Abstract
Transitions to new levels of biological complexity often require cooperation among component individuals, but individual selection among those components may favor a selfishness that thwarts the evolution of cooperation. Biological systems with elements of cooperation and conflict are especially challenging to understand because the very direction of evolution is indeterminate and cannot be predicted without knowing which types of selfish mutations and interactions can arise. Here, we investigated the evolution of two bacteriophages (f1 and IKe) experimentally forced to obey a life cycle with elements of cooperation and conflict, whose outcome could have ranged from extinction of the population (due to selection of selfish elements) to extreme cooperation. Our results show the de novo evolution of a conflict mediation system that facilitates cooperation. Specifically, the two phages evolved to copackage their genomes into one protein coat, ensuring cotransmission with each other and virtually eliminating conflict. Thereafter, IKe evolved such extreme genome reduction that it lost the ability to make its own virions independent of f1. Our results parallel a variety of conflict mediation mechanisms existing in nature: evolution of reduced genomes in symbionts, cotransmission of partners, and obligate coexistence between cooperating species.
Keywords: cooperation, major evolutionary transition, partner fidelity feedback
Cooperation is a prominent feature of life; it is also a puzzling one. Organisms compete to procreate, and much of natural selection consequently stems from individual-based competition, be it among replicating molecules, unicellular organisms, or complex eukaryotes. Cooperation between replicating entities pits the selfish interests of the individual against the good of its partners, and, consequently, cooperation can evolve and persist only under restrictive conditions (1–10). Factors that promote the evolution of cooperation may be fortuitous consequences of a species' ecology, but there is growing awareness that these factor may themselves evolve: that not only does cooperation evolve, but so do the factors that promote its persistence. “Conflict mediation” is one such example: the evolution of characteristics that reduce competing selfish interests and thereby enhance the evolution of cooperation (11).
Conflict mediation, and the cooperative outcomes it engenders, may be critically important to major transitions in evolution (12). Protoreplicator cooperation is a predicted condition for the origin of the gene networks necessary for cellular life (13–15) as well as for the origin of chromosomes (12, 16). Similarly, cooperation between once autonomous cells permitted the origins of eukaryotes (17) and multicellularity (8, 11). Proposed mechanisms of conflict mediation are diverse, including partner specialization, partner fidelity feedback (shared fate or coupled fitness between individuals; see refs. 4 and 10), and policing of uncooperative individuals (7). However, there is little understanding of the genotypic or phenotypic details inherent to conflict mediation or whether common themes govern the evolutionary pathways to cooperation.
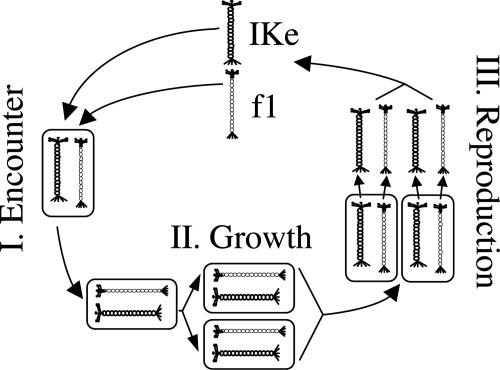
Here, we experimentally investigated the evolution of cooperation and conflict between two divergent filamentous bacteriophages (phages) of Escherichia coli, f1 and IKe. Unlike many phages, f1 and IKe establish permanent nonlethal infections in bacteria and reproduce by extrusion through the cell wall without lysis (18). The experimental evolution protocol was a life history cycle of three steps: (i) encounter, (ii) growth, and (iii) reproduction, iterated 50 times sequentially. Encounter consisted of coinfecting f1 and IKe into naïve E. coli, growth consisted of growing coinfected cells in culture for 18 h such that cotransmission of both phage genomes within the host was enforced, and reproduction consisted of incubating coinfected cells for 1 h to generate phage progeny for the next cycle (see Fig. 1 and Materials and Methods). Each phage was engineered to contain a distinct antibiotic resistance gene to enforce this protocol. By manipulating vertical and infectious transmission of phages, our experimental system created a life history in which pairs of phage genomes were periodically selected for cooperation (encounter and growth), interrupted by episodes in which each phage genome was potentially selected for selfish reproduction at the expense of its partner (reproduction). Natural cooperative systems commonly have life histories in which partners experience alternating phases of interaction and independence such that selection for cooperation and conflict varies temporally. Likewise, our experimental system provides the potential to evolve in multiple directions, to greater or lesser cooperation.
Fig. 1.
The iterated three-step life history. I, encounter: coinfection of f1 and IKe in E. coli. II, growth: enforcement of paired vertical transmission. III, reproduction: production of infectious bacteriophage progeny to complete the cycle. Fifty cycles were performed.
Materials and Methods
Bacteriophages and Cell Lines. f1 and IKe virions are composed of circular, single-stranded DNA encased in flexible protein capsids. The wild-type phages interfere with each other within coinfected cells, lowering phage production in coinfections relative to single infections (19). This discord is likely driven by competition for host resources and by means of nonproductive binding interactions between interphage DNAs and proteins (19, 20). The wild-type f1 genome has 6,407 nt (GenBank accession no. V00606; ref. 21), and the wild-type IKe has 6,883 nt (GenBank accession no. X02139; ref. 22). Capsid length scales with genome size, allowing genetic manipulations including insertion of non-phage DNA (23). Although f1 and IKe share only 55% nucleotide identity, they contain the same 10 genes and share features indicating common ancestry (18, 22). Infection by f1 requires hosts with the F-episome (expressing the F pilus) whereas infection by IKe requires the IncN episome (expressing the N pilus; ref. 24). Hence, each phage uses different means to enter cells, and interference during entry is unlikely. Both f1 and IKe halt superinfection by their own species but not by the other type (18, 25, 26).
Three E. coli K12 cell lines were obtained from Marjorie Russel (The Rockefeller University, New York). Strain A527 contains the tetracycline-resistant IncN conjugative plasmid encoding N pili, K19 is an HfrC strain encoding F pili, and K1037 is HfrC and contains the IncN plasmid, expressing both N and F pili (19). Only K1037 can be infected by both phages.
Genetic Manipulations of Phages. Each phage was engineered to include gene inserts encoding antibiotic resistance. The Tn903 kanamycin (Kn) resistance gene (ref. 27; GenBank accession no. X06404; nt 399-1680) was inserted into the IKe genome at the EcoRI site, and Tn9, a chloramphenicol (Cm) resistance gene (ref. 28; GenBank accession no. J01841; nt 31–930; PCR-amplified to carry PstI ends), was inserted into the f1 genome (the f1 used was CGF3, a wild-type f1 with a PstI site engineered into the intergenic region; ref. 29). Inserts were cloned into the intergenic regions of f1 and IKe between genes IV and II (f1 insert at site 5615 and IKe insert at site 0). Engineered phages were passaged individually before the experiment to adapt them to the gene inserts so that the mutations we scored during our adaptation were unlikely to be compensatory mutations to the insert DNA. Preadaptation of each phage followed the below protocol for five cycles using single antibiotics. We refer to preadapted f1 and IKe (GenBank accession nos. AY842153 and AY842155, respectively) as “ancestral” to distinguish them from wild type.
Passaging Protocol. Encounter (i.e., infection). Cells were incubated in oribital water baths (Labline 3545, Scientific Products) at 200 rpm and 37°C in LB medium (10 g/liter NaCl/10 g/liter bactotryptone/5 g/liter yeast extract/2 μM CaCl2). Phages were added to 10-ml log-phase cultures at ≈2 × 108 cells per ml at low multiplicities of infection (MOI < 0.1). Infection proceeded for 16 min before addition of 37.5 μg/ml Kn and 25.0 μg/ml Cm. Antibiotic concentrations were set at twice the empirically determined minimal inhibitory concentration to minimize selection of high copy number of the phage genomes. Ancestral phages were used for the initial coinfection; subsequent infections used phage mixtures as obtained from the previous cycle. Growth of coinfected cells. Once antibiotics were added, only coinfected cells could grow (enforcing paired vertical transmission of the two phage genomes). Coinfected cells grew for 18 h at 200 rpm (Labline 3545, Scientific Products) and 37°C in the original media.
Reproduction of phage progeny. The 10-ml cell cultures were twice centrifuged and washed with fresh, prewarmed media containing both antibiotics. After the second suspension, in 10 ml, the culture was grown for 1 h to produce phages. Cells were then pelleted and killed by a 20-min exposure to 65°C. The phage supernatant was diluted and used to infect naïve cells for the next passage at MOIs of 0.1–0.01. Only phages could evolve under this protocol, because cells were completely replaced at the beginning of each cycle. Coinfected cells and phage supernatants of every passage were archived. Cells were stored in LB (25% glycerol) at -80°C, and phage supernatants were filter-sterilized and stored at 4°C. This procedure was iterated for 50 cycles.
Fitness Measures. Fitness during growth. To assess the level of cooperation (between phages through their effects on the host) that existed during growth of coinfected cells, cells infected with both evolved phages were competed with cells infected with both ancestral phages. Competitions took place in one flask, with one of the competing cell lines marked with nalidixic acid resistance (NR). Competing cell lines were mixed at low concentrations in prewarmed media with both antibiotics present and grown for 18 h. Relative frequencies of NR and wild-type cells were estimated before and after growth by replica plating, with ancestors arbitrarily assigned a fitness of one. Using changes in relative frequencies, we estimated fitness of cells infected with the evolved phages as W = P1(1 - P0)/P0(1 - P1), where P0 is the initial frequency and P1 is the final frequency of the cell type with evolved phages. NR cells differed slightly (<10%) in fitness from wild type, so assays were done reciprocally and were averaged to correct for fitness differences in the marked cell lines.
Phage output. Phage output from doubly infected cells and ancestral baselines from singly infected cells were estimated from the supernatant titer after the hour-long phage production step. Serially diluted supernatants were infected into K1037 cells at a MOI of <0.01 and plated separately on Kn and Cm to measure densities of each of the phage types.
Sequencing. Both phage genomes were sequenced completely from beginning and end points with at least 2× coverage, and regions with mutations were sequenced over multiple passages to estimate when the mutations ascended to a detectable level. Sequences were obtained from PCR products of phage genomes from supernatant phage populations and from PCR products of phage isolates when results were ambiguous. For base changes that ultimately achieved high frequencies, multiple peaks on a sequence electropherogram (SeqMan 3.6.0, DNASTAR, Madison, WI) at that nucleotide position were assumed to represent polymorphism, and fixation was assumed once multiple peaks resolved to one base. However, consensus sequences only approximate mutant frequencies (30).
Protein Coat Packaging Assays. The frequencies of cross-packaging (one genome packaged in the heterologous coat) and copackaging (both genomes packaged into one coat) were measured because they had important consequences to evolution in this system. Phage supernatants from time points in the evolution were infected into each of the cell lines at a low MOI (<0.001). A527 cells (N pili) could be infected only by phages with IKe coats, K19 cells (F pili) could be infected only by phages with f1 coats, and K1037 cells (both pili) could be infected with either. Resultant colonies growing on Kn contain an IKe genome, colonies growing on Cm contain an f1 genome, and colonies growing on both contain both genomes. Infections were done at low multiplicities, so errors introduced by coinfections of both single genome phages should have been negligible (Table 1).
Table 1. Cross-packaging rates.
| Cells/phage coat | Kn | Cm | Kn, Cm |
|---|---|---|---|
| A527/IKe coats | A | B | C |
| K19/f1 coats | D | E | F |
| K1037/either coat | G | H | I |
Cross-packaging rates were calculated with infections of the three cell types and three antibiotic treatments using the following formulae: f1 genomes in IKe coats = B/(A + B), and IKe genomes in f1 coats = D/(D + E). Rates of copackaging were calculated for each coat type: both genomes in IKe coat = C/(A + B + C), and both genomes in f1 coat = F/(D + E + F). Total copackaging rates were calculated in cells with both pili as the proportion of infections transmitting both genomes at MOIs <10-4, I/(G + H + I).
To test hypotheses about whether mutations in one or both phages were responsible for evolution of copackaging and cross-packaging, packaging assays were also conducted on experimentally doubly infected cells with one evolved and one ancestral phage. Single phage clones from evolved lines were isolated as singly infected colonies and then grown in 10-ml cultures with antibiotics to produce phage supernatants. Mixtures of these supernatants were used to create coinfected cells for the packaging assays.
Results
Chronicle of f1 and IKe Evolution. Both f1 and IKe were initially autonomous, but by the end of the experiment both genomes were frequently packaged together in single f1 coats and IKe had lost all but three of its genes (Fig. 2). The evolution of copackaging preceded the loss of IKe genes. The copackaging ensured a high level of cotransmission between IKe and f1 genomes, reducing and possibly eliminating the conflict that was present initially. The minimized IKe genome (IKe-min) relied on f1 for all coat proteins and morphogenetic processes. The complexities underlying this evolution are most easily explained from the perspective of the encounter, growth, and reproduction phases.
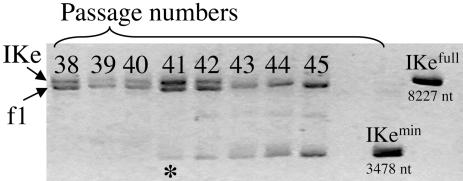
Fig. 2.
IKe-min emerges in passage 41 and spreads to fixation by passage 43 (extinction of IKe-full). Shown is agarose electrophoresis of f1 and IKe genomic preparations from coinfected cells of passages 38–45. Passages are indicated above each lane, and two standards are on the right: IKe-min is a genomic preparation of the minimal IKe phage from passage 50, whereas IKe-full is the IKe used to initiate the experiment. The asterisk indicates the passage in which the minimal IKe genome is first detectable by electrophoresis.
Encounter. In the encounter phase, a pool of the two phages from the previous cycle was given a brief opportunity to infect cells. Addition of antibiotics at 16 min prevented the growth of all cells that were not infected with both phage genomes. There is no expectation of conflict between IKe and f1 at this stage, because each phage needs the other in the same cell. Natural selection should enhance coinfection rates if possible.
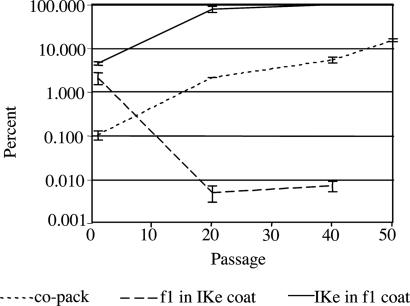
The most striking outcome affecting the encounter phase was the evolution of copackaging: single virions (encapsidated phage particles) containing separate IKe and f1 genomes that could deliver both genomes in a single infection. The initial frequency of virions with both IKe and f1 genomes was 0.1 ± 0.02% of all phages. By passage 20, copackaging had increased to 2.1 ± 0.01% with >99.9% of these in f1 coats. By the end of the evolution the copackaging rate reached 15 ± 1.2%, a 150-fold increase from the ancestral condition (Fig. 3). The evolution of copackaging is phenotypically similar to recombination of the two genomes into one, except that the genomes remained physically separate within the phage protein coat.
Fig. 3.
IKe and f1 cross-packaging rates evolve in opposite directions, and copackaging rates increase 150-fold. Percentage and standard errors of cross-packaging and copackaging rates are shown for f1 and IKe at passages 1, 20, and 40. At passage 50, only copackaging (co-pack) rates are shown, because all genomes were packaged in f1 coats.
Growth. In the growth phase, coinfected cells were incubated 18 h at 37°C in LB with antibiotics, growing to saturation. The expected outcome of selection on this phase is cooperation: improved growth rate and increased density of infected cells at saturation. Coinfected cell concentration at the end of the growth phase increased ≈3.3 times over the course of the evolution (ANOVA, P = 0.01; data not shown). The growth rate increased significantly as well. A competition assay with mixtures of cells infected with passage-0 phages and cells infected with passage-38 or passage-50 phages yielded a relative fitness of 2.9 ± 0.9 for passage 38 and a relative fitness of 8.6 ± 3.0 for passage 50 (Kruskal–Wallis nonparametric one-way test comparing passages 0 and 50, P = 0.01). Passage 38 was tested because it immediately preceded the emergence of IKe-min. The chief benefit of IKe-min was likely an improved growth rate during the growth phase, because the cell would have fewer types of phage proteins to contend with and possibly lower phage protein load.
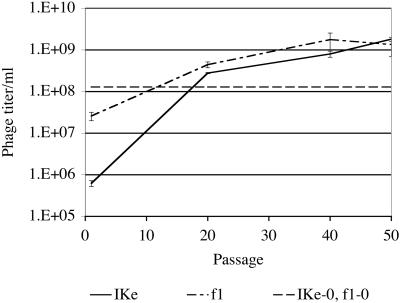
Reproduction. In the reproductive phase, the saturated culture was washed twice to discard free phages and then incubated for 1 h in fresh media to produce new phages. This phase is the most interesting from the perspective of natural selection, because extreme phenotypes can be favored. In general, selection will favor any f1 or IKe variant that produces above-average progeny. Conflict exists because the cell resources are finite, so output by one phage depresses the maximum output from the other phage type. Fecundity during the reproductive phase increased for both phages throughout the experiment: f1 titer increased >50 times from 2.6 × 107 per ml at passage 1 to 1.4 × 109 per ml at passage 50 (ANOVA, P = 0.016), and IKe titer increased >1,000 times from 6.2 × 105 per ml at passage one to 1.8 × 109 per ml at passage 50 (ANOVA, P < 0.001; Fig. 4). Phage titers from the culture illustrate output of the population rather than per-cell and thus combine cell density at the end of the growth phase with phage output per cell during reproduction (thus providing a fitness measure over the growth and reproduction phases).
Fig. 4.
Phage titers increased significantly for both phages over the course of the experiment. Mean and standard errors of phage produced per hour are indicated for passages 1, 20, 40, and 50. The IKe-0, f1–0 line indicates the individual phage production rates of the two ancestral phages.
Another reproductive phenotype that changed during the evolution was cross-packaging: the packaging of a genome into the heterologous protein coat. The ancestral genomes were infrequently cross-packaged: 2.1 ± 1.2% of f1 genomes were found in IKe coats, and 4.5 ± 3.1% of IKe genomes were found in f1 coats. Cross-packaging rates evolved so that each genome increasingly became packaged in f1 coats. Whereas IKe experienced a dramatic increase in cross-packaging by passage 20 to 78.3 ± 12.5%, in the same period the proportion of f1 genomes in IKe coats rate decreased to 0.05 ± 0.002%. Thereafter, IKe's cross-packaging rate continued to increase, reaching 99.2 ± 0.007% by passage 40. Subsequently, the full genome version of IKe was displaced by IKe-min, and all genomes were packaged in f1 coats (Fig. 3). If only single genomes were produced, cross-packaging would be an unambiguous manifestation of conflict, because one phage does not benefit by producing progeny of the other genome. However, copackaging increased in parallel with cross-packaging (possibly as a pleiotropic effect), and by passage 50 >30% of IKe genomes were copackaged with f1 genomes (all in f1 coats).
Molecular Evolution. Over the course of evolution, f1 accrued eight point mutations and IKe had nine point mutations (including two reversions) and two large deletions. The GenBank accession nos. of evolved phage genomes are AT842154 (f1) and AY842156 (IKe). Each phage genome accrued three missense mutations. Functions of the genes/regions in which mutations occurred are described for possible insight to the roles of those mutations (see Table 2, which is published as supporting information on the PNAS web site).
By analyzing the molecular evolution in this experiment, we can assess the possible cooperative nature of the evolution from a more exact approach. In particular, consider packaging evolution, which exhibited significant changes by passage 20 (Fig. 3). To determine the mutational and evolutionary bases of these changes, experimental coinfections mixed the evolved and ancestral phage genomes in different combinations (f120IKe0 and f10IKe20) as well as control combinations (f10IKe0 and f120IKe20); subscripts indicate the passage number of the isolate. The only combination that produced packaging rates similar to passage 20 was the double f120IKe20. Hence, mutations in both IKe and f1 were necessary to explain the packaging evolution. Because f1 mutations contributed to cross-packaging (which was almost entirely IKe in f1 coats, and thus potentially detrimental to f1), f1 must have benefited from copackaging IKe (and, likewise, IKe must have benefited as well). Thus, the molecular evidence indicates that the evolution of cross-packaging (and, with it, copackaging) was beneficial to both phages. Of the mutations that fixed during the first 20 passages, f1 nt 957, a missense mutation in f1 gene V, is the best candidate for driving cross-packaging evolution. Gene V protein binds genomes in preparation for packaging (18, 31). The f1 mutation interacted in some unknown way with a 206-nt deletion of non-protein-coding DNA in IKe, the sole IKe mutation fixing in this period (see Table 2).
Discussion
The Biological Basis of Conflict Mediation. Our design attempted to mimic natural systems of cooperation with elements of conflict, whereby the evolutionary outcome was indeterminate. The result in our system was evolution of a conflict mediation system that facilitated cooperation between the two phage genomes, IKe and f1. Conflict mediation evolved by copackaging of both genomes into one protein coat.
The benefit of copackaging is easily argued. To be successful, each f1 phage must find a host, and that host must also be infected with IKe. Any f1 that copackages itself with IKe need only find a host to be successful. Thus, as long as IKe is uncommon, copackaging benefits f1. The argument also goes the other way: IKe should benefit by copackaging f1 if infection by f1 is not assured. The protocol maintained the phage:cell ratio at 0.1–0.01, ensuring that both f1 and IKe infections were relatively rare, hence that copackaging would be beneficial to both phages. However, there will often be an asymmetry in the strength of this selection: if IKe is rarer than f1, for example, f1 gains more by being copackaged than does IKe. In turn, mutations arising in f1 that facilitate copackaging are more strongly selected than mutations in IKe facilitating copackaging. At the level of detection of our assays, changes in both phages were necessary to achieve high levels of copackaging, so evolution in both phages contributed to this phenotype, and we cannot say whether the evolution was driven by selection on f1 or on IKe.
In principle, the copackaging could have evolved in either direction (IKe in f1 coats or f1 in IKe coats), regardless of which phage benefited more. A mutation in f1 that enhanced copackaging might evolve either by increasing the incorporation of IKe genomes in f1 coats or by increasing the packaging of f1 genomes in IKe coats. The better studied wild-type f1 is known to produce 2–5% polyphages (i.e., virions with multiple genomes) under some conditions (32), so possibly there was a predisposition toward including IKe genomes in f1 coats. Once IKe was packaged mostly in f1 coats, the loss of IKe coat protein genes became feasible, whereas loss of f1 genes was not feasible.
The increase in copackaging, in fact, should have resulted in a substantial fitness increase for both phages during the encounter phase. By passage 20, although copackaged virions were merely 2% of the total, that small number potentially accounted for 30–80% of the successful infections (if the MOI ranged between 0.1 and 0.01, with IKe constituting half of the single virions). Put another way, by passage 20, copackaging should have roughly doubled the number of successful infections in the encounter phase. Overall, the effect of the combined changes supports the evolution of cooperation.
However, we do not suggest that any of the outcomes described are inevitable. We did not replicate the evolution, because our focus was to study one evolutionary trajectory of the ancestral phages under the selective conditions described. Hence, our design is also without a control. An interesting example of the latter might be if we disassociated the genomes from the virions and transfected them into cells separately, thus disallowing the mechanism of cotransmission that evolved under our design.
Conflict Mediation and Evolution of New Selective Forces. The evolution of copackaging did more than merely increase the rate of coinfected cells; it altered the transmission dynamics of the two genomes and potentially eliminated the conflict that was present at the outset. Enhancing cotransmission eliminates the benefit to one phage out-producing its partner, because its partner contributes to its success in the next generation. During the reproduction step each phage is selected for maximal output at the potential expense of the other, provided that they infect independently in the next cycle. However, copackaging the genomes couples their fitness interests, and this creates a form of partner fidelity feedback (4, 10). Partner fidelity feedback stabilizes cooperation between longstanding spatial neighbors (5, 9), in vertically transmitted symbionts and organelles (3, 4, 26, 33–35), and in any case where repeated or long-term interactions couple fitness between interactants (6, 10). Partner fidelity feedback between f1 and IKe is directly tied to the copackaging rate, which increased over the evolution (Fig. 3). Thus, copackaging not only is cooperative (insofar as it increases fitness of both genomes), but it also creates the conditions for further cooperation.
Natural Systems. Cooperative systems are vulnerable to the evolution of selfish individuals that can thwart cooperation. This potential for conflict varied among different stages of the life cycle under our design, and this is often true of cooperative systems in nature. We illustrate this point with two examples, lichens and coviruses, both of which exhibit phases of encounter, growth, and reproduction. This parallel can be further extended to mutualisms that are acquired horizontally (36) or any interaction with varying spatial structure (5, 9).
In lichens, which constitute a symbiosis between an alga and a fungus, the two partner species exist as independent propagules; their encounter leads to the formation of the lichen thallus. Subsequently, the partners undergo a period of shared growth in which their fitness interests may overlap in the goal of enhancing thallus size. Finally, during reproduction, each partner independently disseminates propagules. During reproduction the fitness interests of the partners have the potential to diverge and thus manifest conflict (37).
Coviruses, viral parasites mostly of plants, have the property that the genome is segmented, and segments are encapsidated into separate particles (38). Encounter (coinfection) of both component particles in the same host cell initiates a stage of shared growth within their host's tissues. During reproduction, each type of particle carrying a genome segment is produced and disseminated independent of the host; thus, there is potential for conflict during this phase.
In our experiment, conflict mediation evolved by copackaging, and this enhanced cotransmission of partners. Some natural systems of cooperation have evolved parallel mechanisms of cotransmission. Lichens reveal multiple evolutionary origins of copropagation, where partners codisperse during vegetative or sexual stages (39). In coviruses, mechanisms of copropagation have not been studied per se, but there would seem to be obvious advantages for some means of enhancing coinfection. Copropagation has also evolved in algal–Cnidarian symbioses of jellyfish. In most cases these symbioses are formed when the Cnidarian acquires the algae horizontally from the seawater. The copropagation has evolved by means of “closed” transmission systems in which the algae are acquired directly from the mother and “semiclosed” transmission in which a high proportion of offspring become infected with maternal algae (40). Social bacteria offer a within-species parallel of conflict mediation. Two experimental cases have demonstrated cooperative evolution that enhanced stickiness between individuals through adhesive polymers (41) and socially dependent swarming (42). Mechanisms that enhance partner fidelity feedback thus appear to evolve under diverse natural settings.
Evolutionary Parallels. Two broad evolutionary parallels are found in the evolution of cooperative systems: reduced genomes and obligate dependence of one partner by the other. Reduced genomes are a common feature in vertically transmitted symbionts, as in bacteria that form ancient associations with eukaryotic cells (43, 44). Genome reduction can result from selection for streamlining, as may have driven genome reduction in IKe, or from stochastic loss. Streamlining may evolve cooperatively, to minimize costs to hosts with which the symbiont shares fitness interests (10, 33) or may directly benefit the symbiont by removing metabolic pathways that are redundant with the host (44). Drift is also implicated in the minimization of “resident” genomes, because they are exposed to repeated bottlenecks and replication in small populations (45). However, because the genome reduction in IKe was associated with a 3-fold fitness gain during growth, it was unlikely to be selectively neutral.
Obligate coexistence of symbionts may stem from genome streamlining. Loss of genes in symbionts whose function is shared with the host may accumulate until independent existence becomes impossible. Gene loss in IKe rendered it unable to independently produce infectious progeny, because it lost all packaging and morphogenetic functions. IKe did retain the ability to be vertically transmitted on its own in a plasmid-like state, because it contained the necessary coding genes (II and X) and replication origins (18).
Conclusion. An experimental system of two genomes, with elements of conflict and cooperation, evolved conflict mediation by enhancing the cotransmission of genomes across the life cycle. The conflict mediation resulted from selection for cooperation at one step in the life cycle, but the mediation effect was to remove conflict at a different step in the life cycle. Conflict mediation has been proposed to represent a central hurdle to the evolution of major transitions in evolution (12). The ease and rapidity with which conflict mediation evolved in this system, and from parallels observed in natural systems, raises the question of just how difficult it is to overcome.
Supplementary Material
Acknowledgments
M. Russel provided phages and cell stocks for these experiments. K. Foster, W. Harcombe, R. Heineman, and I. Molineux made helpful comments on the manuscript. This work was supported, in part, by National Institutes of Health Grant GM57756 (to J.J.B.) and National Science Foundation Grant DEB-0308780 (to J.L.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Cm, chloramphenicol; IKe-min, minimized IKe genome, Kn, kanamycin; MOI, multiplicity of infection.
Data deposition: The sequenced genomes (ancestral and evolved) have been deposited in the GenBank database (accession nos. AY842153–AY842156).
References
- 1.Hamilton, W. D. (1964) J. Theor. Biol. 7, 1-52. [DOI] [PubMed] [Google Scholar]
- 2.Trivers, R. L. (1971) Q. Rev. Biol. 46, 35-57. [Google Scholar]
- 3.Axelrod, R. & Hamilton, W. D. (1981) Science 211, 1390-1396. [DOI] [PubMed] [Google Scholar]
- 4.Bull, J. J. & Rice, W. R. (1991) J. Theor. Biol. 149, 63-74. [DOI] [PubMed] [Google Scholar]
- 5.Nowak, M. A. & May, R. M. (1992) Nature 359, 826-829. [Google Scholar]
- 6.Frank, S. A. (1994) J. Theor. Biol. 170, 393-400. [DOI] [PubMed] [Google Scholar]
- 7.Frank, S. A. (1995) Nature 377, 520-522. [DOI] [PubMed] [Google Scholar]
- 8.Michod, R. E. (1997) Am. Nat. 149, 607-645. [Google Scholar]
- 9.Doebeli, M. & Knowlton, N. (1998) Proc. Natl. Acad. Sci. USA 95, 8676-8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sachs, J. L., Mueller, U. G., Wilcox, T. P. & Bull, J. J. (2004) Q. Rev. Biol. 79, 135-160. [DOI] [PubMed] [Google Scholar]
- 11.Michod, R. E. (2003) in Genetic and Cultural Evolution of Cooperation, ed. Hammerstein, O. (MIT Press, Cambridge, MA), pp. 291-307.
- 12.Maynard-Smith, J. & Szathmary, E. (1995) The Major Transitions in Evolution (Oxford Univ. Press, Oxford).
- 13.Eigen, M. (1971) Naturwissenschaften 58, 465-523. [DOI] [PubMed] [Google Scholar]
- 14.Michod, R. E. (1983) Am. Zool. 23, 5-14. [Google Scholar]
- 15.Szathmary, E. & Demeter, L. (1987) J. Theor. Biol. 128, 463-486. [DOI] [PubMed] [Google Scholar]
- 16.Maynard-Smith, J. & Szathmary, E. (1993) J. Theor. Biol. 164, 437-446. [DOI] [PubMed] [Google Scholar]
- 17.Margulis, L. (1981) Symbiosis in Cell Evolution (Freeman, New York).
- 18.Model, P. & Russel, M. (1988) in The Bacteriophages, ed. R. Calendar, (Plenum, New York), pp. 375-456.
- 19.Russel, M. (1992) J. Mol. Biol. 227, 453-462. [DOI] [PubMed] [Google Scholar]
- 20.Peeters, B. P. H., Schoenmakers, J. G. G. & Konings, R. N. H. (1987) DNA 6, 139-147. [DOI] [PubMed] [Google Scholar]
- 21.Beck, E. & Zink, B. (1981) Gene 16, 35-58. [DOI] [PubMed] [Google Scholar]
- 22.Peeters, B. P. H., Peters, R. M., Schoenmakers, J. G. G. & Konings, R. N. H. (1985) J. Mol. Biol. 181, 27-39. [DOI] [PubMed] [Google Scholar]
- 23.Messing, J. (1979) Recomb. DNA Technol. Bull. 2, 43-48. [Google Scholar]
- 24.Bradley, D. E. (1979) Plasmid 2, 632-636. [DOI] [PubMed] [Google Scholar]
- 25.Dotto, G. P., Enea V. & Zinder, N. D. (1981) Virology 114, 463-473. [DOI] [PubMed] [Google Scholar]
- 26.Messenger, S. L., Molineux, I. J. & Bull, J. J. (1999) Proc. R. Soc. London Ser. B 266, 397-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor, L. A. & Rose, R. E. (1988) Nucleic Acids Res. 16, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alton, N. K. & Vapnek, D. (1979) Nature 282, 864-869. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger, T. C., Fulford, W. D. & Zabin, H. B. (1988) Nucleic Acids Res. 16, 9027-9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badgett, M. R., Auer, A., Carmichael, L. E., Parrish, C. R. & Bull, J. J. (2002) J. Virol. 76, 10524-10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salstrom, J. S. & Pratt, D. (1971) J. Mol. Biol. 61, 489-501. [DOI] [PubMed] [Google Scholar]
- 32.Russel, M. & Model, P. (1989) J. Virol. 63, 3284-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fine, P. E. M. (1975) Ann. N.Y. Acad. Sci. 266, 173-194. [DOI] [PubMed] [Google Scholar]
- 34.Bull, J. J., Molineux, I. J. & Rice, W. R. (1991) Evolution (Lawrence, Kans.) 45, 875-882. [DOI] [PubMed] [Google Scholar]
- 35.Bull, J. J. & Molineux, I. J. (1992) Evolution (Lawrence, Kans.) 46, 882-895. [DOI] [PubMed] [Google Scholar]
- 36.Wilkinson, D. M. (2001) Oikos 92, 377-384. [Google Scholar]
- 37.Richardson, D. H. S. (1999) Mycol. Res. 103, 641-650. [Google Scholar]
- 38.Nee, S. (2000) Philos. Trans. R. Soc. London B 355, 1607-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanders, W. B. & Lucking, R. (2002) New Phytol. 155, 425-435. [DOI] [PubMed] [Google Scholar]
- 40.Montgomery, M. K. & Kremer, P. M. (1995) Mar. Biol. (Berlin) 124, 147-155. [Google Scholar]
- 41.Rainey, P. B. & Rainey, K. (2003) Nature 425, 72-74. [DOI] [PubMed] [Google Scholar]
- 42.Velicer, G. J. & Yu, Y. N. (2003) Nature 425, 75-78. [DOI] [PubMed] [Google Scholar]
- 43.Palmer, J. D. (1997) Science 275, 790-791. [DOI] [PubMed] [Google Scholar]
- 44.Moran, N. A. & Wernegreen, J. J. (2000) Trends Ecol. Evol. 15, 321-326. [DOI] [PubMed] [Google Scholar]
- 45.Andersson, S. G. E. & Kurland, C. G. (1998) Trends Microbiol. 6, 263-268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.