Abstract
The occurrence of high extracellular DNA concentrations in aquatic sediments (concentrations that are 3 to 4 orders of magnitude greater than those in the water column) might play an important role in biogeochemical cycling, as well as in horizontal gene transfer through natural transformation. Since isolation of extracellular DNA from sediments is a difficult and unsolved task, in this study we developed an efficient procedure to recover simultaneously DNA associated with microbial cells and extracellular DNA from the same sediment sample. This procedure is specifically suitable for studying extracellular DNA because it avoids any contamination with DNA released by cell lysis during handling and extraction. Applying this procedure to different sediment types, we obtained extracellular DNA concentrations that were about 10 to 70 times higher than the intracellular DNA concentrations. Using specific targeted prokaryotic primers, we obtained evidence that extracellular DNA recovered from different sediments did not contain amplifiable 16S rRNA genes. By contrast, using DNA extracted from microbial cells as the template, we always amplified 16S rRNA genes. Although 16S rRNA genes were not detected in extracellular DNA, analyses of the sizes of extracellular DNA indicated the presence of high-molecular-weight fragments that might have contained other gene sequences. This protocol allows investigation of extracellular DNA and its possible participation in natural transformation processes.
All aquatic sediments are characterized by high DNA concentrations (concentrations that are 3 to 4 orders of magnitude greater than those found in the water column), mostly (up to 90%) due to extracellular DNA (3, 5, 7, 8, 23). Previous studies reported that complex refractory organic molecules and/or inorganic particles are able to bind, adsorb, and stabilize free DNA in sediments (17, 21, 28). Dell'Anno et al. (8) reported that the free extracellular DNA fraction represented less than 5% of the total extracellular DNA pool. The adsorption of extracellular DNA in the sediment might reduce its degradability, and indeed only about 50% of this DNA can be hydrolyzed by nucleases (8). Consequently, the residence time of extracellular DNA in sediments can be much longer than the residence time in the water column (17, 22). The presence and persistence of large amounts of extracellular DNA in the deeper sediment layers (2, 5) might have important implications for bacterial metabolism, providing a source of nitrogen and phosphorous and/or exogenous nucleotides (3, 7, 14, 27, 32), and may also contribute to horizontal gene transfer through natural transformation (17, 24).
In the last few years, several protocols for extraction of DNA from soils and sediments have been developed and improved (13, 15, 29, 31, 35). In fact, application of culture-independent nucleic acid techniques involving DNA extraction and molecular analyses has allowed the detection and identification of microorganisms in natural environments (10). At the same time, much effort has also been devoted to improving DNA extraction efficiency and to minimizing biases due to DNA contamination (13, 15, 18, 19). The most commonly utilized technique for extraction of DNA from sediments is based on direct in situ cell lysis by physical procedures (e.g., bead mill homogenization, ultrasonication, and freeze-thawing) and/or chemical procedures (e.g., the use of sodium dodecyl sulfate [SDS] or Sarkosyl) (13, 19). Although in situ lysis has the potential to circumvent problems caused by biased representation of the microbial community (i.e., by ensuring that the cells from all groups of microorganisms are lysed in equal proportions [35]), in this procedure extracellular DNA is coextracted with nucleic acids released from the lysed cells, which could lead to misinterpretation of the composition of the target community derived from molecular analysis (9).
Discrimination between intracellular and extracellular DNA in marine sediments is therefore essential for carrying out simultaneous molecular studies of the two DNA fractions. However, isolation of extracellular DNA from sediments is an unsolved task, because the procedures available for extraction of nucleic acids adsorbed on organic and inorganic particles disrupt living cells (9, 17, 35). For example, when the procedure of Ogram et al. (25) is used with freshwater sediments, the extracellular DNA yields are at least 1 order of magnitude lower than those obtained with the nuclease-based procedure of Dell'Anno and Danovaro (6). Moreover, the protocol developed by Ogram and coworkers has not been tested for possible contamination by intracellular DNA due to cell lysis during sediment handling.
In this study we developed an efficient procedure for isolating the intracellular DNA from the extracellular fraction from the same sample. This protocol avoids any bias due to DNA release from cell lysis during handling and extraction and provides a DNA yield and purity adequate for molecular studies, thus allowing workers to perform molecular studies with ancient DNA that may be preserved in deeper sediment layers (2, 8, 33).
MATERIALS AND METHODS
Sediment sampling.
In order to ensure that the protocol was suitable for extraction of extracellular and intracellular DNA from a wide variety of sediment samples, three sediment types were selected. The first type was characterized by shallow sand collected in the Adriatic Sea at a depth of 11 m (this is one of the most organically enriched areas of the Mediterranean Sea [8]); the second sample type was represented by deep-sea mud collected in the western Mediterranean Sea at a depth of 2,850 m; and the third type was represented by a sand-mud basaltic sediment collected in the southern Pacific Ocean at a depth of 3,000 m. Sediment samples were collected by using a box corer, a multiple corer, and a piston corer in different independent deployments. At each sampling site, three to six sediment samples were collected, and subsamples (the top 1 cm) were immediately stored at −20°C after recovery.
Recovery of DNA from microbial cells and extracellular DNA.
The glassware utilized for nucleic acid analysis was carefully cleaned by soaking it in 1 N NaOH, 10% HCl, and MilliQ water to remove organic matter contamination and was subsequently treated at 250°C to avoid nuclease contamination. All the solutions were prepared with MilliQ water and then autoclaved. The protocol described here is an adaptation of the procedures extensively utilized previously for extraction of microbial cells from sediments (1, 12, 30). To date, such procedures have been used only for extracting intact microbial cells from sedimentary matrix, and there has been no consideration of the presence of a large fraction of extracellular DNA which may be coextracted. In order to recover simultaneously extracellular DNA and DNA associated with microbial cells (referred to below as intracellular DNA), 2.5 g (wet weight) of sediment was added to 7.5 ml of 0.1 M sodium phosphate buffer (pH 8.0) with 0.5 g of acid-washed polyvinylpolypyrrolidone, prepared as described by Holben et al. (12). Samples were homogenized with a horizontal shaker at a low speed (150 horizontal shakes per min) for three 1-min cycles, with 1 min of cooling on ice between cycles. Subsequently, SDS (final concentration, 0.1%) was added, and the samples were shaken again for 10 s. Then the samples were chilled on ice and centrifuged at 500 × g for 10 min at 4°C, and the supernatants were transferred to sterile tubes. The sediment pellets were washed two more times by adding 7.5 ml of 0.1 M sodium phosphate buffer (pH 8.0) and centrifuged as described above (but in this case there was no addition of SDS). Supernatants were combined and centrifuged for 20 min at 10,000 × g (at 4°C). After centrifugation, the supernatants containing extracellular DNA were filtered through 0.02-μm-pore-size membrane filters (Anotop 25; Whatman) to ensure removal of any contaminating viruses or bacterial cells, whereas the pellets containing bacterial cells were processed as described by Steffan et al. (30) in order to extract intracellular DNA. The extracellular DNA was precipitated adding 1 volume of a cetyltrimethylammonium bromide (CTAB) solution (1% CTAB in 50 mM Tris-10 mM EDTA, pH 8.0). Samples were incubated at 65°C for 30 min and then centrifuged at 5,000 × g for 10 min at 4°C. The supernatants were discharged, and each pellet was resuspended in high-salt TE buffer (10 mM Tris-HCl, 0.1 mM EDTA, 1 M NaCl; pH 8.0). Then 0.6 volume of cold isopropanol was added to each sample, and the samples were incubated for 1 h on ice and centrifuged at 10,000 × g for 15 min at 4°C. The pellets were resuspended in 10 mM Tris-HCl-0.1 mM EDTA (pH 8.0), an equal volume of equilibrated phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol) was added to each preparation, and the preparations were centrifuged at 10,000 × g for 5 min. Each supernatant was mixed with an equal volume of chloroform-isoamyl alcohol (24:1, vol/vol) and centrifuged again. The supernatant was then precipitated with cold ethanol (final concentration, 70%) and sodium chloride (final concentration, 0.2 M), incubated at −20°C for 1 h, and centrifuged at 10,000 × g for 15 min. Finally, the pellet was washed two times with ethanol, dried under a vacuum, and resuspended in MilliQ water. The purity of DNA was checked by determining the ratio of absorbance at 260 nm to absorbance at 280 nm, and the DNA was quantified fluorometrically by using SYBR Green I (Molecular Probes) as described by Zipper et al. (36). The size of extracellular DNA was analyzed by gel electrophoresis.
Efficiency of extracellular DNA recovery.
To assess the efficiency of extracellular DNA recovery, sediment subsamples that had been precombusted in a muffle furnace at 450°C for 2 h were mixed with 20 μg of DNA from Escherichia coli and processed by using the procedure described above.
Testing for the absence of cell lysis.
In order to test the robustness and reliability of the protocol for extracellular DNA extraction (excluding any contamination due to cell lysis), we carried out additional experiments by mixing pure cultures of the marine bacterium Vibrio mediterranei with sediment subsamples that had been pretreated in the muffle furnace at 450°C for 2 h. V. mediterranei was grown in marine broth (Difco) and collected in the exponential growth phase (about 5 × 108 cells ml−1, as measured by optical density at 600 nm). Since preliminary experiments demonstrated that V. mediterranei released extracellular DNA during exponential growth, the bacterial suspension was pretreated with 10 U of DNase I (Sigma) ml−1 for 15 min at 37°C. After incubation, the bacterial suspension was filtered through 0.2-μm-pore-size Nuclepore filters and washed three times with 5 ml of artificial seawater to remove the DNase. The filters, which retained V. mediterranei, were mixed with sediment subsamples (two replicates) that had been treated in the muffle furnace, and then the filters were processed for simultaneous recovery of extracellular and intracellular DNA.
The pellet (containing bacterial cells), separated from the supernatant, was used to extract DNA from intact cells. In order to test if handling and chemical treatments determined bacterial cell lysis, we checked for the presence of extracellular DNA in the supernatant. To do this, an aliquot of the supernatant was precipitated with CTAB (1% CTAB in 50 mM Tris-10 mM EDTA [pH 8.0]) and subsequently processed by using the procedure described above. Moreover, in order to verify the reliability of the procedure for extracellular DNA recovery, another aliquot of the supernatant was supplemented with 1 μg of purified DNA from V. mediterranei before CTAB precipitation. All samples were then analyzed by PCR as described below.
PCR and gel electrophoresis.
16S rRNA was amplified from intracellular and extracellular DNA by using universal primers 27f (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492r (5′-GGT TAC CTT GTT ACG ACT T-3′) (16) and universal primers 530f (5′-GTG CCA GCM GCC GCG G-3′) and 1492r (5′-GGT TAC CTT GTT ACG ACT T-3′) (16). PCRs were performed with a thermocycler (Hybaid) by using 50-μl (final volume) reaction mixtures containing 2.5 U of Taq DNA polymerase (Sigma), each primer at a concentration of 1 μM, each deoxynucleoside triphosphate at a concentration of 0.1 mM, and 1.5 mM MgCl2. The template was 10 to 25 ng of DNA. We used 30 PCR cycles consisting of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, preceded by 3 min of denaturation at 94°C and followed by a final extension for 10 min at 72°C. Negative controls containing all of the components of the PCR mixture except DNA templates were included. The PCR products were examined on a 1% agarose gel in Tris-borate-EDTA buffer (89 mM Tris base, 89 mM boric acid, 2 mM EDTA) containing ethidium bromide for DNA staining and visualization.
Dot blot hybridization.
Dot blot analyses were carried out with extracellular and intracellular DNA by using two different eubacterial probes that hybridize with the 16S rRNA gene. The first probe was prepared by PCR amplification of the 16S rRNA gene from genomic DNA of E. coli by using primers 8f (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 519r (5′-GTA TTA CCG CGG CTG CTG G-3′). The PCR product was purified and labeled with digoxigenin (DIG)-11-dUTP by using a DIG-High Prime random labeling kit according to the manufacturer's instructions (Roche Molecular Biochemicals). The second probe that we utilized was DIG-labeled eubacterial probe 341 (5′-CCT ACG GGA GGC AGC AG-3′) (20), which was synthesized by MWG Biotech. Purified DNA was denatured at 100°C for 10 min, and aliquots containing from 500 to 50 ng of extracellular and intracellular DNA were spotted on positively charged nylon membranes (Roche). Nucleic acids were cross-linked to the air-dried membranes by baking at 60°C for 30 min. The membranes were prehybridized for 2 h at 45°C in a buffer consisting of 5× SSC (1× SSC is 150 mM NaCl plus 15 mM sodium citrate, pH 7), 0.02% (wt/vol) SDS, 0.1% (wt/vol) N-lauryl sarcosine, and 1% blocking reagent (Roche). Hybridization was carried out overnight at 45°C in 15 ml of the same buffer containing 10 pmol of the first denatured probe or 200 pmol of the second probe. The membranes were washed twice with 2× SSC-0.1% SDS for 5 min at room temperature and twice with 0.5× SSC-0.1% SDS for 15 min at 45°C. Chemiluminescent detection of the hybridized probe with anti-DIG-alkaline phosphatase Fab fragments and CSPD (disodium 3-(4-methoxyspiro {1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl) and subsequent removal of bound probe were carried out according to the instructions of the manufacturer (Roche).
RESULTS AND DISCUSSION
A protocol which allows quantitative estimation of levels of extracellular DNA in sediments not biased by DNA contamination due to cell lysis was recently developed (6, 8). However, since this technique is based on nuclease digestion (cleavage of extracellular DNA into nucleotides), it does not allow recovery of DNA for subsequent molecular studies. The recovery of extracellular DNA in water samples has been addressed by several studies and solved (4, 26, 27), but previously only a single attempt was made to isolate extracellular DNA from aquatic sediments (25).
The most difficult tasks for isolation of extracellular DNA in sediments are (i) avoiding extracellular DNA contamination by intracellular DNA, (ii) separating intracellular from the extracellular DNA pool, and (iii) obtaining DNA that is pure enough for subsequent molecular studies. In addition, a wide array of factors (including DNA sorption to sediment colloids, coextraction of enzymatic inhibitors, and degradation or shearing of DNA) affect the recovery of DNA from sediment (13, 19). Therefore, a procedure for extraction of extracellular DNA from sediments should also meet the following criteria: (i) the efficiency of recovery should be high, and the DNA should be representative of the total extracellular DNA pool in the sample; (ii) the DNA extracted should be suitable for accurate quantification (e.g., by fluorescent staining) and for molecular analyses (e.g., by PCR), and (iii) the extracellular and intracellular DNA should be extracted simultaneously from the same sample so that comparative studies (intracellular DNA versus extracellular DNA) can be performed.
All these conditions and requirements have been met with the protocol developed here. This protocol, which was tested with different sediment types characterized by different grain sizes and mineralogical compositions, is based on three washes of the samples with an isotonic solution of sodium phosphate buffer supplemented with polyvinylpolypyrrolidone and low SDS concentrations (concentrations 10 times lower than those generally utilized for in situ lysis) to improve the efficiency of extraction of both extracellular and intracellular DNA pools. By using this procedure, we obtained high concentrations of extracellular DNA (6.7 to 24.3 μg g−1) (Table 1) comparable to those obtained by using the efficient nuclease-based procedure described by Dell'Anno et al. (8) (7.8 to 22.5 μg g−1) and to those obtained in dried soils by a grinding technique by Frostegard et al. (9) (4 to 36 μg g−1). Since the recovery of an internal DNA standard added to sediments ranged from 34 to 60%, the extracellular DNA contents in marine sediments may be even higher than those measured.
TABLE 1.
Extracellular and intracellular DNA concentrations and ratios of extracellular DNA to intracellular DNA in sediment samples (top 1 cm) collected at the three sampling sites
| Sampling location | Sediment type | Water depth (m) | Concn (μg/g [dry wt] of sediment)a
|
Extracellular DNA/intracellular DNA | |
|---|---|---|---|---|---|
| Extracellular DNA | Intracellular DNA | ||||
| Adriatic Sea (Mediterranean Sea) | Shallow sand | 11 | 24.3 ± 9.1 | 0.67 ± 0.04 | 36.3 |
| Western Mediterranean Sea | Deep-sea mud | 2,850 | 21.0 ± 11.3 | 0.31 ± 0.11 | 67.8 |
| Southern Pacific Ocean | Basaltic sandy mud | 3,045 | 6.7 ± 0.1 | 0.57 | 11.8 |
The values are means ± standard deviations.
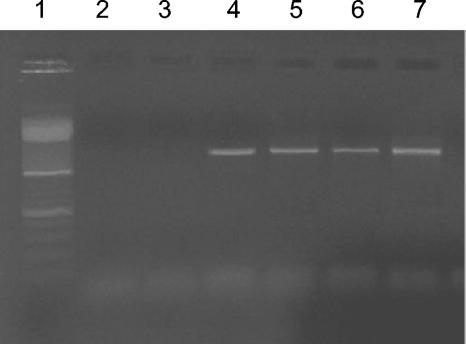
Our procedure was highly specific for extracellular DNA extraction and recovery from sediment samples and was not biased by DNA contamination due to cell lysis (Fig. 1). Experiments carried out with V. mediterranei resulted in detection of 16S rRNA genes only in samples containing DNA extracted from V. mediterranei or samples utilized as positive controls (i.e., samples supplemented with purified DNA from bacterial cells).
FIG. 1.
Gel electrophoresis of the 16S rRNA gene amplified with primers 530f and 1492r and with primers 27f and 1492r from samples utilized to test for DNA contamination due to cell lysis. Lane 1, DNA marker (100 bp; Promega); lanes 2 and 3, PCR products from extracellular DNA recovered from muffle furnace-treated sediment samples inoculated with V. mediterranei; lanes 4 and 5, PCR products from samples containing an internal standard of purified DNA from V. mediterranei; lanes 6 and 7, PCR products from intracellular DNA recovered from muffle furnace-treated sediment samples inoculated with V. mediterranei.
The concentrations of DNA associated with living cells in different sediment types were almost negligible compared with the extracellular DNA concentrations (Table 1), but they were close to theoretical estimates based on DNA contents derived from cell counts (0.97 × 108 and 2.1 × 108 cells extracted from sediments collected in the Adriatic Sea and the deep western Mediterranean Sea, respectively). The fact that the intracellular DNA concentrations were about 10 to 70 times lower than the concentrations of extracellular DNA cannot be explained by the limited cell extraction efficiency (which for our samples ranged from 15 to 22%), indicating that the extracellular DNA pool is by far the largest DNA fraction in marine sediments (7, 8).
The intracellular DNA concentrations displayed limited spatial variability (ratio of highest concentration to lowest concentration, 2.2), whereas the extracellular DNA content varied widely among sampling areas (ratio of highest concentration to lowest concentration, 3.6), and the highest values were obtained for sediments characterized by high organic carbon contents (3). Moreover, the concentrations of the extracellular and intracellular DNA pools did not covary, suggesting that the two components can be subject to different controlling factors in marine sediments. This seems reasonable if we assume that the dynamics of extracellular DNA in aquatic sediments do not depend only on the release and uptake of DNA from dead and living cells but also depend on nuclease degradation (8, 17). At the same time, the lack of information on degradation and turnover rates of extracellular DNA in marine sediments does not allow us to fully understand the diagenetic processes of extracellular DNA in marine sediments.
Recent studies have reported that extracellular DNA may remain preserved in sedimentary deposits over geological time scales (from 1,000 to more than 100,000 years) (2, 33) and thus represent a potential genetic record of functions and compositions of past microbial communities. The identification of prokaryotic genes in the extracellular DNA fraction is therefore the first step toward obtaining insight into the origin of extracellular DNA and toward gaining information on potentially preserved genes (33).
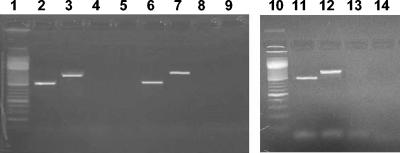
In order to evaluate the presence of prokaryotic 16S rRNA genes in the extracellular DNA pool, we amplified DNA recovered from all sediment samples using specific targeted prokaryotic primers. The results of PCR amplification revealed that extracellular DNA recovered from the surface sediments investigated did not contain amplifiable 16S rRNA genes (Fig. 2), thus confirming the lack of any DNA contamination due to cell lysis. By contrast, when DNA extracted from microbial cells was used as the template, we always amplified 16S rRNA genes.
FIG. 2.
Gel electrophoresis analysis of the 16S rRNA gene amplified with primers 530f and 1492r and with primers 27f and 1492r from extracellular and intracellular DNA extracted from sediment samples collected in the southern Pacific Ocean, in the deep western Mediterranean Sea, and in the Adriatic Sea. Lanes 1 and 10, DNA marker (100 bp; Promega); lanes 2 and 3, intracellular DNA from southern Pacific Ocean sediments; lanes 4 and 5, extracellular DNA from southern Pacific Ocean sediments; lanes 6 and 7, intracellular DNA from western Mediterranean Sea sediments; lanes 8 and 9, extracellular DNA from western Mediterranean Sea sediments; lanes 11 and 12, intracellular DNA from Adriatic Sea sediments; lanes 13 and 14, extracellular DNA from Adriatic Sea sediments.
Although the lack of amplifiable prokaryotic 16S rRNA genes in the extracellular DNA fraction can be due to the low purity of the extracted DNA (34), the analysis of the ratios of absorbance at 260 nm to absorbance at 280 nm indicated that the purity of extracellular DNA recovered from all sediment samples was sufficiently high (the ratios ranged from 1.45 to 1.50). Moreover, no PCR products were obtained after additional extracellular DNA purification with the Wizard DNA Clean-up system (Promega) or by conventional cesium chloride density gradient centrifugation.
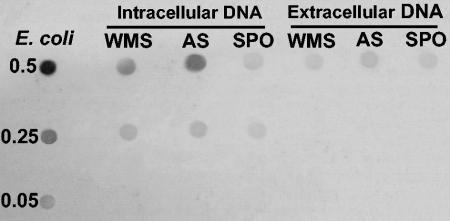
Dot blot analyses carried out with a digoxigenin-labeled probe (prepared by PCR amplification of the 16S rRNA gene of E. coli) indicated a lack of hybridization with extracellular DNA recovered from all sediment samples (data not shown). The lack of amplifiable 16S rRNA genes or hybridization of extracellular DNA could be related to structural integrity. In this regard, additional dot blot analyses carried out with the eubacterial 341 probe allowed detection of a hybridization signal in the extracellular DNA fraction in all samples investigated (Fig. 3), indicating that short sequences of 16S rRNA genes were preserved in the sediment. Further studies are needed to elucidate the quantitative relevance and integrity of gene pools present in the extracellular DNA fraction.
FIG. 3.
Dot blot analysis of extracellular and intracellular DNA with the eubacterial 341 oligonucleotide probe. Equal amounts of the DNA standard from E. coli or of sediment samples were blotted in each row (the amounts [in micrograms] are indicated on the left). The results are for intracellular and extracellular DNA fractions from sediments collected in the deep western Mediterranean Sea (WMS), in the Adriatic Sea (AS), and in the southern Pacific Ocean (SPO).
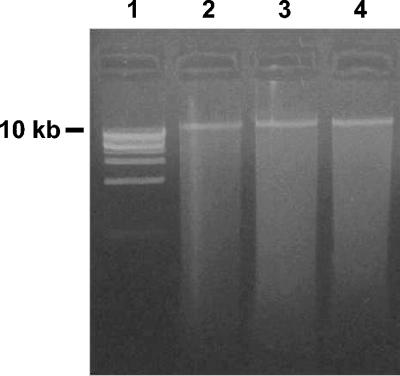
The analysis of the sizes of extracellular DNA fragments by agarose gel electrophoresis indicated the presence of a clear band at a size greater than 10 kb (Fig. 4). Similar results were reported by Ogram et al. (25) for extracellular DNA extracted from freshwater sediments and by De Flaun and Jeffrey (4) for extracellular DNA in the dissolved fraction of seawater samples. Therefore, although the presence of intact 16S rRNA genes in extracellular DNA could not be confirmed for our sediment samples, we could not exclude the possibility that other intact gene sequences could be present and might participate in natural transformation processes (11).
FIG. 4.
Gel electrophoresis of extracellular DNA extracted from different sediment samples. Lane 1, DNA marker (1 kb; Promega); lanes 2, 3, and 4, DNA from sediments collected in the southern Pacific Ocean, Adriatic Sea, and deep western Mediterranean Sea, respectively.
The protocol described here can be considered the most effective method for simultaneous recovery of intracellular and extracellular DNA from marine sediments and provides a basis for progress in the search for gene sequences potentially preserved in the extracellular DNA fraction.
Acknowledgments
We thank the crew of the R/V Abate Molina and Urania and N. Della Croce (DIPTERIS, University of Genoa).
This research was financially supported by the Programme IOC (Oceanic Islands of Chile) and EU Programme ADIOS.
REFERENCES
- 1.Atlas, R. M. 1993. Extraction of DNA from soils and sediments, p. 261-266 In P. F. Kemp, B. F. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, Fla.
- 2.Coolen, M. J., and J. Overmann. 1998. Analysis of subfossil molecular remains of purple sulfur bacteria in a lake sediment. Appl. Environ. Microbiol. 64:4513-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danovaro, R., A. Dell'Anno, A. Pusceddu, and M. Fabiano. 1999. Nucleic acid concentrations (DNA, RNA) in the continental and deep-sea sediments of the Eastern Mediterranean: relationships with seasonal varying organic inputs and bacterial dynamics. Deep-Sea Res. 46:1077-1094. [Google Scholar]
- 4.DeFlaun, M. F., and W. H. Jeffrey. 1987. The distribution and molecular weight of dissolved DNA in subtropical estuarine and oceanic environments. Mar. Ecol. Prog. Ser. 38:65-73. [Google Scholar]
- 5.Dell'Anno A., M. Fabiano, M. L. Mei, and R. Danovaro. 1999. Pelagic-benthic coupling of nucleic acids in an abyssal location of the northeastern Atlantic Ocean. Appl Environ Microbiol 65:4451-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dell'Anno, A., and R. Danovaro. 2001. Nucleic acid turnover in aquatic environments. 1. Determination of total and extracellular DNA in marine sediments, p. 1-9. In A. D. L. Akkermans, J. D. van Elsas, and F. J. De Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 7.Dell'Anno, A., M. Fabiano, G. C. A. Duineveld, A. Kok, and R. Danovaro. 1998. Nucleic acid (DNA, RNA) quantification and RNA/DNA ratio determination in marine sediments: comparison of spectrophotometric, fluorometric, and high-performance liquid chromatography methods and estimation of detrital DNA. Appl. Environ. Microbiol. 64:3238-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dell'Anno, A., S. Bompadre, and R. Danovaro. 2002. Quantification, base composition and fate of extracellular DNA in marine sediments. Limnol. Oceanogr. 47:899-905. [Google Scholar]
- 9.Frostegard, A., S. Courtois, V. Ramisse, S. Clerc, D. Bernillon, F. Le Gall, P. Jeannin, X. Nesme, and P. Simonet. 1999. Quantification of bias related to the extraction of DNA directly from soils. Appl. Environ. Microbiol. 65:5409-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity and ecology: a decade of RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 11.Hermansson, M., and C. Linberg. 1994. Gene transfer in the marine environment. FEMS Microbiol. Ecol. 15:47-54. [Google Scholar]
- 12.Holben, W. E., J. K. Jansson, B. K. Chelm, and J. M. Tiedje. 1988. DNA probe method for the detection of specific microorganisms in the soil bacterial community. Appl. Environ. Microbiol. 54:703-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurt, R. A., X. Qiu, L. Wu, Y. Roh, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 67:4495-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jørgensen, N. O. G., and C. S. Jacobsen. 1996. Bacterial uptake and utilization of dissolved DNA. Aquat. Microb. Ecol. 11:263-270. [Google Scholar]
- 15.Juniper, S. K., M.-A. Cambon, F. Lesonger, and G. Barbier. 2001. Extraction and purification of DNA from organic rich subsurface sediments (ODP Leg 169S). Mar. Geol. 174:241-247. [Google Scholar]
- 16.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, N.Y.
- 17.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58:563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin-Laurent, F., L. Philippot, S. Hallet, R. Chaussod, J. C. Germon, G. Soulas, and G. Catroux. 2001. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 67:2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, D. N., J. E. Bryant, E. L. Madsen, and W. C. Ghiorse. 1999. Evaluation and optimization of DNA extraction and purification for soils and sediment samples. Appl. Environ. Microbiol. 65:4715-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen, K. M., A. M. Bones, K. Smalla, and J. D. van Elsas. 1998. Horizontal gene transfer from transgenic plants to terrestrial bacteria—a rare event. FEMS Microbiol. Rev. 22:79-103. [DOI] [PubMed] [Google Scholar]
- 22.Novitsky, J. A. 1986. Degradation of dead microbial biomass in a marine sediment. Appl. Environ. Microbiol. 52:504-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novitsky, J. A., and D. M. Karl. 1985. Influence of deep ocean sewage outfalls on the microbial activity of the surrounding sediments. Appl. Environ. Microbiol. 50:1464-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ochman, H., J. G., Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 25.Ogram A, G. S. Sayler, D. Gustin, and R. J. Lewis. 1987. The extraction and purification of microbial DNA from sediments. J. Microbiol. Methods 7:57-66. [Google Scholar]
- 26.Paul, J. H., W. H. Jeffrey, and M. F. DeFlaun. 1987. Dynamics of extracellular DNA in the marine environment. Appl. Environ. Microbiol. 53:170-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul, J. H., A. W. David, M. F. DeFlaun, and L. H. Cazares. 1989. Turnover of extracellular DNA in eutrophic and oligotrophic freshwater environments of southwest Florida. Appl. Environ. Microbiol. 55:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romanowski, G., M. G. Lorenz, and W. Wackernagel. 1991. Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Appl. Environ. Microbiol. 57:1057-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rose-Amseleg, C. L., E. Garnier-Sillam, and M. Harry. 2001. Extraction and purification of microbial DNA from soil and sediment samples. Appl. Soil Ecol. 18:47-60. [Google Scholar]
- 30.Steffan, R. J., J. Goksoyr, A. K. Bej, and R. M. Atlas. 1988. Recovery of DNA from soils and sediments. Appl. Environ. Microbiol. 54:2908-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai, Y. L., and B. H. Olson. 1991. Rapid method for direct extraction of DNA from soil and sediments. Appl. Environ. Microbiol. 57:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turk, V., A.-S. Rehnstam, E. Lundberg, and A. Hagstrom. 1992. Release of bacterial DNA by marine nanoflagellates, an intermediate step in phosphorus regeneration. Appl. Environ. Microbiol. 58:3744-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willerslev, E. A. J. Hansen, J. Binladen, T. B. Brand, M. T. P. Gilbert, B. Shapiro, M. Bunce, C. Wiuf, D. A. Gilichinsky, and A. Cooper. 2003. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science 300:791-795. [DOI] [PubMed] [Google Scholar]
- 34.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zipper, H., C. Buta, K. Lämmle, H. Brunner, J. Bernhagen, and F. Vitzthum. 2003. Mechanisms underlying the impact of humic acids on DNA quantification by SYBR Green I and consequences for the analysis of soils and aquatic sediments. Nucleic Acids Res. 31:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]