Abstract
Hematopoietic stem cells (HSCs) have extensive regenerative capacity to replace all blood cell types, an ability that is harnessed in the clinic for bone marrow transplantation. Finding appropriate donors remains a major limitation to more extensive usage of HSC‐based therapies. Derivation of patient‐specific HSCs from pluripotent stem cells offers great promise to remedy this problem if scientists could crack the code on how to make robust, transplantable HSCs in a dish. Studies delving into the native origins of HSC production during embryonic development should supply the necessary playbook. This review presents recent discoveries from animal models, with a focus on zebrafish, and discusses the implications of these new advances in the context of prior knowledge. The focus is on the latest research exploring the role of epigenetic regulation, signaling pathways, and niche components needed for proper HSC formation. These studies provide new directions that should be explored for de novo generation and expansion of HSCs for regenerative therapies. Stem Cells Translational Medicine 2017;6:60–67
Keywords: Hematopoietic stem cells, Developmental biology, Bone marrow transplant, Embryo
Significance Statement.
Hematopoietic stem cells (HSCs) have extensive regenerative capacity to replace all blood cell types, an ability that is harnessed in the clinic for bone marrow transplantation. Finding appropriate donors remains a major limitation to more extensive usage of HSC‐based therapies. Derivation of patient‐specific HSCs from pluripotent stem cells offers great promise to remedy this problem if scientists could crack the code on how to make HSCs in a dish. This review presents data from studies delving into the native origins of HSC production during embryonic development that provide new directions that should be explored for de novo generation and expansion of HSCs for regenerative therapies.
Introduction
Hematopoiesis, the process of blood production, is maintained throughout the lifetime of an organism by long‐lived hematopoietic stem cells (HSCs) that are capable of self‐renewing and generating all types of mature blood cells needed to carry oxygen, fight infection, and prevent bleeding. Clinically, the success of bone marrow and cord blood transplantation therapies is because of the ability of HSCs in those tissues to repopulate the blood system of the new host. Limitations in the number of suitable donors for HSC‐based therapies greatly hamper broader usage of transplantation. Generation of patient‐specific HSCs from pluripotent stem cells holds great promise to remedy this problem if researchers could faithfully derive fully functional, transplantable HSCs in a dish. Although HSCs are present in adults, they are only generated de novo during embryonic development. Although our understanding of the origins of bona fide HSCs is still quite primitive, great strides have been made over the past several decades using animal models to uncover many novel and surprising steps in HSC ontogeny.
Embryonic hematopoiesis occurs in sequential waves, which can be divided into primitive and definitive 1. Primitive waves form erythroid and myeloid cells to help the developing embryo through its first growth steps. The definitive waves generate erythromyeloid progenitors and long‐term HSCs 2 3 4. Generation of definitive HSCs occurs in a region termed the aorta‐gonad‐mesonephros (AGM). Within this region, HSCs specifically arise from specialized hemogenic endothelium found in the ventral wall of the dorsal aorta (DA) in a process termed the endothelial‐to‐hematopoietic transition that is conserved across vertebrates, including zebrafish (Danio rerio), mice, and humans (Fig. 1) 3, 5 6 7. After their specification, HSCs travel to an intermediate niche (fetal liver/placenta in mammals and caudal hematopoietic tissue [CHT] in zebrafish), where they proliferate and expand, and then ultimately colonize their final home, where they will remain throughout adulthood (bone marrow in mammals and kidney marrow in zebrafish) 1, 8 9 10. At each stage, the niche sends environmental signals that regulate HSC behaviors, including proliferation and differentiation 1.
Figure 1.
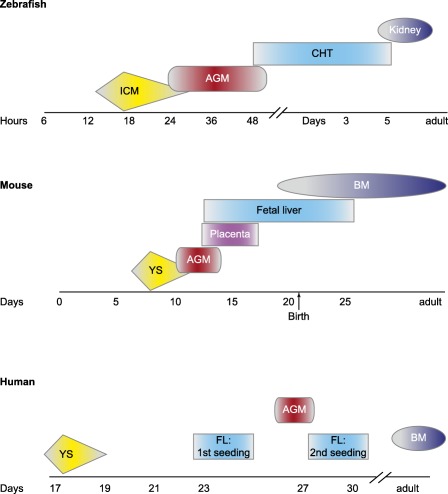
Comparison of timing of hematopoietic development across vertebrates. Time lines showing when and where primitive and definitive hematopoietic induction occurs in zebrafish (top), mice (middle), and humans (bottom). Abbreviations: AGM, aorta‐gonad‐mesonephros; BM, bone marrow; CHT, caudal hematopoietic tissue; FL, fetal liver; ICM, intermediate cell mass; YS, yolk sac.
Hematopoiesis is highly conserved among vertebrates, and most of our knowledge comes from animal models. Françoise Dieterlen‐Lièvre was among the first scientists to demonstrate the intraembryonic origins of definitive hematopoiesis within the DA through studies using chick‐quail chimeras 11. Later work confirmed that mice, Xenopus, and zebrafish form HSCs in a highly similar fashion 3, 12, 13.
Zebrafish provide an excellent model for the study of hematopoiesis because of its numerous advantages. External embryonic development allows for easy access to early embryonic stages, and embryonic transparency permits easy visualization. This latter characteristic allowed scientists to directly visualize in vivo HSC emergence in real time, revealing without a doubt the endothelial origins of HSCs 3, 6. In addition, rapid embryonic development, numerous genetic manipulation tools, and fluorescent transgenic animals make zebrafish highly amenable for exploring the ontogeny of HSCs. Recent studies in zebrafish have yielded novel findings in HSC specification, including contributions from the Notch signaling pathway, inflammatory signaling, and the HSC niche. This review summarizes these new discoveries and how they contribute to our knowledge of definitive hematopoiesis.
Insights Into the Early Stages of HSC Specification
The earliest HSC precursors arise from the posterior lateral plate mesoderm (PLM) that migrates to the medial region of the embryo and gives rise to a vascular cord that will become the dorsal aorta 14 15 16. Once the DA is formed and specified, HSCs emerge from specialized hemogenic endothelial cells that reside within its ventral floor. Once these cells take on a hematogenic fate, they bud out of the aorta, enter circulation, and seed the next developing niche (Fig. 2) 3, 6, 8. HSC specification and emergence require integration of intrinsic factors with input from different signaling pathways that interact to guide tissue development from the earliest mesoderm precursors until the final mature HSCs. Some of the principal signaling pathways that regulate HSC emergence identified from experiments in animal models are the vascular endothelial growth factor (VEGF) pathway, which stimulates endothelial cell differentiation and migration; Sonic hedgehog (SHH) and bone morphogenetic protein (BMP) signaling, which regulate arterial wall polarization necessary for HSC specification; and the Notch pathway 17 18 19. The Notch pathway is involved in many defining steps throughout HSC formation and specification that will be discussed in greater detail later in this review 19.
Figure 2.
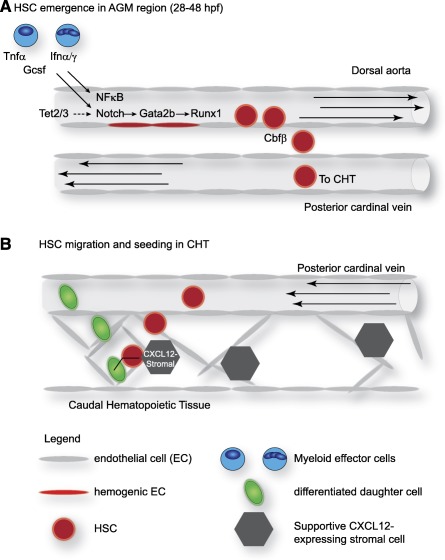
Diverse signals regulate embryonic HSC behaviors in zebrafish. (A): Inflammatory signals from myeloid effector cells promote HSC emergence through activation of Notch and NFκB signaling. Tet2/3 also regulate Notch signaling, leading to gata2b and runx1 expression in hemogenic endothelial cells. Cbfβ is subsequently required to promote the extravasation of emerging HSC out of the dorsal aorta. (B): Nascent HSCs seeding the CHT induce endothelial remodeling to form a “microniche” comprising an HSC surrounded by endothelial cells adjacent to a CXCL12‐expressing supportive stromal cell. The orientation of the division plane of the HSC is dictated by the position of the stromal cell. An open arrow on the HSC (red) and daughter cell (green) shows the angle of the division plane. Arrows within the vessels show the direction of blood flow. Abbreviations: AGM, aorta‐gonad‐mesonephros; CHT, caudal hematopoietic tissue; EC, endothelial cell; hpf, hours postfertilization; HSC, hematopoietic stem cell; Ifnα/γ, interferon α/γ; Tnfα, tumor necrosis factor α.
Transcriptional master regulators that coordinate with and sometimes instruct the epigenetic landscape are essential to define cellular fate. Key transcription factors for HSCs are Gata2, Scl, Runx1, Lmo2, and C‐myb 20. Despite our knowledge of these factors, their precise role in each step of HSC formation is still open to debate.
GATA2 has been well studied in hematopoiesis and is known to act downstream of Notch signaling during HSC specification 21. The importance of GATA2 in hematopoiesis was first demonstrated in mice, where it was shown that null embryos died at approximately embryonic day 10.5 with severe primitive and definitive hematopoietic defects 22. Moreover, studies of endothelial‐ and hematopoietic‐specific mouse knockouts of Gata2 demonstrated a requirement for GATA2 both in the endothelial‐to‐hematopoietic transition and in HSC maintenance 23. Because GATA2 has a function in vasculature, it could act in a cell‐autonomous and/or a non‐cell‐autonomous fashion to regulate the endothelial‐to‐hematopoietic transition. Unlike mammals, zebrafish have two gata2 genes, gata2a and gata2b. During evolution, the various functions of Gata2 became divided between these paralogs: Gata2a plays a role in vasculature formation, whereas Gata2b is required for HSC formation 24. This separation of function in Gata2 paralogs allows more refined dissection of the genetic control of Gata2 during vessel formation and HSC specification. In a recent study examining zebrafish gata2b function, Butko et al. found that hemogenic induction can be detected earlier during embryonic development than previously appreciated 24. gata2b expression starts in the PLM around the midline at 18 hours postfertilization (hpf) (14–19 somites) before the formation of the vascular cord, is later detected in the ventral wall of the DA at 25 hpf, and persists in hematopoietic cells in the CHT at 72 hpf. This novel finding opens the door to studying the earliest steps of hemogenic endothelium before DA formation.
Runx1 is another transcription factor that is indispensable for HSC formation, acting downstream of Notch signaling under the control of gata2b 19, 25. Runx1 interacts with the protein core binding factor subunit β (Cbfβ), and it is thought that they function together during HSC formation because knockout mice for both proteins show similar hematopoietic defects 26 27 28. New data from Bresciani et al. challenge the assumption that Runx1 and Cbfβ coordinate during HSC emergence 29. Their study in cbfb‐null mutant zebrafish showed that, like runx1, cbfb expression responds to Notch1 signaling. However, their work demonstrates that Cbfβ has a separate role from its partner Runx1 during HSC development. Similar to runx1 mutants, the ultimate outcome in cbfb mutants is the lack of definitive hematopoiesis, but the stage of development where the defect occurs in the two mutants is distinct. Zebrafish runx1 mutants fail to induce hematopoietic gene expression at early stages of HSC formation, and therefore HSCs fail to specify. Loss of cbfb does not affect initial HSC formation, but, rather, impairs their ability to detach from the DA and enter circulation (Fig. 2A). Further pharmacological studies inhibiting Runx1‐Cbfβ interactions confirmed that the role of both proteins during HSC development could be uncoupled. This study implied that both Runx1 and Cbfβ are needed at different times during HSC development: Runx1 acts during specification, and Cbfβ acts afterward at the time of HSC extravasation from the DA. The door remains open regarding alternative transcription factor partners for Runx1 and Cbfβ during HSC ontogeny.
Epigenetic factors add an additional layer of complexity to gene expression and cell state control. One epigenetic process that is critical for HSCs is DNA methylation. The Tet family of methylcytosine dioxygenases, comprising Tet1, Tet2, and Tet3, convert 5‐methylcytosine (5‐mC) (typically a mark of repressed gene expression) to 5‐hydroxymethylcytosine (5‐hmC), ultimately leading to DNA demethylation and changes in gene expression. Proper regulation of the Tet family proteins is required for normal adult hematopoiesis. Mutations in TET1 and TET2 are prevalent in leukemias and myeloid malignancies 30, 31. In addition, loss of either TET1 or 2 in mice leads to clonal hematopoiesis, a precursor to leukemia 32 33 34. Despite this knowledge, the role of Tet factors in embryonic hematopoiesis was mostly unknown. Li et al. recently determined that Tet2 and Tet3 are redundantly required during HSC emergence 35. Zebrafish embryos mutant for both tet2 and tet3, but not either single mutant, showed diminished production of HSCs. The tet2/3 double mutants have diminished gata2b expression, and restoring gata2b RNA to embryos rescued HSC formation (Fig. 2A). Further analysis revealed that Notch signaling was decreased specifically in the ventral wall of the DA, suggesting that DNA methylation has a particularly important role in Notch pathway regulation during the endothelial‐to‐hematopoietic transition. This study showed that the Tet proteins are important for both embryonic and adult hematopoiesis, expanding our current understanding of developmental epigenetic regulation. It is also interesting to note that Tet redundancy allows individual mutants to develop normally, which could be interpreted as genetic protection of the important process of HSC formation.
Muscling Up HSCs
As mentioned before, hemogenic endothelium, nonhemogenic vasculature, and HSCs all originate from the PLM (also known as splachnic mesoderm) 14, 15. In zebrafish, it was thought that all cells of the aorta arose from this splachnic mesoderm, but new data challenged this dogma. Nguyen et al. recently showed that somite‐derived endothelial cells contribute to DA formation 36, similar to what happens in mouse and chick 37. These experiments revealed that the somitic contribution comes from the endotome, a specific central region within somites specified by the homeobox factor Meox1 (Fig. 3) 36. These cells are in fact key players in HSC induction: through chemokine signaling, endotome‐derived cells can induce HSC fate in their DA neighbors. Because somitic‐derived endothelial cells are also present in the mammalian DA, it will be interesting to see whether this mechanism is conserved in mammals.
Figure 3.
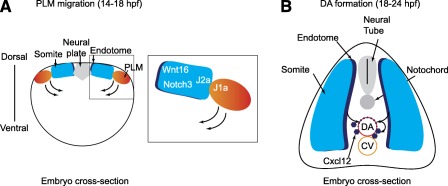
Somitic signals regulate hematopoietic stem cell (HSC) formation in zebrafish. (A): Cross‐sectional view of a zebrafish embryo at 14 hpf. The posterior lateral mesoderm migrates medially, sliding under the somites, where it receives Wnt16 and Notch3 signals that promote hematopoietic induction. The close proximity of PLM and somite cells is achieved by heterotypic interactions between Jam1a on PLM and Jam2a on somites, which helps to strengthen the Notch signal. (B): Cross‐sectional view of a zebrafish embryo at 24 hpf. Anterior portions of somites give rise to the endotome, which contributes cells to the dorsal aorta that promote HSC formation, in part by Cxcl12 production. Curved arrows denote the movement of cells. Abbreviations: CV, caudal vein; DA, dorsal aorta; hpf, hours postfertilization; J1a, Jam1a; J2a, Jam2a; PLM, posterior lateral mesoderm.
Previous studies in zebrafish implicated a role for muscle‐derived signals in HSC formation. Clements et al. demonstrated that expression of the Notch ligands deltaC (dlc) and deltaD (dld) in somites is necessary for HSC specification 38. However, the details of the interactions between somites and precursor hemogenic cells were not known. Recent work from Kobayashi et al. established that HSC fate begins during PLM migration 39. They found that the migrating PLM and the somites they move across are each expressing partner junctional adhesion molecules: PLM cells express jam1a, whereas somite cells express jam2a. Jam1a and Jam2a interact, allowing both types of cells to have close physical contact with each other, which is required for proper intercellular Notch signal transduction (Fig. 3).
This novel involvement of muscle in HSC specification illustrates the dependence of HSC emergence on neighboring tissues. The works from Nguyen et al. 36 and Kobayashi et al. 39 show without a doubt that interactions between embryonic muscle and endothelium are more complex than previously thought and are indispensable for HSC formation. Furthermore, the Kobayashi et al. 39 findings are striking because they also demonstrate that hemogenic endothelium and HSC fate are acquired much earlier than previously thought, in agreement with the work by Butko et al. 24. It seems that the stage of PLM migration is of great importance for HSC specification, and more research focusing on this developmental window is greatly warranted.
Notch at the Many Crossroads of HSC Birth
Notch signaling occurs between adjacent cells through interaction between ligands and receptors. In mammals, four receptors (Notch1–Notch4) and five canonical ligands (Jagged1, Jagged2, delta‐like 1 [Dll1], Dll3, and Dll4) have been identified. Upon ligand binding, the intracellular domain of the Notch receptor (NICD) is cleaved and translocates to the nucleus, where it acts as a transcription factor 40.
Signaling via the Notch pathway is critical for multiple steps of HSC formation, from induction of endothelial tissue to HSC emergence 19. It acts early during development promoting the determination and proliferation of endothelial precursors within the PLM, and later it directly regulates expression of genes acting more proximal to HSC formation, such as gata2, which in turn drives runx1, a necessary step for HSC emergence 19. In addition, somitically expressed ligands Dld and Dlc are required to induce hemogenic fate in endothelial cells 38, 39.
Of the four mammalian receptors, NOTCH1 was shown to be required cell‐autonomously for HSC specification, whereas NOTCH2 seems to be dispensable 41. However, the rest of the receptors were not thoroughly characterized in murine HSC development. Recently, Kim et al. analyzed the role of four Notch receptors (the Notch1 paralogs notch1a and notch1b, notch2, and notch3) during HSC induction 42. In agreement with the murine studies, zebrafish Notch1a and Notch1b are required cell‐autonomously in the endothelium (approximately 20 hpf) for HSC formation. Zebrafish Notch2, like murine NOTCH2, is dispensable for HSC specification, suggesting that Notch receptor functions are conserved in vertebrates. A novel finding from this work is that Notch3 signaling is required for HSC specification, but in a non‐cell‐autonomous fashion. The notch3 gene is expressed in PLM and somites early during development at 13 hpf and then becomes restricted to endothelium including the DA at 24 hpf. Surprisingly, it is somitic Notch3, and not endothelial Notch3, that is required for HSC formation at approximately 14 hpf (Fig. 3). It also appears that Notch3 is acting downstream of the previously known somitic Notch ligands Dlc and Dld. All together, this body of work has given us tremendous insight into the specific roles of different Notch receptors, showing that they perform nonredundant functions in muscle, endothelium, and HSCs during HSC formation, and that their roles are temporally and spatially regulated.
The complex role of Notch in HSC induction is highly conserved in mammals. Jang et al. demonstrated that overexpression of the Notch1 intracellular domain in murine embryonic stem cells resulted in increased hematopoietic differentiation during embryoid body differentiation 43. Microarray analysis of gene expression after induction of NICD overexpression allowed identification of several upregulated genes that could be involved in the specification of hemogenic endothelium, including the transcription factor Foxc2, previously known to be involved in arterial specification 44. Subsequent in vivo knockdown of the Foxc2 paralogs foxc1a and foxc1b in zebrafish embryos showed a loss of HSCs that was not rescued by NICD overexpression, suggesting that Foxc2 acts downstream of Notch. Foxc2‐null mice also have defective HSC formation, suggesting that the requirement of FOXC2 for definitive hematopoiesis is conserved in vertebrates. The discoveries from this study offer vital pieces of information on vertebrate Notch signaling and are especially valuable for improving the current state of in vitro HSC generation for future clinical applications.
Inflammatory Signaling
Studies in the last few years have shown that HSCs have a role in the early immune response to infection 45 46 47 48 49. HSCs proliferate in response to systemic infection to replace effector immune cells 50. This response is not simply secondary to diminished downstream immune cell numbers, but, rather, a direct response to systemically elevated cytokines signaling through cytokine receptors on HSCs 51. It was also found that HSCs could respond to bacterial and viral components directly through toll‐like receptors (TLRs) via the TLR‐nuclear factor κ‐light chain enhancer of activated B (TLR‐NFκB) axis 48, 49, 52.
More recently, similar cytokine signaling pathways have been found to play a critical role during embryonic HSC production independent of infection, a process termed sterile inflammation 53 54 55 56 57. The innate immune system is established early during primitive hematopoiesis with the formation of the myeloid effector cells neutrophils and macrophages. The molecular effectors produced by these cells are various cytokines, including tumor necrosis factors (TNFs) like TNF‐α and ‐β, interleukins (ILs) like IL‐6, and interferons (IFNs) like IFN‐α and ‐γ. Various studies in the last few years have explored a critical role for these proinflammatory cytokines in HSC development.
Through expression profiling studies, Li et al. showed that HSCs from the AGM regions of midgestation mouse embryos have a robust innate inflammatory gene signature 55. In particular, they observed a strong IFN signature, hinting that embryonic HSCs are responding to inflammatory cues. Combined studies in murine and zebrafish systems further demonstrated that IFN signaling is important for HSC development. Mouse or zebrafish embryos lacking IFN‐α or ‐γ signaling had significantly fewer AGM HSCs 55 (Fig. 2A). Sawamiphak et al. made a similar observation demonstrating that zebrafish lacking Ifn‐γ or its receptor Crfb17 act cell‐autonomously to positively regulate HSC development by modulating the endothelial‐to‐hematopoietic transition 56.
Other groups found supportive roles for other proinflammatory cytokines. TNF‐α signaling was shown to be important for the development of embryonic vasculature and HSCs 53, 58. By using zebrafish mutants and morpholino‐mediated knockdown, it was shown that depletion of tnfa and its receptor tnfr2 resulted in reduced numbers of HSCs in the floor of the dorsal aorta, which was not due to a secondary effect of disrupted vascular development 53, 54. Combined knockdown of tnfa and ifn‐γ resulted in more severe reduction of runx1‐positive HSCs than loss of either cytokine individually, suggesting nonredundant and cooperative functions of multiple proinflammatory cytokines during HSC development 55.
Granulocyte‐colony‐stimulating factor (Gcsf) signaling to endothelial cells was first demonstrated during embryogenesis by Liongue et al. where they showed that Gcsf/Gcsfr signaling was required during the emergency granulopoietic response in embryos 59. Stachura et al. recently demonstrated a role for Gscf in HSC emergence and expansion in zebrafish embryonic development 57. This cytokine was also recently implicated to act in the inflammatory signaling pathway during HSC development 54. Tlr4bb and its downstream effectors Myd88 and NFκB act downstream of both Gcsf and Tnf‐α within hemogenic endothelial cells to promote HSC production 54 (Fig. 2A).
As previously discussed, Notch is a well‐established pathway controlling HSC formation 38, 39, 41 42 43, 60. In the above studies, Notch was found to be a common effector of proinflammatory signaling 53, 54, 56. Tnf‐α/Tnfr2 signaling activates Notch by inducing expression of the Notch ligand jagged1a (jag1a) in endothelial cells in the DA 53. Jag1a then activates the Notch1a receptor on the neighboring hemogenic endothelium resulting in activation of the NFκB transcription factor and subsequent promotion of the endothelial‐to‐hematopoietic transition 53. In contrast, Ifn‐γ acts downstream of Notch signaling through atypical activation of Stat3 in the DA 56.
Based on the above studies, it is clear that sterile inflammatory signaling has a fundamental role in HSC emergence. Interestingly, the cellular source of cytokine production important for HSC emergence appears to be the neutrophils and macrophages generated from the first wave of embryonic hematopoiesis, indicating a new level of dependency of definitive HSCs on primitive hematopoietic cells 53 54 55, 61. Work is still needed to resolve the precise molecular and cellular mechanisms used by each myeloid cell type and each cytokine signaling pathway during HSC emergence. Recapitulation of this complex in vivo milieu is likely necessary to improve production of HSCs in vitro.
Defining the Developmental Niche
Ray Schofield was among the first to propose the concept of the niche as a place where HSCs reside 62. The niche that we now know is not only anatomic, but has a large functional dimension 63, 64. HSC niches are specialized microenvironments that maintain and regulate the HSCs, modulating their proliferation, differentiation, and self‐renewal. The interplay between HSCs and their niche is complex and dynamic, involving cell‐to‐cell interactions and secreted growth factors and chemokines, as well as physical parameters. During development, HSCs travel between niches to establish hematopoiesis (Fig. 1). After arising from hemogenic endothelium in the dorsal aorta, HSCs seed an intermediate hematopoietic niche—fetal liver in mammals and caudal hematopoietic tissue in zebrafish 65. HSCs expand rapidly in this intermediate niche and then move out and seed the adult marrow—found within bone in mammals and kidney in zebrafish. The adult niche in mice is well defined and comprises many different components, including mesenchymal stem cells, endothelial cells, osteoblasts, arterioles, and sympathetic nerves, as well as some hematopoietic elements, including megakaryocytes and macrophages 63. The secrets of developmental niches are largely elusive, but recent advances are beginning to shine some light on the problem.
To uncover the molecular underpinnings of various developmental niches, Charbord et al. recently completed a comprehensive transcriptome meta‐analysis of HSC‐supportive cells and defined the core molecular networks of the niche from the embryo to adults 66. The data support the view that niches have different characteristics emblematic of the needs of their tissue of origin. They showed that the bone marrow‐supportive cells exhibited a mesenchymal phenotype, AGM‐supportive cells expressed genes implicated in blood vessel formation, and fetal liver‐supportive cells expressed genes related to cell cycle regulation 66. These data highlight the plastic nature of niche cells and the influence of the environment on their function.
Because of the complexity of cell types that comprise the in vivo niche, cell culture systems have been invaluable to defining which cell types are capable of supporting HSC maintenance, expansion, or differentiation. Experiments using explant cultures of AGM regions helped to define the importance of BMP, VEGF, and SHH signaling from this development niche 37, 67, 68. The zebrafish is an established in vivo model, but new developments in zebrafish cell culture are now providing similar tools to mammalian systems that will permit the discovery of HSC niche components 69, 70. These discoveries can then be easily tested for functionality in vivo. Campbell et al. recently described the creation of a stromal line, zebrafish embryonic stromal trunk (ZEST) cells, which are derived from tissue surrounding the embryonic dorsal aorta 71. Similar to the findings from Charbord et al. 66 regarding mammalian AGM niche cells, ZEST cells have endothelial characteristics. Through the expression of various cytokines, ZEST cells can support both HSC proliferation and multilineage differentiation 71. This model will be a useful tool in studying the niche components important for HSC emergence and differentiation.
The next developmental niche in zebrafish is the caudal hematopoietic tissue—the functional equivalent to the mammalian fetal liver—where HSCs expand and differentiate. Recent work from Tamplin et al. explored the spatial relationship between HSCs, endothelial cells, and stromal cells within the CHT 72. They found that HSCs trigger a dynamic remodeling of endothelial cells during colonization of the CHT 72. When an HSC arrives in the perivascular niche of the CHT, a group of endothelial cells remodels to form a surrounding pocket (Fig. 2B). During cell division, the HSC is then anchored to a perivascular stromal cell, which dictates the orientation of the division plane. Interestingly, they observed similar cellular behaviors of HSCs and niche components in the murine fetal liver, indicating that this process is highly conserved. They also performed a chemical genetic screen in zebrafish to identify molecules that control the process of HSC engraftment into the CHT. They identified lycorine as a novel compound that promoted HSC lodgement during development and ultimately led to a sustained increase in the size of the stem cell pool into adulthood 72. Improving HSC lodgement after bone marrow transplantation with lycorine could represent a new therapeutic modality for improving engraftment and patient health after transplantation.
Together, these studies demonstrate the changing form and function of each developmental niche. Understanding how the AGM niche promotes HSC specification can lead to improvements in HSC production from pluripotent stem cells, while studies of how the CHT/fetal liver niche promotes HSC proliferation can aid in the expansion of HSCs in vitro to improve current bone marrow transplantation approaches. Extension of the above approaches to understand the development of the bone and kidney niches will provide insight into the origins of the ultimate HSC niche. Insights into how microenvironments adjust to the requirements of their HSC residents could reveal new therapies for treating diseases such as leukemia and improving outcomes in stem cell transplantation settings.
Conclusion
Our understanding of the origins and regulation of definitive hematopoietic stem cells has come a long way since the groundbreaking work of Dieterlen‐Lièvre and others. In particular, recent advances have identified new cellular constituents that were heretofore unknown to constitute the developing microenvironment important for HSC induction, expansion, differentiation, and migration. Additionally, signaling pathways that were previously thought to only function in adult stress hematopoiesis, such as inflammatory signaling, have now been revealed to play important roles during HSC formation. Yet, many questions still remain. Recent work in mammalian systems has challenged the hierarchical nature of HSCs claiming that most of steady‐state adult hematopoiesis is maintained by lineage‐restricted and multipotent progenitors once thought to be short‐lived 73, 74. Recently, several groups made the surprising finding that adult tissue‐resident macrophages and microglia are derived from yolk sac cells that arise before definitive HSCs, demonstrating that embryonic cells once thought to be restricted in self‐renewal are rather long‐lived and persist into adulthood 75, 76. Therefore, it is now important to understand the developmental origins and lineal relationships between embryonic HSCs and progenitor populations and these long‐lived adult progenitors. As scientists attempt to make transplantable HSCs in a dish, it is imperative to understand the repertoire and heterogeneity of long‐lived progenitors that an embryo generates. New single‐cell approaches to lineage‐tracing and gene expression profiling in the zebrafish could provide the high resolution required to dissect not only the complexity of hematopoiesis, but also the developing niche that regulates HSC and progenitor cell properties 77 78 79 80.
Author Contributions
A.D.L.G. and A.S.: conception and design, manuscript writing; T.V.B.: conception and design, financial support, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
Acknowledgments
This work was funded by Gabrielle's Angel Foundation and NIH Grant 6R03DK102975‐02 (to T.V.B.).
References
- 1. Jagannathan‐Bogdan M, Zon LI. Hematopoiesis. Development 2013;140:2463–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bertrand JY, Kim AD, Violette EP et al. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development 2007;134:4147–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bertrand JY, Chi NC, Santoso B et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 2010;464:108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen MJ, Li Y, De Obaldia ME et al. Erythroid/myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell 2011;9:541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Medvinsky A, Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 1996;86:897–906. [DOI] [PubMed] [Google Scholar]
- 6. Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 2010;464:112–115. [DOI] [PubMed] [Google Scholar]
- 7. Boisset JC, van Cappellen W, Andrieu‐Soler C et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 2010;464:116–120. [DOI] [PubMed] [Google Scholar]
- 8. Murayama E, Kissa K, Zapata A et al. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity 2006;25:963–975. [DOI] [PubMed] [Google Scholar]
- 9. Zanjani ED, Ascensao JL, Tavassoli M. Liver‐derived fetal hematopoietic stem cells selectively and preferentially home to the fetal bone marrow. Blood 1993;81:399–404. [PubMed] [Google Scholar]
- 10. Mikkola HK, Orkin SH. The journey of developing hematopoietic stem cells. Development 2006;133:3733–3744. [DOI] [PubMed] [Google Scholar]
- 11. Dieterlen‐Lievre F. On the origin of haemopoietic stem cells in the avian embryo: An experimental approach. J Embryol Exp Morphol 1975;33:607–619. [PubMed] [Google Scholar]
- 12. Medvinsky AL, Samoylina NL, Müller AM et al. An early pre‐liver intraembryonic source of CFU‐S in the developing mouse. Nature 1993;364:64–67. [DOI] [PubMed] [Google Scholar]
- 13. Walmsley M, Ciau‐Uitz A, Patient R. Adult and embryonic blood and endothelium derive from distinct precursor populations which are differentially programmed by BMP in Xenopus. Development 2002;129:5683–5695. [DOI] [PubMed] [Google Scholar]
- 14. Medvinsky A, Rybtsov S, Taoudi S. Embryonic origin of the adult hematopoietic system: Advances and questions. Development 2011;138:1017–1031. [DOI] [PubMed] [Google Scholar]
- 15. Paik EJ, Zon LI. Hematopoietic development in the zebrafish. Int J Dev Biol 2010;54:1127–1137. [DOI] [PubMed] [Google Scholar]
- 16. Ellertsdóttir E, Lenard A, Blum Y et al. Vascular morphogenesis in the zebrafish embryo. Dev Biol 2010;341:56–65. [DOI] [PubMed] [Google Scholar]
- 17. Wilkinson RN, Pouget C, Gering M et al. Hedgehog and Bmp polarize hematopoietic stem cell emergence in the zebrafish dorsal aorta. Dev Cell 2009;16:909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jin SW, Beis D, Mitchell T et al. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 2005;132:5199–5209. [DOI] [PubMed] [Google Scholar]
- 19. Butko E, Pouget C, Traver D. Complex regulation of HSC emergence by the Notch signaling pathway. Dev Biol 2016;409:129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ciau‐Uitz A, Monteiro R, Kirmizitas A et al. Developmental hematopoiesis: Ontogeny, genetic programming and conservation. Exp Hematol 2014;42:669–683. [DOI] [PubMed] [Google Scholar]
- 21. Robert‐Moreno A, Espinosa L, de la Pompa JL et al. RBPjkappa‐dependent Notch function regulates Gata2 and is essential for the formation of intra‐embryonic hematopoietic cells. Development 2005;132:1117–1126. [DOI] [PubMed] [Google Scholar]
- 22. Tsai FY, Keller G, Kuo FC et al. An early haematopoietic defect in mice lacking the transcription factor GATA‐2. Nature 1994;371:221–226. [DOI] [PubMed] [Google Scholar]
- 23. de Pater E, Kaimakis P, Vink CS et al. Gata2 is required for HSC generation and survival. J Exp Med 2013;210:2843–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Butko E, Distel M, Pouget C et al. Gata2b is a restricted early regulator of hemogenic endothelium in the zebrafish embryo. Development 2015;142:1050–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. North TE, de Bruijn MF, Stacy T et al. Runx1 expression marks long‐term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity 2002;16:661–672. [DOI] [PubMed] [Google Scholar]
- 26. Adya N, Castilla LH, Liu PP. Function of CBFbeta/Bro proteins. Semin Cell Dev Biol 2000;11:361–368. [DOI] [PubMed] [Google Scholar]
- 27. Wang Q, Stacy T, Miller JD et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell 1996;87:697–708. [DOI] [PubMed] [Google Scholar]
- 28. Wang Q, Stacy T, Binder M et al. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA 1996;93:3444–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bresciani E, Carrington B, Wincovitch S et al. CBFβ and RUNX1 are required at 2 different steps during the development of hematopoietic stem cells in zebrafish. Blood 2014;124:70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cimmino L, Abdel‐Wahab O, Levine RL et al. TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell 2011;9:193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shih AH, Abdel‐Wahab O, Patel JP et al. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer 2012;12:599–612. [DOI] [PubMed] [Google Scholar]
- 32. Cimmino L, Dawlaty MM, Ndiaye‐Lobry D et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol 2015;16:653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moran‐Crusio K, Reavie L, Shih A et al. Tet2 loss leads to increased hematopoietic stem cell self‐renewal and myeloid transformation. Cancer Cell 2011;20:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ko M, Bandukwala HS, An J et al. Ten‐Eleven‐Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA 2011;108:14566–14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li C, Lan Y, Schwartz‐Orbach L et al. Overlapping requirements for Tet2 and Tet3 in normal development and hematopoietic stem cell emergence. Cell Reports 2015;12:1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nguyen PD, Hollway GE, Sonntag C et al. Haematopoietic stem cell induction by somite‐derived endothelial cells controlled by meox1. Nature 2014;512:314–318. [DOI] [PubMed] [Google Scholar]
- 37. Jaffredo T, Lempereur A, Richard C et al. Dorso‐ventral contributions in the formation of the embryonic aorta and the control of aortic hematopoiesis. Blood Cells Mol Dis 2013;51:232–238. [DOI] [PubMed] [Google Scholar]
- 38. Clements WK, Kim AD, Ong KG et al. A somitic Wnt16/Notch pathway specifies haematopoietic stem cells. Nature 2011;474:220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kobayashi I, Kobayashi‐Sun J, Kim AD et al. Jam1a‐Jam2a interactions regulate haematopoietic stem cell fate through Notch signalling. Nature 2014;512:319–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Andersson ER, Sandberg R, Lendahl U. Notch signaling: Simplicity in design, versatility in function. Development 2011;138:3593–3612. [DOI] [PubMed] [Google Scholar]
- 41. Kumano K, Chiba S, Kunisato A et al. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity 2003;18:699–711. [DOI] [PubMed] [Google Scholar]
- 42. Kim AD, Melick CH, Clements WK et al. Discrete Notch signaling requirements in the specification of hematopoietic stem cells. EMBO J 2014;33:2363–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jang IH, Lu YF, Zhao L et al. Notch1 acts via Foxc2 to promote definitive hematopoiesis via effects on hemogenic endothelium. Blood 2015;125:1418–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kume T. Foxc2 transcription factor: A newly described regulator of angiogenesis. Trends Cardiovasc Med 2008;18:224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baldridge MT, King KY, Boles NC et al. Quiescent haematopoietic stem cells are activated by IFN‐gamma in response to chronic infection. Nature 2010;465:793–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Feng CG, Weksberg DC, Taylor GA et al. The p47 GTPase Lrg‐47 (Irgm1) links host defense and hematopoietic stem cell proliferation. Cell Stem Cell 2008;2:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Essers MA, Offner S, Blanco‐Bose WE et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature 2009;458:904–908. [DOI] [PubMed] [Google Scholar]
- 48. Kobayashi H, Kobayashi CI, Nakamura‐Ishizu A et al. Bacterial c‐di‐GMP affects hematopoietic stem/progenitors and their niches through STING. Cell Reports 2015;11:71–84. [DOI] [PubMed] [Google Scholar]
- 49. Takizawa H, Regoes RR, Boddupalli CS et al. Dynamic variation in cycling of hematopoietic stem cells in steady state and inflammation. J Exp Med 2011;208:273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Takizawa H, Boettcher S, Manz MG. Demand‐adapted regulation of early hematopoiesis in infection and inflammation. Blood 2012;119:2991–3002. [DOI] [PubMed] [Google Scholar]
- 51. King KY, Goodell MA. Inflammatory modulation of HSCs: Viewing the HSC as a foundation for the immune response. Nat Rev Immunol 2011;11:685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao JL, Ma C, O'Connell RM et al. Conversion of danger signals into cytokine signals by hematopoietic stem and progenitor cells for regulation of stress‐induced hematopoiesis. Cell Stem Cell 2014;14:445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Espín‐Palazón R, Stachura DL, Campbell CA et al. Proinflammatory signaling regulates hematopoietic stem cell emergence. Cell 2014;159:1070–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. He Q, Zhang C, Wang L et al. Inflammatory signaling regulates hematopoietic stem and progenitor cell emergence in vertebrates. Blood 2015;125:1098–1106. [DOI] [PubMed] [Google Scholar]
- 55. Li Y, Esain V, Teng L et al. Inflammatory signaling regulates embryonic hematopoietic stem and progenitor cell production. Genes Dev 2014;28:2597–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sawamiphak S, Kontarakis Z, Stainier DY. Interferon gamma signaling positively regulates hematopoietic stem cell emergence. Dev Cell 2014;31:640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stachura DL, Svoboda O, Campbell CA et al. The zebrafish granulocyte colony‐stimulating factors (Gcsfs): 2 paralogous cytokines and their roles in hematopoietic development and maintenance. Blood 2013;122:3918–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Espín R, Roca FJ, Candel S et al. TNF receptors regulate vascular homeostasis in zebrafish through a caspase‐8, caspase‐2 and P53 apoptotic program that bypasses caspase‐3. Dis Model Mech 2013;6:383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liongue C, Hall CJ, O'Connell BA et al. Zebrafish granulocyte colony‐stimulating factor receptor signaling promotes myelopoiesis and myeloid cell migration. Blood 2009;113:2535–2546. [DOI] [PubMed] [Google Scholar]
- 60. Bigas A, Guiu J, Gama‐Norton L. Notch and Wnt signaling in the emergence of hematopoietic stem cells. Blood Cells Mol Dis 2013;51:264–270. [DOI] [PubMed] [Google Scholar]
- 61. Hall C, Crosier P, Crosier K. Inflammatory cytokines provide both infection‐responsive and developmental signals for blood development: Lessons from the zebrafish. Mol Immunol 2016;69:113–122. [DOI] [PubMed] [Google Scholar]
- 62. Schofield R. The relationship between the spleen colony‐forming cell and the haemopoietic stem cell. Blood Cells 1978;4:7–25. [PubMed] [Google Scholar]
- 63. Mendelson A, Frenette PS. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med 2014;20:833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Boulais PE, Frenette PS. Making sense of hematopoietic stem cell niches. Blood 2015;125:2621–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bowman TV, Zon LI. Lessons from the niche for generation and expansion of hematopoietic stem cells. Drug Discov Today Ther Strateg 2009;6:135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Charbord P, Pouget C, Binder H et al. A systems biology approach for defining the molecular framework of the hematopoietic stem cell niche. Cell Stem Cell 2014;15:376–391. [DOI] [PubMed] [Google Scholar]
- 67. Durand C, Robin C, Bollerot K et al. Embryonic stromal clones reveal developmental regulators of definitive hematopoietic stem cells. Proc Natl Acad Sci USA 2007;104:20838–20843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Crisan M, Solaimani Kartalaei P, Neagu A et al. BMP and Hedgehog Regulate Distinct AGM Hematopoietic Stem Cells Ex Vivo. Stem Cell Rep 2016;6:383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Stachura DL, Traver D. Cellular dissection of zebrafish hematopoiesis. Methods Cell Biol 2011;101:75–110. [DOI] [PubMed] [Google Scholar]
- 70. Svoboda O, Stachura DL, Machonova O et al. Ex vivo tools for the clonal analysis of zebrafish hematopoiesis. Nat Protoc 2016;11:1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Campbell C, Su T, Lau RP et al. Zebrafish embryonic stromal trunk (ZEST) cells support hematopoietic stem and progenitor cell (HSPC) proliferation, survival, and differentiation. Exp Hematol 2015;43:1047–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tamplin OJ, Durand EM, Carr LA et al. Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell 2015;160:241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sun J, Ramos A, Chapman B et al. Clonal dynamics of native haematopoiesis. Nature 2014;514:322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Busch K, Klapproth K, Barile M et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 2015;518:542–546. [DOI] [PubMed] [Google Scholar]
- 75. Gomez Perdiguero E, Klapproth K, Schulz C et al. Tissue‐resident macrophages originate from yolk‐sac‐derived erythro‐myeloid progenitors. Nature 2015;518:547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ginhoux F, Greter M, Leboeuf M et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010;330:841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chen CH, Puliafito A, Cox BD et al. Multicolor cell barcoding technology for long‐term surveillance of epithelial regeneration in zebrafish. Dev Cell 2016;36:668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pan YA, Freundlich T, Weissman TA et al. Zebrabow: Multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development 2013;140:2835–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Satija R, Farrell JA, Gennert D et al. Spatial reconstruction of single‐cell gene expression data. Nat Biotechnol 2015;33:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wu CC, Kruse F, Vasudevarao MD et al. Spatially resolved genome‐wide transcriptional profiling identifies bmp signaling as essential regulator of zebrafish cardiomyocyte regeneration. Dev Cell 2016;36:36–49. [DOI] [PubMed] [Google Scholar]


