylovorus enzyme would make the starch binding stronger, but our results showed no substantial difference between the adsorption of the two amylases (Kad of 0.53 versus. 0.67) even if, unlike the L. amylovorus α-amylase, at low protein concentration almost 100% of the L. plantarum amylase is adsorbed onto starch.
The presence of carbohydrate binding domains is usually associated with the attachment of the catalytic domain to its polymeric insoluble substrate, which increases effective substrate concentration at the active site, but it is not clear whether binding domains have other functions. For example, the SBD may target the enzyme to particular sites or it may disrupt the saccharide surface of the granule, as reported for the A. niger glucoamylase (23), where the SBD binds starch strands in an approximately perpendicular orientation, thus disordering the starch structure (40).
In the case of the two lactobacillus enzymes studied here, we have previously shown that the SBD is necessary for raw starch hydrolysis and adsorption and that it may play a role in soluble starch hydrolysis because different rates of reaction have been observed in L. amylovorus α-amylase with or without SBD (35). Similar results have been observed with other amylases (41). Abe suggests (1) that the SBD forms two types of starch-binding sites since the structure of starch is thought to vary from a rigid helical structure in the crystal state of amylose to a loose helical structure in solution; consequently, it is expected that the SBD plays a role in helping the enzyme to approach starch by recognizing the surfaces of its relatively rigid helical structures, like a starch granule binding site. On the other hand, the SBD is expected to recognize specifically the loose helical structure of starch in order to help the catalytic site to interact with this structure, because the rigid helical region of starch cannot bind to the catalytic site of the enzyme.
It has been proposed that a linker sequence is likely to be necessary in the four-domain amylases (α-amylases, maltotetrao-, and maltopentao-hydrolases, etc.) to connect domain C to the SBD (19), especially because the flexible nature of the linker may allow the catalytic domain to access large areas of the starch granule surface (39). Even though these linkers are vital for correct folding, secretion, and, consequently, activity (15), there are a few cases in which the linkers are absent and the amylases are able to degrade raw starch (18, 42). When linkers are present they are characteristically rich in glycine, serine, and threonine (19), as are the IRs linking the RU in L. plantarum α-amylase. The presence of several SBDs and linkers may affect substrate retention, even though the observed differences in adsorption do not explain the catalytic differences.
In the case of the β-1,4-glucanase cellulose-binding domain (CBD) from Cellulomonas fimi (22), two CBDs are juxtaposed without an intervening linker. The authors observed that the affinity of each domain for cellotetraose is equivalent whether the domains are joined or isolated, but for phosphoric acid-swollen cellulose, CBDN1N2, binding is approximately twofold greater than CBDN1 binding. The interpretation of these results is that the two domains comprising CBDN1N2 are structurally constrained, due to the lack of a flexible linker, so that they cannot bind simultaneously to adjacent regions of a single polymer chain. However, Sauer et al. (37) explain that there is no strict linker structure-dependent cooperation between the two domains; rather, the linker may hold the catalytic and binding domains in correct positions relative to each other, allowing specific interdomain stabilizing contacts. In the case of α-amylase, it could be extrapolated that every IR acts as a linker, allowing the interaction of each RU with starch and retarding product liberation, while the L. amylovorus SBD interacts as a unit.
Although it is clear that the sequence and structural features of the SBD are responsible for the different efficiencies observed for the two α-amylases, the mechanism that governs binding is not necessarily evident. When protein binding involves single, well-defined binding sites, the interpretation of a single measured parameter is relatively straightforward; however, in cases involving multiple binding interactions, no approach can directly examine the interplay of different structural modules and the impact of their effects on binding and catalysis. The adsorption ability of the RUs as separate units and the impact of the IRs on starch hydrolysis are currently under study.
Acknowledgments
This work was supported in part by CONACyT, Mexico, grants 38966-B and 41222-Z.
We are grateful to Guillermo Aguilar for helpful discussion. Elizabeth Langley and Isabel Perez Montfort corrected the English version of the manuscript.
REFERENCES
- 1.Abe, A., T. Tonozuka, Y. Sakano, and S. Kamitori. 2004. Complex structures of Thermoactinomyces vulgaricus R-47 α-amylase 1 with malto-oligosaccharides demonstrate the role of domain N acting as a starch binding domain. J. Biol. Chem. 335:811-822. [DOI] [PubMed] [Google Scholar]
- 2.Ashikari, T., N. Nakamura, Y. Tanaka, N. Kiuchi, Y. ShiBano, T. Tanaka, T. Amachi, and H. Yoshizumi. 1986. Rhizopus raw-starch degrading glucoamylase: its cloning and its expression in yeast. Agric. Biol. Chem. 50:957-964. [Google Scholar]
- 3.Blakesly, R. W., and J. A. Boezi. 1977. A new staining technique for protein in polyacrylamide gels using Coomassie Brilliant Blue G-250. Ann. Biochem. 82:580-582. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye-binding. Ann. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Candussio, A., G. Schmid, and A. Böck. 1990. Biochemical and genetic analysis of a maltopentaose-producing amylase from an alkaliphilic Gram-positive bacterium. Eur. J. Biochem. 191:1177-1185. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L., P. M. Coutinho, Z. Nikolov, and C. Ford. 1995. Deletion analysis of the starch-binding domain of Aspergillus glucoamylase. Protein Eng. 8:1049-1055. [DOI] [PubMed] [Google Scholar]
- 7.Christensen, T., B. Svensson, and B. W. Sigurskjold. 1999. Thermodynamics of reversible and irreversible unfolding and domain interactions of glucoamylase from Aspergillus niger studied by differential scanning and isothermal titration calorimetry. Biochemistry 38:6300-6310. [DOI] [PubMed] [Google Scholar]
- 8.De Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 9.Frandsen, T. P., M. M. Palcic, and B. Svensson. 2002. Substrate recognition by three family 13 yeast α-glucosidases. Eur. J. Biochem. 269:728-734. [DOI] [PubMed] [Google Scholar]
- 10.Gilkes, N. R., B. Henrissat, D. G. Kilburn, R. C. Miller, and R. A. J. Warren. 1991. Domains in microbial β-1,4-glycanases: sequence conservation, function and enzymes families. Microbio. Rev. 55:303-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giraud, E., and G. Cunny. 1997. Molecular characterization of the α-amylase genes of Lactobacillus plantarum A6 and Lactobacillus amylovorus reveals an unusual 3′ end structure with direct tandem repeats and suggests a common evolutionary origin. Gene 198:149-157. [DOI] [PubMed] [Google Scholar]
- 12.Giraud, E., B. Gosselin, B. Marin, J. L. Parada, and M. Raimbault. 1993. Purification and characterization of an extracellular amylase activity from Lactobacillus plantarum strain A6. J. Appl. Bacteriol. 75:276-282. [Google Scholar]
- 13.Giraud, E., A. Brauman, S. Keleke, B. Lelong, and M. Raimbault. 1991. Isolation and physiological study of an amylolytic strain of Lactobacillus plantarum. Appl. Microbiol. Biotechnol. 36:379-383. [Google Scholar]
- 14.Goto, M., M. Tsukamoto, I. Kwon, K. Ekino, and K. Furukawa. 1999. Functional analysis of O-linked oligosaccharides in threonine/serine-rich region of Aspergillus glucoamylase by expression in mannosyltransferase-disruptants of yeast. Eur. J. Biochem. 260:596-602. [DOI] [PubMed] [Google Scholar]
- 15.Goto, M., N. Shinoda, T. Oka, Y. Sameshima, K. Ekino, and K. Furukawa. 2004. Thr/Ser-rich domain of Aspergillus glucoamylase is essential for secretion. Biosci. Biotechnol. Biochem. 68:961-963. [DOI] [PubMed] [Google Scholar]
- 16.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henrissat, B., and A. Bairoch. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hostinová, E., A. Solovicová, R. Dvorský, and J. Gasperík. 2003. Molecular structure prediction of the first raw-starch-degrading glucoamylase without a separate starch-binding domain. Arch. Biochem. Biophys. 411:189-195. [DOI] [PubMed] [Google Scholar]
- 19.Janec̆ek, S., B. Svensson, and A. MacGregor. 2003. Relation between domain evolution, specificity, and taxonomy of the α-amylase family members containing a C-terminal starch-binding domain. Eur. J. Biochem. 270:635-645. [DOI] [PubMed] [Google Scholar]
- 20.Janec̆ek, S., E. Lévêque, A. Belardi, and B. Haye. 1999. Close evolutionary relatedness of α-amylases from archaea to plants. J. Mol. Evol. 48:421-426. [DOI] [PubMed] [Google Scholar]
- 21.Janec̆ek, S., B. Svensson, and B. Henrissat. 1997. Domain evolution in α-amylase family. J. Mol. Evol. 45:322-331. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, P. E., E. Brun, L. F. MacKenzie, S. G. Withers, and L. McIntosh. 1999. The cellulose-binding domains from Cellulomonas fimi β-1,4-glucanase CenC bind nitroxide spin-labeled cellooligosaccharides in multiple orientations. J. Mol. Biol. 287:609-625. [DOI] [PubMed] [Google Scholar]
- 23.Juge, N., M. F. Le-Gal-Coëffet, C. S. M. Furniss, A. P. Gunning, B. Kramhoft, V. J. Morris, G. Williamson, and B. Svensson. 2002. The starch binding domain of glucoamylase from Aspergillus niger: overview of its structure, function, and role in raw-starch hydrolysis. Biologia 57(Suppl. 11):239-245. [Google Scholar]
- 24.Lacks, S. A., and S. S. Springhorn. 1980. Renaturation of enzymes after polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate. J. Biol. Chem. 255:7467-7473. [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Leloup, V. M., P. Colonna, and S. G. Ring. 1991. α-Amylase adsorption on starch crystallites. Biotechnol. Bioeng. 38:127-134. [DOI] [PubMed] [Google Scholar]
- 27.Libby, C. B., C. A. Cornett, P. J. Reilly, and C. Ford. 1994. Effect of amino acid deletions in the O-glycosylated region of Aspergillus awamori glucoamylase. Protein Eng. 7:1109-1114. [DOI] [PubMed] [Google Scholar]
- 28.Linder, M., and T. T. Teeri. 1997. The roles and function of cellulose binding domains. J. Biotechnol. 57:15-28. [DOI] [PubMed] [Google Scholar]
- 29.MacGregor, E. A., S. Janecek, and B. Svensson. 2001. Relationship of sequence and structure to specificity in the α-amylase family of enzymes. Biochim. Biophys. Acta 1546:1-20. [DOI] [PubMed] [Google Scholar]
- 30.Mikami, B., M. Adachi, T. Kage, E. Sarikaya, T. Nanmori, R. Shinke, and S. Utsumi. 1999. Structure of raw starch-digesting Bacillus cereus β-amylase complexed with maltose. Biochemistry 38:7050-7061. [DOI] [PubMed] [Google Scholar]
- 31.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Ann. Biochem. 31:426-428. [Google Scholar]
- 32.Morlon-Guyot, J., F. Mucciolo-Roux, R. Rodríguez-Sanoja, and J. P. Guyot. 2001. Characterization of the L. manihiotivorans α-amylase gene. DNA Seq. 12:27-37. [DOI] [PubMed] [Google Scholar]
- 33.Penninga, D., B. A. van der Veen, R. M. A. Knegtel, S. A. F. T. van Hijum, H. J. Rozeboom, K. H. Kalk, B. W. Dijstra, and L. Dijkhuizen. 1996. The raw starch binding domain of cyclodextrin glycosyltransferase from Bacillus circulans strain 251. J. Biol. Chem. 271:32777-32784. [DOI] [PubMed] [Google Scholar]
- 34.Quentin, M., M. Ebbelaar, J. Derksen, C. Mariani, and H. van der Valk. 2002. Description of a cellulose-binding domain and a linker sequence from Aspergillus fungi. Appl. Microbiol. Biotechnol. 58:658-662. [DOI] [PubMed] [Google Scholar]
- 35.Rodríguez-Sanoja, R., J. Morlon-Guyot, J. Jore, J. Pintado, N. Juge, and J. P. Guyot. 2000. Comparative characterization of complete and truncated forms of Lactobacillus amylovorus α-amylase and role of the C-terminal direct repeats in raw starch binding. Appl. Environ. Microbiol. 66:3350-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rumbak, E., D. E. Rawlings, G. G. Lindsey, and D. R. Woods. 1991. Cloning, nucleotide sequence and enzymatic characterization of an α-amylase from ruminal bacterium Butyrivibrio fibrisolvens H17c. J. Bacteriol. 173:4203-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauer, J., T. Christensen, T. P. Frandsen, E. Mirgorodskaya, K. A. McGuire, H., Driguez, P. Roepstorff, B. W. Sigurskjold, and B. Svensson. 2001. Stability and function of interdomain linker variants of glucoamylase I from Aspergillus niger. Biochemistry 40:9336-9346. [DOI] [PubMed] [Google Scholar]
- 38.Semimaru, T., M. Goto, K. Furukawa, and S. Hayashida. 1995. Functional analysis of the threonine and serine rich Gp-I domain of glucoamylase I from Aspergillus awamori var. kawachi. Appl. Environ. Microbiol. 61:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorimachi, K., M. F. Le Gal-Coëffet, G. Williamson, D. B. Archer, and M. P. Williamson. 1997. Solution structure of the granular starch binding domain of Aspergillus niger glucoamylase bound to β-cyclodextrin. Structure 5:647-661. [DOI] [PubMed] [Google Scholar]
- 40.Southall, S. M., P. J. Simpson, H. J. Gilbert, G. Williamson, and M. P. Williamson. 1999. The starch binding domain from glucoamylase disrupts the structure of starch. FEBS Lett. 447:58-60. [DOI] [PubMed] [Google Scholar]
- 41.Sumitani, J., T. Tottori, T. Kawaguchi, and M. Arai. 2000. New type of starch binding domain: the direct repeat motif in the C-terminal region of Bacillus sp. no. 195 α-amylase contributes to starch binding and raw starch degrading. Biochem. J. 350:477-484. [PMC free article] [PubMed] [Google Scholar]
- 42.Tibbot, B. K., D. W. S. Wong, and G. H. Robertson. 2002. Studies on the C-terminal region of barley α-amylase 1 with emphasis on raw starch-binding. Biologia 57(Suppl. 11):229-238. [Google Scholar]
- 43.Vihinen, M., and P. Mäntsälä. 1989. Microbial amylolytic enzymes. Crit. Rev. Biochem. Mol. Biol. 24:329-418. [DOI] [PubMed] [Google Scholar]
- 44.Wilcox, E. R., and J. R. Whitaker. 1984. Some aspects of the mechanism of complexation of red kidney bean α-amylase inhibitor and α-amylase. Biochemistry 23:1783-1791. [DOI] [PubMed] [Google Scholar]
- 45.Williamson, G., N. J. Belshaw, and M. P. Williamson. 1992. O-glycosylation in Aspergillus glucoamylase. Biochem. J. 282:423-428. [DOI] [PMC free article] [PubMed] [Google Scholar]