Free-living amoebae are important predators that control microbial communities. They are ubiquitous and have been isolated from various natural sources such as soil, freshwater, salt water, dust, and air. Although their abundance in soil is only limited, they have been implicated in the stimulation of phosphorus and nitrogen turnover and thus play an important role in soil ecosystems (84). Free-living amoebae are also frequently isolated from anthropogenic ecosystems such as tap water, air-conditioning units, and cooling towers, feeding on the microbial biofilm present in those systems. However, several bacteria have developed mechanisms to survive phagocytosis by free-living amoebae and are able to exploit them as hosts.
Transient association with amoebae have been reported for a number of different bacteria including Legionella pneumophila, many Mycobacterium spp., Francisella tularensis, and Escherichia coli O157, among others (3, 21, 77, 85). As most of these bacteria are pathogens of humans, amoebae have been suggested to represent their environmental reservoirs, acting as “Trojan horses” of the microbial world (10). To date, only the interaction of L. pneumophila with free-living amoebae has been studied in greater detail. L. pneumophila is an intracellular pathogen of humans causing Legionnaires' disease. L. pneumophila multiplies within its host cell (human macrophages and amoebae) but can also grow outside its host cell and can be cultured by routine methodologies (for reviews, see references 19 and 46). Therefore, it is considered as a facultative intracellular pathogen.
In addition to those facultative intracellular bacteria, various obligate endosymbionts have been observed in free-living amoebae. In this context, it should be noted that the term endosymbiont implies neither a mutualistic nor a parasitic relationship between the bacteria and their host cells. According to the broad concept of symbiosis suggested originally by Anton de Bary (30a) and consistent with the recent awareness of the variability of symbiotic interactions (81), we consider bacteria as endosymbionts when they manage to establish their replicative niche within eukaryotic host cells. Approximately 20% of isolates of ubiquitous Acanthamoeba spp. recovered from clinical and environmental sources are found to harbor such bacterial endosymbionts (37). These endosymbionts have been shown to maintain a stable interaction with their hosts (37), but in contrast to L. pneumophila, they cannot be cultured outside their host cells, a primary reason why their identification and analysis have been hampered.
This review examines the impact of the interaction between bacteria and amoebae on the evolution of intracellular bacterial pathogens of humans (i.e., bacteria capable of causing disease) by focusing on recent findings on the stable association of free-living amoebae with their obligate endosymbionts and the transient interaction between L. pneumophila and amoebae as a model system.
DIVERSITY OF BACTERIAL SYMBIONTS OF FREE-LIVING AMOEBAE
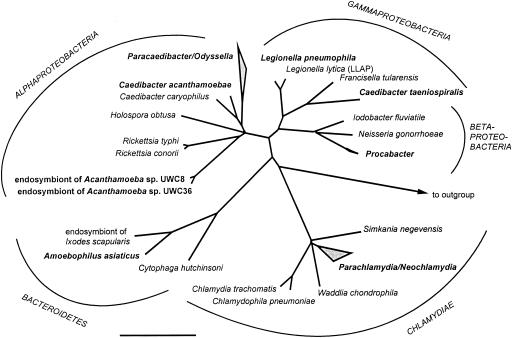
Bacterial endosymbionts of amoebae were microscopically observed at least three decades ago (83). They have been shown to maintain a stable interaction with their hosts and are able to outlast encystations of the amoebae (37). However, identification and analysis of these endosymbionts have been severely hampered by the fact that they cannot be cultured outside their eukaryotic host cells. Only the advent of molecular biological techniques and the establishment of an rRNA-based phylogenetic system for the classification of prokaryotes enabled a more detailed characterization of these elusive bacteria (8). During the past few years, specific PCR-mediated amplification of bacterial rRNA genes, comparative sequence analysis, and fluorescence in situ hybridization led to the identification of five novel evolutionary lineages of bacterial endosymbionts of free-living amoebae (9, 16, 38, 39, 53-55, 57). These lineages have been found to be affiliated with (i) the Alphaproteobacteria, (ii) the Betaproteobacteria, (iii) the Gamaproteobacteria, (iv) the Bacteroidetes, and (v) the Chlamydiales (Fig. 1). Interestingly, co-occurrence of phylogenetically different endosymbionts in a single amoebal isolate has never been observed, and significant differences regarding the host range have been shown for the different endosymbionts (55). Apart from one exception (Neochlamydia hartmannellae [57]), spontaneous loss of endosymbionts from the original amoeba host or successful eradication of the symbionts from their hosts by antibiotics has never been reported, confirming the stable endosymbiosis. Consistent with this finding, evidence for coevolution between endosymbionts and hosts has been reported for one endosymbiont phylogenetic lineage (14). The phylum-level diversity of the endosymbionts suggests that the capability to exploit free-living amoebae by endosymbionts has evolved several times, independently, in different lineages within the domain Bacteria. But there is also unity in variety. For many of these lineages, endosymbionts with very similar or almost identical 16S rRNA gene sequences were found in geographically dispersed regions and different ecosystems, suggesting a ubiquitous distribution of these organisms and their hosts.
FIG. 1.
16S rRNA-based phylogenetic tree showing the affiliation of L. pneumophila and obligate endosymbionts of free-living protozoa. Bar, 10% estimated evolutionary distance.
A common feature of most of the recently identified endosymbionts of free-living amoebae is their close phylogenetic relationship to other obligate intracellular bacteria. For example, the Bacteroidetes-related symbiont “Candidatus Amoebophilus asiaticus” is monophyletic with two different insect symbionts (55). Furthermore, several of the closest relatives of the different amoebal endosymbionts are well-known intracellular pathogens of humans and animals (Fig. 1). For instance, the two alphaproteobacterial lineages (16, 38, 53) form a common branch with the rickettsiae comprising many important intracellular pathogens like Rickettsia typhi, the causative agent of typhus fever. In addition, the endosymbionts of amoebae belonging to the Chlamydiales are the closest relatives of previously known chlamydiae (9, 39, 57), which are a deeply branching evolutionary lineage exclusively containing important intracellular pathogens of animals and humans, including Chlamydia trachomatis and Chlamydophila pneumoniae. The emerging theme is that many obligate intracellular bacteria thriving in free-living protozoa share a common ancestor with intracellular bacteria pathogenic for humans. This theme is further supported by the identification of a gammaproteobacterial endoparasite of the ciliate Paramecium tetraurelia as being related to the Francisella group containing the highly infectious pathogen Francisella tularensis, the causative agent of tularemia (14). F. tularensis has also been shown to survive and replicate within intact trophozoites of Acanthamoeba castellanii and has been found in excreted vesicles and cysts (1). Taken together, these findings implicate the interaction with amoebae during the evolution of intracellular bacterial pathogens. Through interaction with unicellular eukaryotes, the common ancestor of protozoan symbionts and human pathogens might have evolved mechanisms for intracellular survival in eukaryotic cells. Evidence for these “training grounds” for pathogens is supported by numerous studies on the interaction between legionellae and amoebae (10, 46), which are discussed below. Additional support comes from a more detailed analysis of the interaction between the recently identified chlamydia-related symbionts and endosymbionts and their amoeba hosts.
NEOCHLAMYDIA AND PARACHLAMYDIA: ENVIRONMENTAL COUNTERPARTS OF PATHOGENIC CHLAMYDIAE?
The majority of the identified endosymbionts of free-living amoebae belongs to the Chlamydiales, thrives within a host vacuole, and possesses the unique chlamydial biphasic developmental cycle. During this cycle, metabolically active intracellular reticulate bodies undergo binary fission and subsequently differentiate to metabolically inactive elementary bodies which are adapted to extracellular survival and infection of eukaryotic host cells (Fig. 2) (48). Consequently, the chlamydia-related endosymbionts of amoebae, also referred to as environmental chlamydiae, are classified within the new genera Neochlamydia or Parachlamydia of the order Chlamydiales (9, 32, 57). These studies and the finding of other chlamydia-like bacteria (Simkania negevensis, Waddlia chondrophila, and Fritschea bemisiae) as contaminants of a tissue culture (63), within an aborted bovine fetus (49, 89), or as symbionts of insects (98) have dramatically changed our view of chlamydial diversity and ecology. Further evidence for a wide distribution of novel chlamydiae in the environment is provided by PCR-based culture-independent monitoring of environmental and clinical samples (28, 29, 56, 79). Today, several broad-range PCR assays and a comprehensive set of oligonucleotide probes for application in fluorescence in situ hybridization are available for the assessment of chlamydial diversity and occurrence of chlamydia-related bacteria in the environment (70, 82).
FIG. 2.
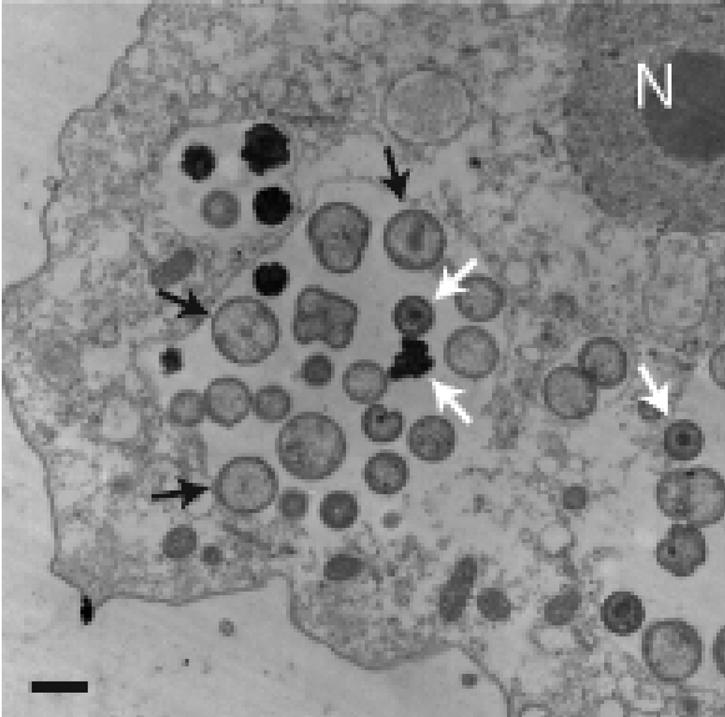
Electron micrograph of environmental chlamydiae within an Acanthamoeba host cell. The two different stages of the developmental cycle are indicated by black (reticulate bodies) and white (elementary bodies) arrows. N, nucleus; bar, 1 μm.
The presence of a developmental cycle similar to the developmental cycle of pathogenic chlamydiae suggests that amoebal symbionts and human pathogens share a common life style. Pathogenic chlamydiae possess a specialized transport protein which catalyzes the import of host cell ATP in exchange for ADP (100). Such transport proteins are very rare and have to date been identified only in chlamydiae, rickettsiae, mitochondria, and plastids (102). Although chlamydiae still contain the functional capacity to produce their own ATP, they use the ATP/ADP translocase in certain stages of the developmental cycle, thereby thriving as energy parasites within their host cells (61). By using a set of degenerate primers and a combination of arbitrary PCR approaches, a gene coding for an ATP/ADP transport protein in the Parachlamydia-related symbiont UWE25 has been identified (91). Detection of transcripts and biochemical characterization of the heterologously expressed protein clearly indicate that these environmental chlamydiae, similar to the pathogenic chlamydiae, are able to utilize the ATP pool of their amoeba host cells. Interestingly, phylogenetic analysis suggested that nonmitochondrial ATP/ADP transport proteins have evolved within the common ancestor of present-day chlamydiae and that they have been subsequently acquired by rickettsiae and plastids or cyanobacteria via lateral gene transfer (44, 91). Additional insights into the lifestyle of symbiotic chlamydiae were recently gained by analysis of the complete genome sequence of the Parachlamydia-related symbiont UWE25 of Acanthamoeba sp. These data reveal striking similarities between the biology of symbiotic and pathogenic chlamydiae and show that the symbiont UWE25 is highly adapted to intracellular life, being dependent on a variety of host metabolites (52).
MEDICAL SIGNIFICANCE OF ENVIRONMENTAL CHLAMYDIAE
Classical chlamydiae are important pathogens of humans and animals. Chlamydophila (formerly classified as Chlamydia) pneumoniae is responsible for about 10% of community-acquired pneumonia (73) and has recently been associated with intrinsic asthma and cardiovascular disease (reviewed in references 23 and 68). Chlamydia trachomatis causes trachoma and is considered to be the world's leading cause of blindness, with about 6 million people affected as a result of this disease. In addition, C. trachomatis is the most frequent sexually transmitted bacterial pathogen, with an estimated 90 million new cases occurring each year worldwide (90). Chlamydophila (formerly classified as Chlamydia) psittaci can cause systemic fatal disease in birds and is also able infect mammals and humans, causing a wide variety of diseases (90). The diverse assembly of recently identified, ubiquitously distributed environmental isolates of chlamydiae represents a large environmental reservoir of bacteria that are closely related to human pathogens, with an as-yet-undefined relevance for public health. Thus, the expanding diversity of the Chlamydiales also deserves attention from a medical point of view. Interestingly, recent data suggest a possible clinical importance for environmental chlamydiae. Kahane and coworkers have reported high antibody prevalence against Simkania negevensis in infants suffering from bronchiolitis (62), and it has been suggested that the same organism may also be associated with community-acquired pneumonia in adults (69). In addition, specific antibodies against “Hall's coccus,” an amoebal endosymbiont belonging to the genus Parachlamydia, have been detected in patients with pneumonia of unknown etiology (18, 74). The thoughts that novel chlamydia-related bacteria are capable of thriving within humans is also supported by the PCR-mediated retrieval of several of their 16S rRNA gene sequences from clinical specimens of respiratory disease patients (28, 29, 79). Although these findings do not yet prove that environmental chlamydiae do actually cause disease, they clearly indicate the need for a more thorough analysis of these newly discovered bacteria and their role in infectious disease.
THE ROLE OF FREE-LIVING AMOEBAE IN THE EVOLUTION OF CHLAMYDIAE
The natural reservoir of pathogenic chlamydiae is still unknown. In this context, the finding that (besides environmental chlamydiae) Chlamydophila pneumoniae is also able to multiply in free-living amoebae (31) indicates that protozoa might serve as an environmental reservoir for chlamydiae. Although the persistence of this interaction during encystment of the amoeba host has not been demonstrated, this observation shows that a single chlamydial organism can thrive in both protozoa and humans. Consistent with this finding, it has recently been reported that the amoeba symbiont Parachlamydia acanthamoebae is able to enter and multiply in human macrophages (43) and thus might have the potential to be a human pathogen. This is also supported by the identification of several genes, like, e.g., a complete type III secretion system, that have been associated with pathogenicity of chlamydiae in the genome of the Parachlamydia-related symbiont UWE25 (52). Moreover, phylogenetic analyses of those genes have indicated that they are already present in the last common ancestor of chlamydia-related symbionts of amoebae and pathogenic chlamydiae (52).
Taken together, there is cumulative evidence to suggest an important role of protozoa in the evolution of chlamydiae. Data from comparative genome analysis and the fact that both the environmental chlamydiae and the recognized pathogenic chlamydiae possess orthologous genes for ATP/ADP translocases, which are exclusively found in intracellular bacteria, can be used to infer that the last common ancestor of both lineages was also characterized by an intracellular lifestyle (52, 91). This chlamydial ancestor evolved around 700 million years ago, long before higher multicellular eukaryotes had evolved (52). Thus, a conceivable scenario for the contribution of protozoa to the evolution of chlamydiae is that a common ancestor of all chlamydiae has developed mechanisms for intracellular survival and multiplication in primitive unicellular eukaryotes. One might speculate that these ancient eukaryotes grazed on microorganisms and digested them in a phagocytic vacuole in a manner similar to that of today's free-living amoebae. The chlamydial ancestor must have evolved to alter its fate within the phagosomes by exporting proteins into the phagosome membrane and possibly also the host cytosol to evade degradation and to exploit the eukaryote as a host. Long-term coexistence of both organisms eventually led to the stable symbiotic interaction observed today between free-living amoebae and environmental chlamydiae. Sometime during this evolutionary process, a chlamydial clone evolved to infect as a parasite of multicellular eukaryotes, making a different ecological niche accessible, thus leading to diversification of chlamydiae. Considering the broad host range of the members of the Chlamydiales, such host changes might have occurred more than once. Therefore, significant diversity exists within the formerly uniform order Chlamydiales. A better understanding of the evolution, ecology, and virulence of Chlamydiales will require a much more encompassing analysis of its recently discovered members. This knowledge not only is important for basic science but might also eventually assist in the selection of suitable antichlamydial drug targets and vaccine strategies.
LEGIONELLAE AS FACULTATIVE SYMBIONTS OF FREE-LIVING AMOEBAE
L. pneumophila is a gram-negative facultative intracellular bacillus responsible for Legionnaires' disease. It replicates within evolutionarily distant eukaryotic host cells such as protozoa and mammalian macrophages (Fig. 3). In aquatic environments, L. pneumophila is ubiquitous and grows within protozoan hosts. At least 13 species of amoebae and 2 species of ciliated protozoa support intracellular replication of L. pneumophila (33). Among the most predominant amoebae in water sources are hartmannellae and acanthamoebae, which have been also isolated from water sources associated with Legionnaires' disease outbreaks (33). Interaction between L. pneumophila and protozoa is considered to be central to the pathogenesis and ecology of L. pneumophila (46, 85). In humans, L. pneumophila reaches the lungs after inhalation of contaminated aerosol droplets (33, 36). The main sources of contaminated water droplets are hot water and air-conditioning systems, but the bacteria have been isolated from fountains, spas, pools, dental and hospital units, and other man-made water systems (36). No person-to-person transmission has ever been described. Once in the lungs, L. pneumophila is ingested by alveolar macrophages, which are thought to be the major site of bacterial replication. This ingestion results in an acute and severe pneumonia. Approximately one-half of the 46 species of Legionella have been associated with human disease. L. pneumophila is responsible for 90% of cases of Legionnaires' disease. However, all the Legionella species may be capable of intracellular growth and infliction of human disease under appropriate conditions. Infections due to less common species of legionellae are not frequently diagnosed and reported and are less studied than L. pneumophila (34).
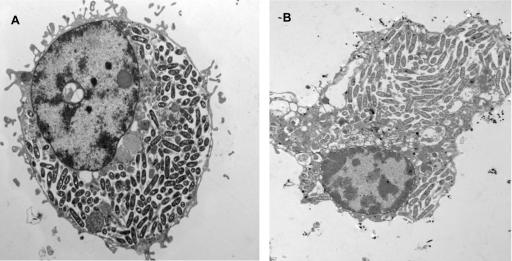
FIG. 3.
Electron micrographs of U937 macrophages (A) and A. polyphaga (B) infected by L. pneumophila (strain AA100) at 24 h. The infection is very similar in both host cells. Legionella is cytoplasmic at 24 h in the two panels.
In addition to recognized Legionella species, a number of Legionella-related bacteria designated Legionella-like amoebal pathogens (LLAPs) (85, 86) have been described (6). Interestingly, many LLAPs have been associated with Legionnaires' disease (17, 74). In contrast to other Legionella species, however, most LLAPs cannot be cultured in vitro on artificial media but are isolated by coculture with protozoa (33). The recent developments in the use of PCR for bacterial identification in environmental samples will facilitate better identification of legionellae and LLAPs. Further cellular and molecular biology studies are needed to better understand the intracellular life of these endosymbionts.
THE ROLE OF FREE-LIVING AMOEBAE IN PERSISTENCE OF LEGIONELLAE IN THE ENVIRONMENT
It is most likely that the association of legionellae with protozoa is a major factor in the continuous presence of the bacteria in the environment. Many strategies have been used to eradicate legionellae from sources of infection in water and plumbing systems that have been associated with disease outbreaks. These strategies include chemical biocides such as chlorine, overheating of the water, and UV irradiation (20, 66, 76). Such interventions have been successful for short periods of time, after which the bacteria reappear in these sources (20, 103). Thus, eradication of L. pneumophila from the environmental sources of infection requires continuous treatment of the water with agents such as monochloramine or copper-silver ions in addition to maintenance of the water temperature above ∼55°C (30, 51, 66, 67). Compared to in vitro-grown L. pneumophila, amoeba-grown bacteria have been shown to be highly resistant to chemical disinfectants and to treatment with biocides (11). Amoeba-grown L. pneumophila has been shown to manifest a dramatic increase in its resistance to harsh environmental conditions such as fluctuation in temperature, osmolarity, pH, and exposure to oxidizing agents (4). Protozoa have been shown to release vesicles containing L. pneumophila that are highly resistant to biocides (15). The ability of L. pneumophila to survive within amoebic cysts further contributes to the resistance of L. pneumophila to physical and biochemical agents used in bacterial eradication (11, 13). It is likely that eradication of the bacteria from the environment should start by preventing protozoan infection, an integral part of the infectious cycle of L. pneumophila. Extracellular L. pneumophila is more susceptible to environmental conditions and is not protected from biocides and disinfectants.
THE ROLE OF FREE-LIVING AMOEBAE IN PATHOGENESIS OF LEGIONELLAE
There are many lines of evidence to suggest that protozoa play major roles in the transmission of L. pneumophila as an infectious particle for Legionnaires' disease (87). First, many protozoan hosts that allow intracellular bacterial replication, the only documented means of bacterial amplification in the environment, have been identified (5, 33, 46). Second, in outbreaks of Legionnaires' disease, amoebae and bacteria have been isolated from the same source of infection, and the isolated amoebae supported intracellular replication of the bacteria (35). Third, following intracellular replication within protozoa, L. pneumophila exhibits a dramatic increase in resistance to harsh conditions including high temperature, acidity, and high osmolarity, which may facilitate bacterial survival in the environment (4). Fourth, intracellular L. pneumophila within protozoa are more resistant to chemical disinfection and biocides compared to in vitro-grown bacteria (11-13). Fifth, protozoa have been shown to release vesicles of respirable size that contain numerous L. pneumophila isolates. The vesicles are resistant to freeze-thawing and sonication, and the bacteria within the vesicles are highly resistant to biocides (15). Sixth, following their release from the protozoan host, the bacteria exhibit a dramatic increase in their infectivity for mammalian cells in vitro (27). In addition, it has been demonstrated that intracellular bacteria within Hartmanella vermiformis are dramatically more infectious and are highly lethal in mice (22). Seventh, the number of bacteria isolated from the source of infection of Legionnaires' disease is usually very low or undetectable, and thus, enhanced infectivity of intracellular bacteria within protozoa may compensate for the low infectious dose (78). Eighth, viable but nonculturable L. pneumophila can be resuscitated by coculture with protozoa (95). This observation may suggest that the failure to isolate the bacteria from environmental sources of infection may be due to this “dormant” phase of the bacteria that cannot be recovered on artificial media. Ninth, there has been no documented case of bacterial transmission between individuals. The only source of transmission is environmental droplets generated from man-made devices such as shower heads, water fountains, whirlpools, and cooling towers of air-conditioning systems (33). These findings indicate a rather sophisticated host-parasite interaction and a tremendous adaptation of legionellae to parasitize protozoa. This host-parasite interaction is central to the pathogenesis and ecology of these bacteria.
ENTRY, INTRACELLULAR TRAFFICKING, SURVIVAL, AND REPLICATION OF L. PNEUMOPHILA WITHIN MACROPHAGES AND AMOEBAE
The attachment and entry mechanisms of L. pneumophila entry into its protozoan hosts and variations in these mechanisms have been reported for both H. vermiformis and Acanthamoeba spp. Attachment of L. pneumophila to H. vermiformis is mediated by adherence to a protozoan receptor characterized as a putative galactose/N-acetylgalactosamine (Gal/GalNAc) lectin (47, 71, 101). Host protein synthesis by A. polyphaga is not required for invasion by L. pneumophila, whereas it is required for invasion of H. vermiformis (47). Following this initial host-parasite interaction, uptake of L. pneumophila by A. castellanii occurs by coiling phagocytosis (3, 21). However, the uptake of L. pneumophila by H. vermiformis occurs mainly through cup-shaped invaginations (or zipper phagocytosis) on the surface of the amoeba, in addition to occasional coiling phagocytosis (3, 21). Human macrophages are able to phagocytose heat- or formalin-killed L. pneumophila by coiling phagocytosis (59), which indicates that coiling phagocytosis does not play a role in the intracellular fate of L. pneumophila. However, the infectivity of A. castellanii and macrophages by L. pneumophila has been shown to be similar (50). In addition, the adherence receptors of macrophages used by L. pneumophila do not seem to affect its intracellular survival profoundly as the bacterium multiplies within phagocytes after entry under different opsonizing or nonopsonizing conditions (26, 60, 72, 80)
Similar to its intracellular fate within macrophages, L. pneumophila is enclosed, after entry into amoebae, in a phagosome surrounded by host cell organelles such as mitochondria, vesicles, and a multilayer membrane derived from the rough endoplasmic reticulum (RER) of amoeba (3, 58, 97, 99) (Fig. 4). The bacterial phagosome is blocked from fusion to the lysosomes (21). The role of the RER in the intracellular infection is not known, but the RER is not required as a source of proteins for the bacteria (2). Interestingly, examination of the intracellular infection of macrophages, alveolar epithelial cells, and protozoa by another Legionella species, Legionella micdadei, showed that within all of these host cells, the bacteria were localized to RER-free phagosomes (42). Whether other Legionella species replicate within RER-free phagosomes is still to be determined. Following formation of this phagosome within protozoan cells, bacterial replication is initiated. The 4-h period prior to initiation of intracellular replication may be the time required for L. pneumophila to recruit host cell organelles that may be required for replication. Alternatively, the 4-h period may be a lag phase of metabolic and environmental adjustment of the bacteria to a new niche.
FIG. 4.
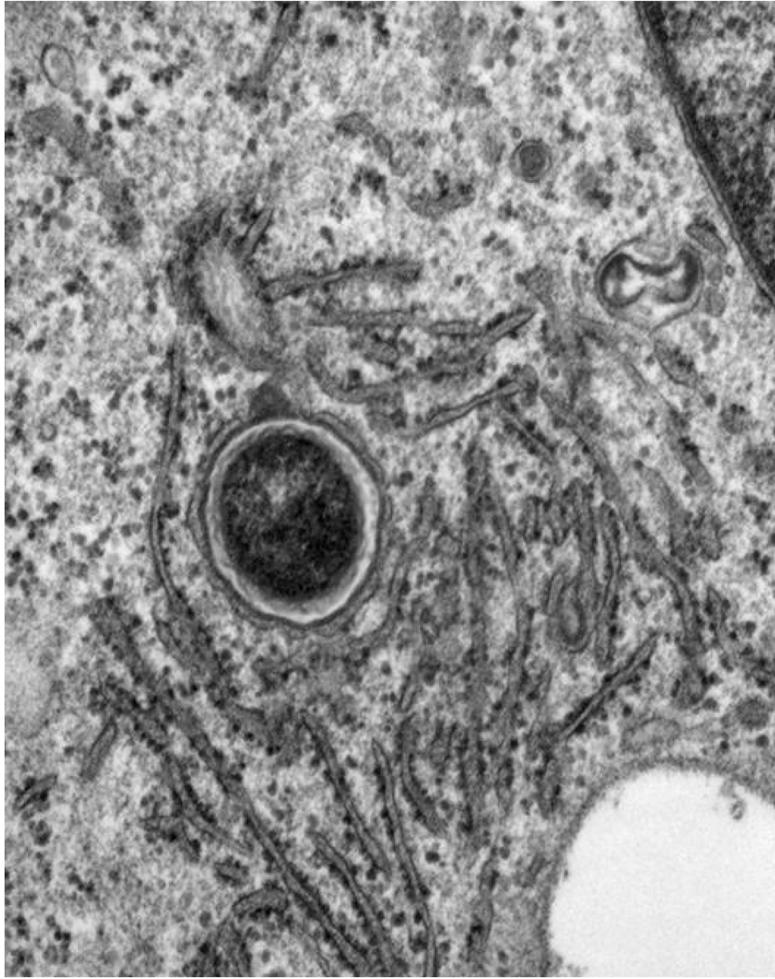
Electron micrograph of U937 cells infected by L. pneumophila at 8 h postinfection. The LCP is intact and is surrounded by the endoplasmic reticulum.
ROLE OF THE dot/icm GENES IN EVASION OF THE ENDOCYTIC PATHWAY
The main virulence system of L. pneumophila is the Dot/Icm type IV secretion system. Because the Dot/Icm secretion system is ancestrally related to type IV secretion systems that mediate conjugal DNA transfer between bacteria (24), L. pneumophila may utilize this transporter to transfer macromolecules into the host cell to evade endocytic fusion (88). The Dot/Icm-mediated transfer is thought to occur through the insertion of a pore in the host cellular membrane through which the effector proteins are transported (64, 65). With few exceptions, the function of individual Dot/Icm proteins is unknown. However, the dot/icm genes were present in all tested Legionella species (7).
Most of the dot/icm genes required for intracellular growth within human cells are also required for intracellular growth in the protozoan host A. castellanii (92). In addition, enhanced phagocytosis of wild-type L. pneumophila by A. castellanii has also been demonstrated to be dependent on dot/icm genes (50). Although some loci have been shown to be essential only for the intracellular growth of L. pneumophila in macrophages (40), numerous loci have been identified as essential for survival and intracellular replication of L. pneumophila in A. polyphaga or H. vermiformis and in macrophages (25, 41, 96). The slime mold Dictyostelium discoideum, a unicellular soil organism, has been shown to support intracellular multiplication of L. pneumophila (45, 93, 94). In the amoebal form, the cells are highly motile and are very active in phagocytosis. This model is interesting because genetic tools are available for analysis of host-pathogen interactions (93, 94). The intracellular fate of L. pneumophila in infected D. discoideum cells is very similar to that in macrophages, including the recruitment of RER and evasion of lysosomal fusion (94) and the dependence of intracellular growth on dot/icm gene functions (94). The similarity between the infections of different protozoa by L. pneumophila supports the idea that the ability of L. pneumophila to parasitize macrophages and hence to cause human disease is a consequence of its prior adaptation to intracellular growth within protozoa.
EGRESS OF L. PNEUMOPHILA FROM AMOEBAE
A detailed ultrastructural analysis of late stages of intracellular replication has been performed to examine egress of L. pneumophila from both macrophages and amoebae by electron microscopy (75). The membrane of the L. pneumophila-containing phagosome (LCP) within both macrophages and Acanthamoeba polyphaga cells is intact up to 8 h postinfection (75). However, at 12 h, the majority of the LCPs are disrupted within both hosts, while the plasma membrane remains intact (75). At 18 and 24 h postinfection, cytoplasmic elements such as mitochondria, lysosomes, vesicles, and amorphous material are dispersed among the bacteria, and these bacteria are considered cytoplasmic (75). Thus, by 18 to 24 h postinfection, the majority of the remaining host cells harbor cytoplasmic bacteria, and this transient cytoplasmic presence of L. pneumophila precedes lysis of the plasma membrane (75). Interestingly, within both macrophages and amoebae, bacterial replication proceeds in the cytoplasm (75).
Therefore, rather than simultaneous lysis of both the phagosomal and the plasma membranes, the phagosomal membrane is disrupted first. These disruptions of the LCP may be the result of multifactorial events linked to apoptosis, the pore-forming activity of the type IV secretion system, and mechanical pressure due to the increase of the phagosomal size (65).
CONCLUSION
The interaction between legionellae and free-living amoebae shows strong similarity with processes that occur during infection of mammalian cells by legionellae. In addition, free-living amoebae seem to play a crucial role for persistence and dispersal of legionellae in the environment, and there is convincing evidence that intracellular multiplication of L. pneumophila in free-living amoebae is a prerequisite for the infection of humans. It thus seems conceivable that these intracellular bacterial pathogens have developed and evolved mechanisms for survival in eukaryotic host cells during the interaction with free-living amoebae. This hypothesis of amoebae acting as a (evolutionary) training ground for intracellular bacterial pathogens is further supported by the analysis of chlamydia-related symbionts of free-living amoebae, which still use strategies for host cell interaction that were developed 700 million years ago in interplay with early unicellular eukaryotes (and in the absence of higher, multicellular organisms). Clearly, although other protozoa might have acted in the same manner, the interaction between intracellular bacteria and prokaryotes other than amoebae has rarely been addressed. Future studies should examine obligate and facultative symbionts of other protozoa in order to elucidate the role of unicellular eukaryotes in the transition of free-living bacteria to intracellular bacteria that eventually become able to infect animals and humans. A better understanding of such processes will help to develop novel strategies and targets for vaccines and antibiotics against intracellular bacterial pathogens.
Acknowledgments
Michael Schweikert is greatly acknowledged for assistance with electron microscopy.
Y.A.K. is supported by Public Health Service awards RO1AI43965 and R21AI038410-06A1 and by the Commonwealth of Kentucky Research Challenge Trust Fund (R.D.S. and J.S.). The original research of M.W. and M.H. was supported by the German Ministry for Education and Science (bmb+f) grants 01KI0104 and PTJ-BIO/03U213B and by the Austrian Science Fund (FWF) grant P16566-B14.
REFERENCES
- 1.Abd, H., T. Johansson, I. Golovliov, G. Sandström, and M. Forsman. 2003. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl. Environ. Microbiol. 69:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Kwaik, Y. 1998. Induced expression of the Legionella pneumophila gene encoding a 20-kilodalton protein during intracellular infection. Infect. Immun. 66:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu Kwaik, Y. 1996. The phagosome containing Legionella pneumophila within the protozoan Hartmanella vermiformis is surrounded by the rough endoplasmic reticulum. Appl. Environ. Microbiol. 62:2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu Kwaik, Y., L.-Y. Gao, O. S. Harb, and B. J. Stone. 1997. Transcriptional regulation of the macrophage-induced gene (gspA) of Legionella pneumophila and phenotypic characterization of a null mutant. Mol. Microbiol. 24:629-642. [DOI] [PubMed] [Google Scholar]
- 5.Abu Kwaik, Y., L.-Y. Gao, B. J. Stone, C. Venkataraman, and O. S. Harb. 1998. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl. Environ. Microbiol. 64:3127-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adeleke, A., J. Pruckler, R. Benson, T. Rowbotham, M. Halablab, and B. S. Fields. 1996. Legionella-like amoebal pathogens—phylogenetic status and possible role in respiratory disease. Emerg. Infect. Dis. 2:225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alli, O. A., S. Zink, N. K. von Lackum, and Y. Abu-Kwaik. 2003. Comparative assessment of virulence traits in Legionella spp. Microbiology 149:631-641. [DOI] [PubMed] [Google Scholar]
- 8.Amann, R., N. Springer, W. Ludwig, H.-D. Gortz, and K.-H. Schleifer. 1991. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature 351:161-164. [DOI] [PubMed] [Google Scholar]
- 9.Amann, R., N. Springer, W. Schonhuber, W. Ludwig, E. N. Schmid, K.-D. Muller, and R. Michel. 1997. Obligate intracellular bacterial parasites of acanthamoebae related to Chlamydia spp. Appl. Environ. Microbiol. 63:115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barker, J., and M. R. Brown. 1994. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology 140:1253-1259. [DOI] [PubMed] [Google Scholar]
- 11.Barker, J., M. R. W. Brown, P. J. Collier, I. Farrell, and P. Gilbert. 1992. Relationships between Legionella pneumophila and Acanthamoeba polyphaga: physiological status and susceptibility to chemical inactivation. Appl. Environ. Microbiol. 58:2420-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barker, J., P. A. Lambert, and M. R. W. Brown. 1993. Influence of intra-amoebic and other growth conditions on the surface properties of Legionella pneumophila. Infect. Immun. 61:3503-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barker, J., H. Scaife, and M. R. W. Brown. 1995. Intraphagocytic growth induces an antibiotic-resistant phenotype of Legionella pneumophila. Antimicrob. Agents Chemother. 39:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beier, C. L., M. Horn, R. Michel, M. Schweikert, H.-D. Görtz, and M. Wagner. 2002. The genus Caedibacter comprises endosymbionts of Paramecium spp. related to the Rickettsiales (Alphaproteobacteria) and to Francisella tularensis (Gammaproteobacteria). Appl. Environ. Microbiol. 68:6043-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berk, S. G., R. S. Ting, G. W. Turner, and R. J. Ashburn. 1998. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl. Environ. Microbiol. 64:279-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birtles, R. J., T. J. Rowbotham, R. Michel, D. G. Pitcher, B. Lascola, S. Alexiou-Daniel, and D. Raoult. 2000. ‘Candidatus Odyssella thessalonicensis ’ gen. nov., sp. nov., an obligate intracellular parasite of Acanthamoeba species. Int. J. Syst. Evol. Microbiol. 50:63-72. [DOI] [PubMed] [Google Scholar]
- 17.Birtles, R. J., T. J. Rowbotham, D. Raoult, and T. G. Harrison. 1996. Phylogenetic diversity of intra-amoebal legionellae as revealed by 16S rRNA gene sequence comparison. Microbiology 142:3525-3530. [DOI] [PubMed] [Google Scholar]
- 18.Birtles, R. J., T. J. Rowbotham, C. Storey, T. J. Marrie, and D. Raoult. 1997. Chlamydia-like obligate parasite of free-living amoebae. Lancet 349:925-926. [DOI] [PubMed] [Google Scholar]
- 19.Bitar, D. M., M. Molmeret, and Y. Abu Kwaik. 2004. Molecular and cell biology of Legionella pneumophila. Int. J. Med. Microbiol. 293:519-527. [DOI] [PubMed] [Google Scholar]
- 20.Biurrun, A., L. Caballero, C. Pelaz, E. Leon, and A. Gago. 1999. Treatment of a Legionella pneumophila-colonized water distribution system using copper-silver ionization and continuous chlorination. Infect. Control Hosp. Epidemiol. 20:426-428. [DOI] [PubMed] [Google Scholar]
- 21.Bozue, J. A., and W. Johnson. 1996. Interaction of Legionella pneumophila with Acanthamoeba castellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect. Immun. 64:668-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brieland, J. K., J. C. Fantone, D. G. Remick, M. LeGendre, M. McClain, and N. C. Engleberg. 1997. The role of Legionella pneumophila-infected Hartmanella vermiformis as an infectious particle in a murine model of Legionnaires' disease. Infect. Immun. 65:4892-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell, L. A., and C. C. Kuo. 2003. Chlamydia pneumoniae and atherosclerosis. Semin. Respir. Infect. 18:48-54. [DOI] [PubMed] [Google Scholar]
- 24.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cianciotto, N. P., and B. S. Fields. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA 89:5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cirillo, J. D., S. L. Cirillo, L. Yan, L. E. Bermudez, S. Falkow, and L. S. Tompkins. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cirillo, J. D., L. S. Tompkins, and S. Falkow. 1994. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 62:3254-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corsaro, D., D. Venditti, A. Le Faou, P. Guglielmetti, and M. Valassina. 2001. A new chlamydia-like 16S rDNA sequence from a clinical sample. Microbiology 147:515-516. [DOI] [PubMed] [Google Scholar]
- 29.Corsaro, D., D. Venditti, and M. Valassina. 2002. New parachlamydial 16S rDNA phylotypes detected in human clinical samples. Res. Microbiol. 153:563-567. [DOI] [PubMed] [Google Scholar]
- 30.Darelid, J., S. Löfgren, and B.-E. Malmvall. 2002. Control of nosocomial Legionnaires' disease by keeping the circulating hot water temperature above 55°C: experience from a 10-year surveillance programme in a district general hospital. J. Hosp. Infect. 50:213-219. [DOI] [PubMed] [Google Scholar]
- 30a.De Bary, A. 1879. Die Erscheinung der Symbiose. Verlag Von Karl J. Trubner, Strassburg, Austria.
- 31.Essig, A., M. Heinemann, U. Simnacher, and R. Marre. 1997. Infection of Acanthamoeba castellanii by Chlamydia pneumoniae. Appl. Environ. Microbiol. 63:1396-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Everett, K. D., R. M. Bush, and A. A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 33.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 34.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fields, B. S., T. A. Nerad, T. K. Sawyer, C. H. King, J. M. Barbaree, W. T. Martin, W. E. Morrill, and G. N. Sanden. 1990. Characterization of an axenic strain of Hartmannella vermiformis obtained from an investigation of nosocomial legionellosis. J. Protozool. 37:581-583. [DOI] [PubMed] [Google Scholar]
- 36.Fliermans, C. B. 1996. Ecology of Legionella: from data to knowledge with a little wisdom. Microb. Ecol. 32:203-228. [DOI] [PubMed] [Google Scholar]
- 37.Fritsche, T. R., R. K. Gautom, S. Seyedirashti, D. L. Bergeron, and T. D. Lindquist. 1993. Occurrence of bacterial endosymbionts in Acanthamoeba spp. isolated from corneal and environmental specimens and contact lenses. J. Clin. Microbiol. 31:1122-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fritsche, T. R., M. Horn, S. Seyedirashti, R. K. Gautom, K.-H. Schleifer, and M. Wagner. 1999. In situ detection of novel bacterial endosymbionts of Acanthamoeba spp. phylogenetically related to members of the order Rickettsiales. Appl. Environ. Microbiol. 65:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fritsche, T. R., M. Horn, M. Wagner, R. P. Herwig, K.-H. Schleifer, and R. K. Gautom. 2000. Phylogenetic diversity among geographically dispersed Chlamydiales endosymbionts recovered from clinical and environmental isolates of Acanthamoeba spp. Appl. Environ. Microbiol. 66:2613-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao, L.-Y., O. S. Harb, and Y. Abu Kwaik. 1998. Identification of macrophage-specific infectivity loci (mil) of Legionella pneumophila that are not required for infectivity of protozoa. Infect. Immun. 66:883-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao, L.-Y., O. S. Harb, and Y. Abu Kwaik. 1997. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant hosts, mammalian and protozoan cells. Infect. Immun. 65:4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao, L.-Y., M. Susa, B. Ticac, and Y. Abu Kwaik. 1999. Heterogeneity in intracellular replication and cytopathogenicity of Legionella pneumophila and Legionella micdadei in mammalian and protozoan cells. Microb. Pathog. 27:273-287. [DOI] [PubMed] [Google Scholar]
- 43.Greub, G., J.-L. Mege, and D. Raoult. 2003. Parachlamydia acanthamoebae enters and multiplies within human macrophages and induces their apoptosis. Infect. Immun. 71:5979-5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greub, G., and D. Raoult. 2003. History of the ADP/ATP-translocase-encoding gene, a parasitism gene transferred from a Chlamydiales ancestor to plants 1 billion years ago. Appl. Environ. Microbiol. 69:5530-5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagele, S., R. Kohler, H. Merkert, M. Schleicher, J. Hacker, and M. Steinert. 2000. Dictyostelium discoideum: a new host model system for intracellular pathogens of the genus Legionella. Cell. Microbiol. 2:165-171. [DOI] [PubMed] [Google Scholar]
- 46.Harb, O. S., L.-Y. Gao, and Y. Abu Kwaik. 2000. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ. Microbiol. 2:251-265. [DOI] [PubMed] [Google Scholar]
- 47.Harb, O. S., C. Venkataraman, B. J. Haack, L.-Y. Gao, and Y. Abu Kwaik. 1998. Heterogeneity in the attachment and uptake mechanisms of the Legionnaires' disease bacterium, Legionella pneumophila, by protozoan hosts. Appl. Environ. Microbiol. 64:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hatch, T. P. 1999. Developmental biology, p. 29-67. In R. S. Stephens (ed.), Chlamydia. American Society for Microbiology, Washington, D.C.
- 49.Henning, K., G. Schares, H. Granzow, U. Polster, M. Hartmann, H. Hotzel, K. Sachse, M. Peters, and M. Rauser. 2002. Neospora caninum and Waddlia chondrophila strain 2032/99 in a septic stillborn calf. Vet. Microbiol. 85:285-292. [DOI] [PubMed] [Google Scholar]
- 50.Hilbi, H., G. Segal, and H. A. Shuman. 2001. Icm/Dot-dependent upregulation of phagocytosis by Legionella pneumophila. Mol. Microbiol. 42:603-617. [DOI] [PubMed] [Google Scholar]
- 51.Hoebe, C. J., and J. L. Kool. 2000. Control of legionella in drinking-water systems. Lancet 355:2093-2094. [DOI] [PubMed] [Google Scholar]
- 52.Horn, M., A. Collingro, S. Schmitz-Esser, C. L. Beier, U. Purkhold, B. Fartmann, P. Brandt, G. J. Nyakatura, M. Droege, D. Frishman, T. Rattei, H. W. Mewes, and M. Wagner. 2004. Illuminating the evolutionary history of chlamydiae. Science 304:728-730. [DOI] [PubMed] [Google Scholar]
- 53.Horn, M., T. R. Fritsche, R. K. Gautom, K. H. Schleifer, and M. Wagner. 1999. Novel bacterial endosymbionts of Acanthamoeba spp. related to the Paramecium caudatum symbiont Caedibacter caryophilus. Environ. Microbiol. 1:357-367. [DOI] [PubMed] [Google Scholar]
- 54.Horn, M., T. R. Fritsche, T. Linner, R. K. Gautom, M. D. Harzenetter, and M. Wagner. 2002. Obligate bacterial endosymbionts of Acanthamoeba spp. related to the beta-Proteobacteria: proposal of ‘Candidatus Procabacter acanthamoebae ’ gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 52:599-605. [DOI] [PubMed] [Google Scholar]
- 55.Horn, M., M. D. Harzenetter, T. Linner, E. N. Schmid, K.-D. Müller, R. Michel, and M. Wagner. 2001. Members of the Cytophaga-Flavobacterium-Bacteroides phylum as intracellular bacteria of acanthamoebae: proposal of ‘Candidatus Amoebophilus asiaticus.’ Environ. Microbiol. 3:440-449. [DOI] [PubMed] [Google Scholar]
- 56.Horn, M., and M. Wagner. 2001. Evidence for additional genus-level diversity of Chlamydiales in the environment. FEMS Microbiol. Lett. 204:71-74. [DOI] [PubMed] [Google Scholar]
- 57.Horn, M., M. Wagner, K.-D. Müller, E. N. Schmid, T. R. Fritsche, K.-H. Schleifer, and R. Michel. 2000. Neochlamydia hartmannellae gen. nov., sp. nov. (Parachlamydiaceae), an endoparasite of the amoeba Hartmannella vermiformis. Microbiology 146:1231-1239. [DOI] [PubMed] [Google Scholar]
- 58.Horwitz, M. A. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158:1319-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horwitz, M. A. 1984. Phagocytosis of the Legionnaires' disease bacterium (Legionella pneumophila) occurs by a novel mechanism: engulfment within a pseudopod coil. Cell 36:27-33. [DOI] [PubMed] [Google Scholar]
- 60.Horwitz, M. A., and S. C. Silverstein. 1981. Interaction of the Legionnaires' disease bacterium (Legionella pneumophila) with human phagocytes. II. Antibody promotes binding of L. pneumophila to monocytes but does not inhibit intracellular multiplication. J. Exp. Med. 153:398-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iliffe-Lee, E. R., and G. McClarty. 1999. Glucose metabolism in Chlamydia trachomatis: the ‘energy parasite’ hypothesis revisited. Mol. Microbiol. 33:177-187. [DOI] [PubMed] [Google Scholar]
- 62.Kahane, S., D. Greenberg, M. G. Friedman, H. Haikin, and R. Dagan. 1998. High prevalence of “Simkania Z,” a novel Chlamydia-like bacterium, in infants with acute bronchiolitis. J. Infect. Dis. 177:1425-1429. [DOI] [PubMed] [Google Scholar]
- 63.Kahane, S., E. Metzer, and M. G. Friedman. 1995. Evidence that the novel microorganism ‘Z’ may belong to a new genus in the family Chlamydiaceae. FEMS Microbiol. Lett. 126:203-207. [DOI] [PubMed] [Google Scholar]
- 64.Kirby, J. E., and R. R. Isberg. 1998. Legionnaires' disease: the pore macrophage and the legion of terror within. Trends Microbiol. 6:256-258. [DOI] [PubMed] [Google Scholar]
- 65.Kirby, J. E., J. P. Vogel, H. L. Andrews, and R. R. Isberg. 1998. Evidence for pore-forming ability by Legionella pneumophila. Mol. Microbiol. 27:323-336. [DOI] [PubMed] [Google Scholar]
- 66.Kool, J. L., J. C. Carpenter, and B. S. Fields. 1999. Effect of monochloramine disinfection of municipal drinking water on risk of nosocomial Legionnaires' disease. Lancet 353:272-277. [DOI] [PubMed] [Google Scholar]
- 67.Kusnetsov, J., E. Iivanainen, N. Elomaa, O. Zacheus, and P. J. Martikainen. 2001. Copper and silver ions more effective against legionellae than against mycobacteria in a hospital warm water system. Water Res. 35:4217-4225. [DOI] [PubMed] [Google Scholar]
- 68.Lemanske, R. F., Jr. 2003. Is asthma an infectious disease? Thomas A. Neff lecture. Chest 123:385S-390S. [DOI] [PubMed] [Google Scholar]
- 69.Lieberman, D., S. Kahane, and M. G. Friedman. 1997. Pneumonia with serological evidence of acute infection with the Chlamydia-like microorganism “Z.” Am. J. Respir. Crit. Care Med. 156:578-582. [DOI] [PubMed] [Google Scholar]
- 70.Loy, A., M. Horn, and M. Wagner. 2003. probeBase: an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mann, B. J., B. E. Torian, T. S. Vedvick, and W. A. J. Petri. 1991. Sequence of a cysteine-rich galactose-specific lectin of Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 88:3248-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marra, A., M. A. Horwitz, and H. A. Shuman. 1990. The HL-60 model for the interaction of human macrophages with the Legionnaires' disease bacterium. J. Immunol. 144:2738-2744. [PubMed] [Google Scholar]
- 73.Marrie, T. J., R. W. Peeling, M. J. Fine, D. E. Singer, C. M. Coley, and W. N. Kapoor. 1996. Ambulatory patients with community-acquired pneumonia: the frequency of atypical agents and clinical course. Am. J. Med. 101:508-515. [DOI] [PubMed] [Google Scholar]
- 74.Marrie, T. J., D. Raoult, B. La Scola, R. J. Birtles, and E. de Carolis. 2001. Legionella-like and other amoebal pathogens as agents of community-acquired pneumonia. Emerg. Infect. Dis. 7:1026-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Molmeret, M., D. M. Bitar, L. Han, and Y. A. Kwaik. 2004. Disruption of the phagosomal membrane and egress of Legionella pneumophila into the cytoplasm during the last stages of intracellular infection of macrophages and Acanthamoeba polyphaga. Infect. Immun. 72:4040-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muraca, P., J. E. Stout, and V. L. Yu. 1987. Comparative assessment of chlorine, heat, ozone, and UV light for killing Legionella pneumophila within a model plumbing system. Appl. Environ. Microbiol. 53:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Newsome, A. L., R. L. Baker, R. D. Miller, and R. R. Arnold. 1985. Interactions between Naegleria fowleri and Legionella pneumophila. Infect. Immun. 50:449-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O'Brein, S. J., and R. S. Bhopal. 1993. Legionnaires' disease: the infective dose paradox. Lancet 342:5-6. [DOI] [PubMed] [Google Scholar]
- 79.Ossewaarde, J. M., and A. Meijer. 1999. Molecular evidence for the existence of additional members of the order Chlamydiales. Microbiology 145:411-417. [DOI] [PubMed] [Google Scholar]
- 80.Payne, N. R., and M. A. Horwitz. 1987. Phagocytosis of Legionella pneumophila is mediated by human monocyte complement receptors. J. Exp. Med. 166:1377-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pennisi, E. 2003. Systems biology. Tracing life's circuitry. Science 302:1646-1649. [DOI] [PubMed] [Google Scholar]
- 82.Poppert, S., A. Essig, R. Marre, M. Wagner, and M. Horn. 2002. Detection and differentiation of chlamydiae by fluorescence in situ hybridization. Appl. Environ. Microbiol. 68:4081-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Proca-Ciobanu, M., G. H. Lupascu, A. Petrovici, and M. D. Ionescu. 1975. Electron microscopic study of a pathogenic Acanthamoeba castellani strain: the presence of bacterial endosymbionts. Int. J. Parasitol. 5:49-56. [DOI] [PubMed] [Google Scholar]
- 84.Rodriguez-Zaragoza, S. 1994. Ecology of free-living amoebae. Crit. Rev. Microbiol. 20:225-241. [DOI] [PubMed] [Google Scholar]
- 85.Rowbotham, T. J. 1986. Current views on the relationships between amoebae, legionellae and man. Isr. J. Med. Sci. 22:678-689. [PubMed] [Google Scholar]
- 86.Rowbotham, T. J. 1983. Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J. Clin. Pathol. 36:978-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roy, C. R., and L. G. Tilney. 2002. The road less traveled: transport of Legionella to the endoplasmic reticulum. J. Cell Biol. 158:415-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rurangirwa, F. R., P. M. Dilbeck, T. B. Crawford, T. C. McGuire, and T. F. McElwain. 1999. Analysis of the 16S rRNA gene of micro-organism WSU 86-1044 from an aborted bovine foetus reveals that it is a member of the order Chlamydiales: proposal of Waddliaceae fam. nov., Waddlia chondrophila gen. nov., sp. nov. Int. J. Syst. Bacteriol. 49:577-581. [DOI] [PubMed] [Google Scholar]
- 90.Schachter, J. 1999. Infection and disease epidemiology, p. 139-146. In R. S. Stephens (ed.), Chlamydia. American Society for Microbiology, Washington, D.C.
- 91.Schmitz-Esser, S., N. Linka, A. Collingro, C. L. Beier, H. E. Neuhaus, M. Wagner, and M. Horn. 2004. ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to chlamydiae and rickettsiae. J. Bacteriol. 186:683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Solomon, J. M., and R. R. Isberg. 2000. Growth of Legionella pneumophila in Dictyostelium discoideum: a novel system for genetic analysis of host-pathogen interactions. Trends Microbiol. 8:478-480. [DOI] [PubMed] [Google Scholar]
- 94.Solomon, J. M., A. Rupper, J. A. Cardelli, and R. R. Isberg. 2000. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect. Immun. 68:2939-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Steinert, M., L. Emody, R. Amann, and J. Hacker. 1997. Resuscitation of viable but nonculturable Legionella pneumophila Philadelphia JR32 by Acanthamoeba castellanii. Appl. Environ. Microbiol. 63:2047-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stone, B. J., A. Brier, and Y. A. Kwaik. 1999. The Legionella pneumophila prp locus; required during infection of macrophages and amoebae. Microb. Pathog. 27:369-376. [DOI] [PubMed] [Google Scholar]
- 97.Swanson, M. S., and R. R. Isberg. 1995. Formation of the Legionella pneumophila replicative phagosome. Infect. Agents Dis. 2:269-271. [PubMed] [Google Scholar]
- 98.Thao, M. L., L. Baumann, J. M. Hess, B. W. Falk, J. C. Ng, P. J. Gullan, and P. Baumann. 2003. Phylogenetic evidence for two new insect-associated chlamydia of the family Simkaniaceae. Curr. Microbiol. 47:46-50. [DOI] [PubMed] [Google Scholar]
- 99.Tilney, L. G., O. S. Harb, P. S. Connelly, C. G. Robinson, and C. R. Roy. 2001. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J. Cell Sci. 114:4637-4650. [DOI] [PubMed] [Google Scholar]
- 100.Tjaden, J., H. H. Winkler, C. Schwoppe, M. Van Der Laan, T. Mohlmann, and H. E. Neuhaus. 1999. Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for transport of energy. J. Bacteriol. 181:1196-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Venkataraman, C., B. J. Haack, S. Bondada, and Y. Abu Kwaik. 1997. Identification of a Gal/GalNAc lectin in the protozoan Hartmannella vermiformis as a potential receptor for attachment and invasion by the Legionnaires' disease bacterium, Legionella pneumophila. J. Exp. Med. 186:537-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Winkler, H. H., and H. E. Neuhaus. 1999. Non-mitochondrial ATP transport. Trends Biochem. Sci. 24:64-68. [DOI] [PubMed] [Google Scholar]
- 103.Yamamoto, H., T. Ezaki, M. Ikedo, and E. Yabuuchi. 1991. Effects of biocidal treatments to inhibit the growth of legionellae and other microorganisms in cooling towers. Microbiol. Immunol. 35:795-802. [DOI] [PubMed] [Google Scholar]