Abstract
The filamentous nonheterocystous cyanobacterial genus Katagnymene is a common diazotrophic component of tropical and subtropical oceans. To assess the phylogenetic affiliation of this taxon, two partial 16S rRNA gene sequences and 25 partial hetR gene sequences originating from the genera Katagnymene and Trichodesmium collected from open, surface waters of the Atlantic, Indian, and Pacific oceans were compared. Single trichomes or colonies were identified morphologically by using light microscopy and then used directly as templates in hetR PCR analyses. In addition, three cultured strains, identified as Katagnymene pelagica, Katagnymene spiralis, and Trichodesmium sp., were examined. The data show that the genus Katagnymene is in the Trichodesmium cluster and that K. pelagica Lemmermann and K. spiralis Lemmermann are most likely one species, despite their different morphologies. Phylogenetic analyses also unveiled four distinct clusters in the Trichodesmium cluster, including one novel cluster. Our findings emphasize the conclusion that known morphological traits used to differentiate marine nonheterocystous cyanobacteria at the genus and species levels correlate poorly with genetic data, and a revision is therefore suggested.
The world's tropical and subtropical oceans are highly oligotrophic and are therefore a suitable habitat for diazotrophic cyanobacteria. In the open ocean, the nonheterocystous cyanobacterial genus Trichodesmium is very common. Trichodesmium was first discovered in 1830 (9), and in 1961 it was found to be diazotrophic; its ecology and physiology have since been characterized in numerous studies (6, 17). Recent estimates suggest that Trichodesmium alone may account for 40 to 50% of the global nitrogen sequestered through biological nitrogen fixation (17). One key feature of Trichodesmium is its ability to form colonies, but the cells may also exist as free trichomes. The marine nonheterocystous genus Katagnymene occurs only as free trichomes and was recently found to be diazotrophic (22). Like Trichodesmium, its nitrogen fixation activity is restricted to the photophase, and nitrogenase is confined to subsets of cells termed diazocytes (10, 12), which in Katagnymene account for about 7% of the total cells. Thus, Katagnymene and Trichodesmium share a unique diazotrophic behavior (5, 15, 22).
Katagnymene was first described by Lemmermann (19) and was divided into two species, Katagnymene pelagica and Katagnymene spiralis, based on the degree of trichome coiling. Katagnymene occurs in all major tropical and subtropical oceans (18, 19, 22, 24, 30). The trichomes are characterized by cells that are shorter than they are wide and by being surrounded by a distinct mucilaginous sheath. K. spiralis is more or less spirally coiled, and both Katagnymene species may form long trichomes that sometimes are up to 15 mm long. In previous taxonomic studies, Katagnymene was suggested to belong to the oscillatoriacean family, together with Trichodesmium spp.; the former genus was placed in the subfamily Hormoscilloidae, and the latter genus was placed in the Phormidiaceae (1). Previously, Drouet (8) suggested that K. spiralis should be classified as Microcoleus lyngbyaceus based on the thickened cell walls of the trichome end cells and that K. pelagica var. major Wille should be classified as Oscillatoria (Trichodesmium) erythraea. Besides the two marine species, the genus Katagnymene also includes five fresh or brackish water species (1, 2). It has recently been suggested that K. spiralis is closely related to Trichodesmium spp. based on certain gene sequence similarities (22, 26).
Likewise, the taxonomy of Trichodesmium has often been revised, and only one extensive genetic study has been performed (14). This study showed that the genus harbors five marine species in the following three main clusters: (i) a cluster which includes Trichodesmium hildebrandtii and fusiform and spherical colonies of Trichodesmium thiebautii; (ii) a cluster which includes Trichodesmium erythraeum (including the laboratory strain Trichodesmium sp. strain IMS 101); and (iii) a cluster which includes Trichodesmium tenue and Trichodesmium contortum.
In the present study, collected Katagnymene and Trichodesmium specimens first were identified by using light microscopy and then were used as templates in PCR in order to generate sequences that could be directly linked to a specific morphology. A 448-bp fragment of the hetR gene, proposed to be involved in diazocyte differentiation (10), was amplified and analyzed for cyanobacteria identified as Katagnymene spp. or Trichodesmium spp., which were collected in various geographic regions, and for a few cultured specimens. Our aim was to determine the relationship between Katagnymene spp. and Trichodesmium spp. using both 16S rRNA gene and hetR gene sequences isolated from natural populations and cultures of members of both genera.
MATERIALS AND METHODS
Sample collection.
Cyanobacteria were collected during three cruises, in 1998 during a cruise of the R/V Roger Revelle in the southwestern Pacific Ocean from New Zealand to Fiji, in 1999 during a cruise of the R/V Maurice Ewing off northern Australia, and in 2001 during a cruise of the R/V Seward Johnson in the midwestern Atlantic Ocean off Barbados. Samples were collected from the surface down to a depth of 20 m by using plankton tows. The morphological descriptions of Lemmermann (19), Wille (30), and Karsten (18) were used for identification (Table 1). The samples obtained in 1999 and 2000 were identified and photographed with a light microscope (Fig. 1 and Table 2). Then in each case the same trichome was recovered and transferred in a micropipette in 4 droplets of filtered seawater and 1 droplet of Milli-Q water before it was placed in a PCR tube containing 10 μl of Milli-Q water. The samples were frozen, thawed, and used directly as templates in PCR.
TABLE 1.
Morphological characteristics of K. spiralis and K. pelagica given in the original descriptionsa
FIG. 1.
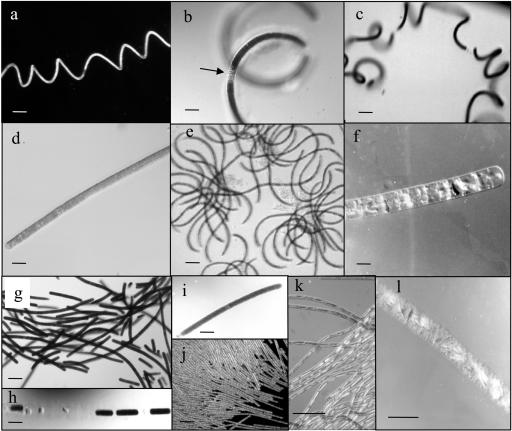
Representative morphologies of some of the species of Katagnymene and Trichodesmium included in the phylogenetic analyses. Further details are given in Table 2. The morphotypes belong to cluster I (a to e), cluster II (f), cluster III (g to j), and cluster IV (k and l). (a) Cultured JWI1 (K. spiralis). Bar, 100 μm. (b) Specimen A27 (K. spiralis), with a pronounced mucilaginous sheath and with less pigmentation and segmentation (arrow). Bar, 50 μm. (c) Specimen B41-1 (K. spiralis). Bar, 100 μm. (d) Cultured PLA1 (K. pelagica). Bar, 50 μm. (e) Specimen A25, puff-shaped colony of Trichodesmium sp. with curved trichomes. Bar, 100 μm. (f) Specimen B49 (Trichodesmium sp.). Note the scattered gas vacuoles appearing as light-reflecting objects. Bar, 50 μm. (g) Cultured PLA2 (Trichodesmium sp.). Bar, 100 μm. (h) Part of a trichome of specimen B41-3 (Trichodesmium sp.) showing segmentation. Bar, 100 μm. (i) Specimen A13 (Trichodesmium sp.). Bar, 100 μm. (j) Part of a colony of T. erythraeum. Bar, 100 μm. (k) Part of a colony of T. tenue. Note the presence of the few light-reflecting large gas vesicles in the narrow cells. Bar, 50 μm. (l) Specimen B46 (T. contortum). Note the presence of several clustered light-reflecting gas vesicles. Bar, 50 μm.
TABLE 2.
Morphological characteristics, collection sites, and accession numbers for the trichomes and colonies used for hetR and 16S rRNA gene sequencing
| Specimena | Diam of trichome (μm) | Filament shape/segments | Colony | Geographic originb | Current affiliation | Cluster | Accession no. |
|---|---|---|---|---|---|---|---|
| hetR sequences | |||||||
| JWI1 | 14-17 | Coiled/+ | − | Zanzibar, IO | K. spiralis | I | AF490673 |
| PLA1 | 15-20 | Straight/+ | − | AO | K. pelagica | I | AF490676 |
| PLA2 | 25-35 | Straight/+ | + | AO | NDc | III | AF490686 |
| A13 | 60 | Straight/− | − | PO/IO | ND | III | AF490675 |
| A25 | 13 | Curled/− | + | PO/IO | Trichodesmium | I | AF490684 |
| A27 | 20 | Coiled/− | − | PO/IO | K. spiralis | I | AF490681 |
| A28 | 15 | Straight/− | − | PO/IO | ND | III | AF490678 |
| A31 | 20 | Straight/− | − | PO/IO | ND | III | AF490688 |
| A32-1 | 15 | Straight/+ | − | PO/IO | K. pelagica | I | AF490690 |
| A32-2 | 18 | Straight/+ | − | PO/IO | ND | III | AF490689 |
| B41-1 | 15 | Coiled/− | − | 10°01′N, 44°57′W | K. spiralis | I | AF490677 |
| B41-2 | 12 | Coiled/− | − | 10°01′N, 44°57′W | K. spiralis | I | AF490687 |
| B41-3 | 35 | Straight/− | − | 10°01′N, 44°57′W | ND | III | AF490674 |
| B46 | 30 | Straight/− | − | 09°00′N, 52°01′W | T. contortum | IV | AF490685 |
| B49 | 20 | Straight/− | − | 10°60′N, 56°17′W | ND | II | AF490680 |
| B51-1 | 20 | Straight/− | − | 11°30′N, 55°00′W | ND | I | AF490679 |
| B51-2 | 20 | Straight/− | − | 11°30′N, 55°00′W | K. pelagica | I | AF490683 |
| B51-3 | 25 | Straight/− | − | 11°30′N, 55°00′W | ND | II | AF490682 |
| F8-1 | 20 | Straight/− | − | 25°57′S, 167°39′E | K. pelagica | I | AF490694 |
| F8-3 | 35 | Straight/− | − | 25°57′S, 167°39′E | K. pelagica | I | AF490695 |
| F8-5 | 17 | Coiled/− | − | 25°57′S, 167°39′E | K. spiralis | I | AF490697 |
| F8-6 | 17 | Coiled/− | − | 25°57′S, 167°39′E | K. spiralis | I | AF490691 |
| F8-8 | 25 | Straight/− | − | 25°57′S, 167°39′E | K. pelagica | I | AF490696 |
| F47-6 | 12-23 | Coiled/− | − | 19°17′S, 175°35′E | K. spiralis | I | AF490693 |
| F45-1 | 17 | Coiled/− | − | 18°57′S, 175°40′E | K. spiralis | I | AF490692 |
| 16S rRNA gene sequences | |||||||
| F34-8 | 30 | Straight/− | − | PO | ND | IV | AF518770 |
| JWI1 | 14-17 | Coiled/− | − | Zanzibar, IO | K. spiralis | I | AF518769 |
The origins of the samples are indicated as follows: F, 1998 southwest Pacific Ocean cruise; A, 1999 cruise off northern Australia (Indian Ocean or Pacific Ocean); and B, 2001 cruise in the midwestern Atlantic Ocean.
Abbreviations: AO, Atlantic Ocean; IO, Indian Ocean; PO, Pacific Ocean.
ND, not determined.
Cultures.
Cultures PLA1 (preliminarily identified as K. pelagica) and PLA2 (no affiliation) were isolated (by P.L.) as single trichomes during the 2001 cruise in the western Atlantic Ocean. The culture of K. spiralis (JWI1) was isolated by J. Waterbury (Woods Hole Oceanographic Institution, Woods Hole, Mass.) from the Zanzibar Channel in Tanzania in 1999. The cultures are maintained in a nitrogen-free amended seawater (Sigma) medium containing trace metals, EDTA, and vitamins as described previously (7) and supplemented with 15 μM phosphoric acid, and they are grown with a daily cycle consisting of 12 h of darkness and 12 h of light (40 μmol of photons · m−2 · s−1) at 27°C.
PCR amplification and sequencing.
Partial 16S rRNA gene sequences were obtained by using the cyanobacterium-specific primers CYA106F and CYA781R (25). Each PCR was performed with an initial denaturation step consisting of 95°C for 4 min, followed by 30 cycles of 1 min at 93°C, 1 min at 50°C (57°C for the 16S rRNA gene), and 1 min at 72°C and a final extension step consisting of 72°C for 4 min. For all of the samples except those obtained in 1998, the trichomes were used directly as templates. Each PCR mixture (total volume, 25 μl) contained each deoxynucleoside triphosphate at a concentration of 200 nM, 0.5 mM MgCl2, 0.5 U of Dynazyme polymerase, Dynazyme reaction buffer, 0.5 μM primer PH1, and 0.5 μM primer PH2. Primers PH1 (5′-TGY GCK ATT TAY ATG ACC TA-3′) and PH2 (5′-ATG AAN GGT ATK CCC CAA GGA-3′) were constructed based on previously published Trichodesmium hetR sequences for amplification of a 448-bp fragment. The PCR fragments were separated on a 1.5% agarose gel. When a band was weak, the band was purified and used as a template in a second PCR. The gel fragments were purified and subjected to one of the following two treatments. The PCR products were either sequenced directly or cloned into the pCR2.1 vector (Invitrogen, San Diego, Calif.) by using a Rapid DNA ligation kit (Boehringer, Mannheim, Germany) and transformed by using One Shot competent cells (Invitrogen). Plates were screened for white colonies. These colonies were checked to make sure that they contained the correct insert by colony PCR, and positive colonies were grown in Luria-Bertani medium overnight. Then DNA preparations were checked for the correct insert by enzymatic restriction. For sequencing of cloned inserts primers T7 and M13 reverse were used. When the purified PCR products were sequenced directly, primers PH1 and PH2 were used. cDNA strands were sequenced by using BigDye terminators (PE Applied Biosystems) with a Perkin-Elmer model ABI 377 automated sequencer.
Sequence analysis.
When necessary, the alignments of sequences were edited manually with Seaview (13). hetR trees were constructed by using PHYLIP (version 3.6a; Department of Genetics, University of Washington, Seattle) (http://evolution.genetics.washington.edu/phylip.html). The sequences of Aphanizomenon sp. strain KAC 15 and Leptolyngbya sp. strain PCC 73110 were used as outgroups. When distance matrices were used to generate the trees, the correction of Jukes and Cantor (16) was employed. For the nucleotide maximum-likelihood tree, the DNAML program was used (11), with the transition/transversion ratio set to 1.69 and the base frequencies set to 0.291 (A), 0.189 (C), 0.214 (G), and 0.305 (T). The transition/transversion ratio and base frequencies were determined by using PhyloWin. For maximum-likelihood analyses of the translated amino acid sequences of hetR, TREE-PUZZLE 5.0 was used (27). The analysis with TREE-PUZZLE was performed with the default settings, except that the model of rate heterogeneity was chosen as a gamma distributed rate.
The 16S rRNA gene sequences were analyzed by calculating distances between pairs of sequences by using the distance correction of Tajima and Nei (28), followed by construction of a phylogenetic tree by the neighbor-joining method and bootstrap resampling with 500 replicates (Fig. 2). This analysis was carried out by using the TREECON software package (29). Leptolyngbya sp. strain PCC 73110 was used as the outgroup in the 16S rRNA gene-based tree.
FIG. 2.
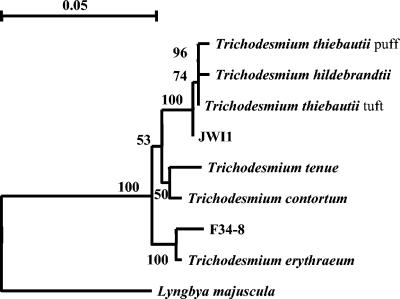
Phylogenetic comparison of F34-5 (Trichodesmium sp.), JWI1 (K. spiralis), and the extant Trichodesmium cluster (14) based on partial 16S rRNA gene sequences. The F34-5 (Trichodesmium sp.) and JWI1 (K. spiralis) sequences both fall in the Trichodesmium cluster with 100% bootstrap support. The sequences were analyzed by calculating the distances between pairs of sequences by using the distance correction of Tajima and Nei (28), followed by construction of a phylogenetic tree by the neighbor-joining method and bootstrap resampling with 500 replicates. The analysis was carried out with the TREECON software package (29). Lyngbya majuscula was used as an outgroup. Scale bar = 0.05 substitution per sequence position.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences determined in the present study are given in Table 2.
RESULTS
Microscopic observations.
Our microscopic observations and the origins of the collected specimens are summarized in Table 2. For identification of Katagnymene we used the following criteria described by Lemmermann (19) for the two marine species: (i) trichomes in which the cell diameter varies from 12 to 35 μm, with cells that are shorter than they are wide; (ii) trichomes surrounded by a mucilaginous sheath; (iii) trichomes coiled to straight; (iv) trichomes often segmented, yet retained in the same sheath; and (v) trichomes solitary (trichomes do not aggregate into colonies). Seven specimens, including the cultured JWI1 specimen, were identified as K. spiralis. The trichomes were coiled, and the cell diameters ranged from 12 to 30 μm (Fig. 1a to d). The degree of coiling varied, and the trichomes were surrounded by a thick mucilaginous sheath. Regions with less pigmentation were often observed; this may have been an initial step in the formation of necrotic cells, leading to segmentation of trichomes (Fig. 1b and c). Six specimens, including the cultured PLA1specimen, were identified as K. pelagica. The trichomes were straight, had widths that ranged from 15 to 35 μm, and were surrounded by a mucilaginous sheath (Table 2 and Fig. 1e and f). Regions where there was less pigmentation and segmentation of the trichome were also observed in K. pelagica (Fig. 1e).
The remainder of the collected but unidentified single cyanobacterial trichomes and one colony were separated into three different groups on the basis of morphological characteristics (i.e., morphotypes). None of the morphotypes observed clearly fit descriptions given previously. These included A25, collected off northern Australia in 1999, which formed loosely aggregated colonies composed of curled trichomes. The trichomes ranged in diameter from 10 to 14 μm (Fig. 1e). Colonies with similar morphologies have also been observed in the Zanzibar Channel in Tanzania (unpublished observations). Another morphotype was observed in the midwestern Atlantic Ocean in 2001. The trichomes were 20 to 25 μm wide (B49 and B51-3) and sheathless and had randomly scattered gas vacuoles. The trichomes were golden and did not form colonies (Fig. 1f). Moreover, on all three cruises yet another morphotype was observed; in this morphotype the trichome diameters varied from 18 to 60 μm, and the cells were shorter than they were wide (they were one-fifth to one-eighth times as long as they were wide). The trichomes were often highly segmented, and colonies were not observed. However, the cultured PLA2 organism is a representative of this morphotype, and it can form loose colonies at least under laboratory conditions (Fig. 1g to i). This morphotype is identical to one described in previous work (15) and does not correspond to the description of either Katagnymene or Trichodesmium. Trichome morphology identical to that of T. contortum (14) was detected in the midwestern Atlantic Ocean in 2001. The trichomes were 30 to 35 μm wide and pale brown (B46) (Fig. 1l). The trichome morphologies of T. erythraeum and T. tenue are included for comparative purposes in Fig. 1 (Fig. 1j and k), while no sequences are reported here for either of these species.
Genetic analyses.
Part (584 bp) of the highly conserved gene sequence of the small-subunit rRNA gene was sequenced for some of the cyanobacteria collected. In the resulting phylogenetic tree the cultured JWI1 specimen (Table 2 and Fig. 1a), assigned to the species K. spiralis, and the morphotype F34-5 (Table 2 and Fig. 1i to k) both fell in the Trichodesmium cluster with 100% bootstrap support (Fig. 2). Furthermore, the phylogenetic analyses revealed that the 16S rRNA gene sequence of JWI1 was closely related to those of T. thiebautii and T. hildebrandtii, while F34-5 (Trichodesmium sp.) was closely related to T. erythraeum.
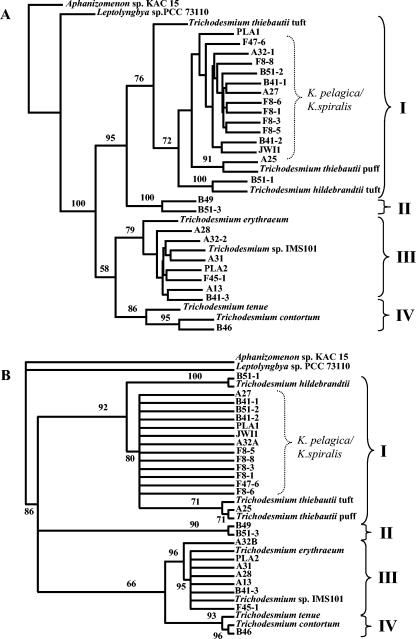
As Trichodesmium spp. display low genetic diversity, the more variable gene hetR was used previously to analyze the intrageneric phylogeny (14). In view of this, we used partial hetR gene sequences (448 bp) to further resolve the relationship between Katagnymene spp. and Trichodesmium spp. The 25 hetR sequences amplified in the present study were all from organisms located in the Trichodesmium cluster with 100% bootstrap support (Fig. 3a). Both the resulting hetR nucleotide and amino acid trees, containing novel and previously described Trichodesmium spp. (the latter sequences were included for comparative purposes), contained four well-defined clusters of related sequences (Fig. 3). Cluster I comprised T. thiebautii, T. hildebrandtii, two of the novel sequences, and all 13 sequences from members of K. spiralis and K. pelagica. Cluster II exclusively encompassed two identical sequences from a novel morphotype. Cluster III comprised T. erythraeum along with seven sequences from the large dark-pigmented trichomes. Cluster IV contained T. tenue and T. contortum, as well as one sequence from T. contortum from this study. As Fig. 3B shows, the 13 hetR sequences derived from trichomes morphologically identified as K. spiralis or K. pelagica formed a distinct radiation within Trichodesmium cluster I. These sequences included sequences from K. pelagica collected from six different sampling sites on the three cruises, and seven sequences were generated from K. spiralis collected on the three cruises and one cultured isolate from the Indian Ocean (Table 2 and Fig. 3). Morphologically, K. spiralis from the Pacific Ocean had a larger cell diameter than the corresponding trichomes from the Atlantic Ocean. Smaller morphological differences were observed for K. pelagica from the different geographic areas sampled (Table 2). The overall topologies of the hetR nucleotide and deduced amino acid trees were similar (Fig. 3). However, there was one exception in cluster I: in the nucleotide-based tree (Fig. 3A) the sequence of the T. thiebautii tuft appeared to be ancestral to all other sequences in cluster I with a weak supporting bootstrap value (72), but in the amino acid tree (Fig. 3B) T. thiebautii was ancestral to the A25 and T. thiebautii puff sequences only. This makes classification of the different T. thiebautii phylotypes and morphotypes difficult, and further studies are needed in order to resolve this issue. The different tree topologies observed for cluster I could be explained by a high degree of homoplasy within the group of sequences, to which the maximum-likelihood method is less sensitive. The distance and parsimony analyses of the hetR nucleotide sequences resulted in similar topologies, but the sequence of the cultured PLA1 specimen was grouped outside the cluster containing the other sequences from K. pelagica and K. spiralis (data not shown).
FIG. 3.
Phylogenetic trees inferred from partial hetR sequences obtained in the present study from natural and cultured populations of Katagnymene and Trichodesmium species and from the extant Trichodesmium cluster (14). Aphanizomenon sp. strain KAC15 and Leptolyngbya sp. strain PCC 73110 were used as outgroups. The brackets with roman numbers indicate the four main clusters in the Trichodesmium cluster. The brackets labeled K. pelagica/K. spiralis indicate the 13 sequences derived from trichomes originally identified as K. pelagica or K. spiralis. (A) Maximum-likelihood analysis of partial hetR nucleotide sequences. (B) Maximum-likelihood analysis of partial hetR amino acid sequences. For the analyses of the amino acid sequences encoded by hetR, TREE-PUZZLE 5.0 (27) was performed with the default settings, except that the model of rate heterogeneity was chosen as a gamma distributed rate. Abbreviations: IMS, culture collection of the Institute of Marine Science, University of North Carolina, Morehead City; KAC, Kalmar Algae Collection; PCC, Pasteur Culture Collection.
DISCUSSION
Our data clearly demonstrate that the classification of the marine cyanobacteria hitherto classified as belonging to the genus Katagnymene is not supported by the 16S rRNA gene and hetR phylogenies presented here. Rather, all the marine nonheterocystous filamentous cyanobacteria collected and analyzed fall in the same major cluster, the Trichodesmium cluster. Our data also for the first time demonstrate that Trichodesmium is divided into four main clusters, thereby extending previous studies (14, 26) that identified three clusters by analysis of the same hetR sequence and identified two clusters with a smaller number of strains by 16S-23S rRNA intergenic transcribed sequence (ITS) analyses.
The large cluster I, besides known Trichodesmium species, contained all the morphologically identified Katagnymene spp. sequences. The partial hetR sequences from the 13 specimens of K. pelagica and K. spiralis appeared to be randomly intermixed in this cluster. Hence, our data suggest that these two species should be merged into one species in spite of the morphological diversity represented by trichomes whose widths range from 12 to 35 μm and the great differences in the degree of coiling. Furthermore, it is clear that phenotypes such as appearing in a noncolonial state, as solitary trichomes, and as coiled trichomes are inconsistent with the genetic data presented here. The original descriptions of Katagnymene and Trichodesmium (9, 19) relied on such phenotypic traits, and revision is therefore needed. Consistent with this, analyses of Baltic Sea Nodularia demonstrated that the distinction between coiled and noncoiled trichomes was not supported by phycocyanin-intergenic sequence, gvpA-IGS, or rDNA-ITS genotypic grouping (3). Furthermore, strains of the coiled cyanobacterium Arthrospira are not closely related to other coiled cyanobacteria based on 16S rRNA gene sequences, and coiling of trichomes can be lost during culturing (20, 23). The capacity to form colonies and the colony shapes were not correlated to the genetic data, since different colony shapes of the colony-forming organisms (Trichodesmium) and the non-colony-forming organisms (Katagnymene) were intermixed in cluster I. Moreover, in cluster III the tuft-shaped T. thiebautii colonies were also shown to be genetically identical to the puff-shaped colonies of the same species when other genetic markers were used (4, 26). However, the only partial hetR sequence from a puff-shaped colony examined here clustered with the sequence reported previously. The close relationship between Katagnymene spp. and Trichodesmium found here was substantiated by previous data obtained by using partial nifH sequencing (22), as well as sequencing of the 16S-23S rRNA ITS combined with hetR denaturing gradient gel electrophoresis analyses and HIP1 fingerprinting (26).
Cluster II, represented by gold-pigmented colonies (Table 2 and Fig. 3), is a novel cluster compared to the three clusters identified by Janson et al. (14). The cluster II specimens most likely represent a new species. However, due to a lack of reference material other than the hetR sequences, a formal description cannot be presented at this time.
Our data and those in previous analyses (14, 26) collectively demonstrate that T. erythraeum is the most distinctly separate cluster (cluster III) in the Trichodesmium radiation. Also included in this cluster are sequences from large (>30-μm) dark-pigmented trichomes with a high degree of segmentation, previously referred to as T. contortum due to the morphological overlap (15). This was unexpected as T. erythraeum is characterized by a cell width of 6 to 12 μm (30) and forms raft-shaped colonies (Fig. 1l), while the dark-pigmented morphotype examined here was typically more than 30 μm wide and colonies were not observed. Like cluster I, cluster III apparently encompasses sequences from a wide array of morphotypes, which, however, showed more than 98% hetR nucleotide sequence similarity.
Furthermore, a hetR sequence was retrieved from T. contortum (sample B-46) (Fig. 1m and 3). The specimen was morphologically and genotypically (100% sequence identity) homologous to the specimen described previously (14), and both matched the original morphological description (30). Our data therefore extended a recent study (26) by demonstrating a morphological and genotypic difference between T. contortum and K. spiralis.
In conclusion, the genus Trichodesmium was resolved into four main clusters, which are composed of morphologically diverse strains. One of these clusters (cluster I) also contains a radiation comprising the two previously described marine Katagnymene species, and cluster II is a novel Trichodesmium lineage. One apparent link between the members of the genus Katagnymene and the members of the genus Trichodesmium is their nitrogen fixation behavior, which is unique among cyanobacteria (5, 15, 21, 22). We therefore propose that the genus description of Trichodesmium must be extended to include species and strains which predominantly live as individual trichomes (organisms classified as Katagnymene) and that K. pelagica and K. spiralis should be united into one species.
Acknowledgments
We are grateful to E. J. Carpenter (San Francisco State University, San Francisco, Calif.) and D. Capone (The Wrigley Institute for Environmental Studies, University of California at Los Angeles, Los Angeles) for inviting us to participate in research cruises. J. Waterbury (Woods Hole Oceanographic Institution, Woods Hole, Mass.) is acknowledged for the cultured JWI1 specimen (K. spiralis).
We also acknowledge financial assistance from STINT (The Swedish Foundation for International Cooperation in Research and Higher Education) and Sida/SAREC (Swedish International Development Cooperation Agency) to B.B. and the Swedish Research Council for financial support provided to B.B. and S.J.
REFERENCES
- 1.Anagnostidis, K., and J. Komarek. 1988. Modern approach to the classification system of cyanophytes. Arch. Hydrobiol. 80:1-4. [Google Scholar]
- 2.Ashtekar, P. V., and N. D. Kamat. 1979. A new species of Katagnymene. Curr. Sci. 48:131. [Google Scholar]
- 3.Barker, G. L. A., P. K. Haye, S. L. O'Mahony, P. Vacharapiyasophon, and A. E. Walsby. 1999. A molecular and phenotypic analysis of Nodularia (cyanobacteria) from the Baltic Sea. J. Phycol. 35:931-937. [Google Scholar]
- 4.Ben-Porath, J., E. J. Carpenter, and J. P. Zehr. 1993. Genotypic relationship in Trichodesmium (Cyanophyceae) based on nifH sequence comparisons. J. Phycol. 29:806-810. [Google Scholar]
- 5.Berman-Frank, I., P. Lundgren, Y.-B. Chen, H. Kupper, Z. Kolber, B. Bergman, and P. Falkowski. 2001. Segregation of nitrogen fixation and oxygenic photosynthesis in the marine cyanobacterium Trichodesmium. Science 294:1534-1537. [DOI] [PubMed] [Google Scholar]
- 6.Capone, D. G., J. P. Zehr, H. W. Paerl, B. Bergman, and E. J. Carpenter. 1997. Trichodesmium, a globally significant marine cyanobacterium. Science 276:1221-1229. [Google Scholar]
- 7.Chen, Y.-B., J. P. Zehr, and M. Melon. 1996. Growth and nitrogen fixation of the diazotrophic filamentous nonheterocystous cyanobacterium Trichodesmium sp. IMS 101 in defined media: evidence for a circadian rhythm. J. Phycol. 32:916-923. [Google Scholar]
- 8.Drouet, F. 1968. Revision of the classification of the Oscillatoriaceae. Monograph 15. Academy of Natural Science of Philadelphia, Philadelphia, Pa.
- 9.Ehrenberg, C. G. 1830. Neue Beobachtungen über blauartige Erscheinungen in Aegypten, Arabien und Sibirien, nebst einer Übersicht und Kritik der früher Bekannten. Ann. Phys. Chem. 18:477-514. [Google Scholar]
- 10.El-Shehawy, R., C. Lugomela, A. Ernst, and B. Bergman. 2003. Diurnal expression of ntcA and hetR and diazocyte formation in the filamentous non-heterocystous cyanobacterium Trichodesmium spp. Microbiology 149:1139-1146. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 12.Fredriksson, C., and B. Bergman. 1997. Ultrastructural characterization of cells specialised for nitrogen fixation in a non-heterocystous cyanobacterium, Trichodesmium spp. Protoplasma 197:76-85. [Google Scholar]
- 13.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 14.Janson, S., B. Bergman, E. J. Carpenter, S. J. Giovannoni, and K. Vergin. 1999. Genetic analysis of natural populations of the marine diazotrophic cyanobacterium Trichodesmium. FEMS Microbiol. Ecol. 30:57-65. [Google Scholar]
- 15.Janson, S., E. J. Carpenter, and B. Bergman. 1994. Compartmentalisation of nitrogenase in a non-heterocystous cyanobacterium: Trichodesmium contortum. FEMS Microbiol. Lett. 118:9-14. [Google Scholar]
- 16.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 17.Karl, D. M., A. Michaels, B. Bergman, D. G. Capone, E. J. Carpenter, R. Letelier, F. Lipschultz, H. W. Paerl, D. Sigman, and L. Stal. 2002. Nitrogen fixation in the world's oceans. Biogeochemistry 57/58:47-98. [Google Scholar]
- 18.Karsten, G. 1907. Das indishe Phytoplankton. Wiss. Ergebn. Valdivia 2:221-548. [Google Scholar]
- 19.Lemmermann, E. 1900. Ergebnisse einer Reise nach dem Pacific. Abh. Natwiss. Ver. Brem. 16:313-406. [Google Scholar]
- 20.Li, R. H., H. J. Debella, and W. W. Carmichael. 2001. Isolates identifiable as Arthrospira maxima and Arthrospira fusiformis (Oscillatoriales, Cyanobacteria) appear identical on the basis of a morphological study in culture and 16S rRNA gene sequences. Phycologia 40:367-371. [Google Scholar]
- 21.Lin, S., S. Henze, P. Lundgren, B. Bergman, and E. J. Carpenter. 1998. Whole-cell immunolocalization of nitrogenase in marine diazotrophic cyanobacteria, Trichodesmium spp. Appl. Environ. Microbiol. 64:3052-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lundgren, P., E. Söderbäck, A. Singer, E. J. Carpenter, and B. Bergman. 2001. Katagnymene: a novel marine diazotroph. J. Phycol. 37:1052-1062. [Google Scholar]
- 23.Nelissen, B., A. Wilmotte, J.-M. Neefs, and R. De Wachter. 1994. Phylogenetic relationships among filamentous helical cyanobacteria investigated on the basis of 16S ribosomal RNA gene sequence analysis. Syst. Appl. Microbiol. 17:206-210. [Google Scholar]
- 24.Norris, R. E. 1961. Observations on phytoplankton organisms collected on the N.Z.O.I. Pacific cruise, September 1958. N. Z. J. Sci. 4:162-188. [Google Scholar]
- 25.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orcutt, K. M., U. Rasmussen, E. A. Webb, J. B., Waterbury, K. Gundersen, and B. Bergman. 2002. Characterization of Trichodesmium spp. by genetic techniques Appl. Environ. Microbiol. 68:2236-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 28.Tajima, F., and M. Nei. 1984. Estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol. 1:269-285. [DOI] [PubMed] [Google Scholar]
- 29.Van de Peer, Y., and R. de Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 30.Wille, N. 1904. Die Schizophyceen der Plankton-expedition. Ergebnisse der Plankton-Expedition der Humboldt-Stiftung IV. Verlag von Lipsius & Tisher, Kiel and Leipzig, Germany.