Abstract
Human placenta is rich in mesenchymal stem/stromal cells (MSC), with their origin widely presumed fetal. Cultured placental MSCs are confounded by a high frequency of maternal cell contamination. Our recent systematic review concluded that only a small minority of placental MSC publications report fetal/maternal origin, and failed to discern a specific methodology for isolation of fetal MSC from term villi. We determined isolation conditions to yield fetal and separately maternal MSC during ex vivo expansion from human term placenta. MSCs were isolated via a range of methods in combination; selection from various chorionic regions, different commercial media, mononuclear cell digest and/or explant culture. Fetal and maternal cell identities were quantitated in gender‐discordant pregnancies by XY chromosome fluorescence in situ hybridization. We first demonstrated reproducible maternal cell contamination in MSC cultures from all chorionic anatomical locations tested. Cultures in standard media rapidly became composed entirely of maternal cells despite isolation from fetal villi. To isolate pure fetal cells, we validated a novel isolation procedure comprising focal dissection from the cotyledonary core, collagenase/dispase digestion and explant culture in endothelial growth media that selected, and provided a proliferative environment, for fetal MSC. Comparison of MSC populations within the same placenta confirmed fetal to be smaller, more osteogenic and proliferative than maternal MSC. We conclude that in standard media, fetal chorionic villi‐derived MSC (CV‐MSC) do not grow readily, whereas maternal MSC proliferate to result in maternal overgrowth during culture. Instead, fetal CV‐MSCs require isolation under specific conditions, which has implications for clinical trials using placental MSC. Stem Cells Translational Medicine 2017;6:1070–1084
Keywords: Fetal stem cells, Placenta, Decidua, Chorionic villi, Mesenchymal stromal cells, Primary cell culture
Significance Statement.
This is the first study to detail the requirements to isolate reproducibly fetal mesenchymal stem/stromal cells (MSC) from human term placental chorionic villi (CV). We confirmed the high frequency of maternal cell contamination as a genuine problem in placental MSC cultures and quantified how maternal MSC overgrowth increases rapidly with time in culture. The placental dissection method, culture media and explant culture method are critical determinants for successful fetal CV‐MSC propagation and elimination of maternal MSC contamination. Culture medium to specifically promote rapid proliferation of pure fetal CV‐MSC cultures was defined and is critical step for future large‐scale clinical production of fetal CV‐MSC for regenerative medicine applications.
Introduction
Mesenchymal stem/stromal cells (MSC) have broad potential as clinical therapies, due to their tissue regeneration, immunosuppressive, anti‐inflammatory, and ex vivo proliferation capacities 1, 2, 3. Fetal tissue‐derived MSCs are particularly advantageous, with enhanced proliferation and differentiation potential compared to adult MSC 4, 5, 6, 7. However, due to ethical and technical challenges using fetal cells from abortal material, MSC derived from term placenta are gaining clinical importance 8, 9, 10. The placenta is essentially a fetally derived organ, and thus many published studies assume that ex vivo cultured placenta‐derived MSC (pMSC) are fetal and so fail to confirm the origin of the cells obtained (reviewed in 11). However, the delivered term placenta also contains small amounts of invaginating decidual tissue of maternal origin. Recently it has emerged that pMSC cultures derived from the human chorion are not uniformly fetal, but often comprise maternal or mixed fetal and maternal populations 8. Notwithstanding this, the International Placental Stem Cell Society (IPLASS) consensus report explicitly excludes maternal MSC derived from placenta 8. It is thus of biological and clinical importance to know the origin of pMSC and understand why various isolation methods produce maternal or fetal pMSC.
A recent systematic review from our group examined the origin of MSC isolated from chorionic villi 11. Inclusion criteria were human chorionic MSC that satisfied the International Society for Cellular Therapy (ISCT) definition for MSC 12. Of 147 published studies on pMSC, only 15 examined the origin of the MSC, with eight reporting MSC of maternal origin 13, 14, 15, 16, 17, 18, 19. This phenomenon of maternal cell contamination (MCC) confounds isolation of MSC from placenta, and their mixed origin confuses the stem cell community. Fetal and maternal pMSC have been compared to MSC isolated from amniotic membrane (fetal) and decidua (maternal). But these anatomical locations have markedly different niches and functions in vivo, whereas none compare fetal and maternal MSC both isolated from the same niche, that is, the chorionic villi. Comparison of their cell properties can only be resolved by studying both fetal and maternal cells isolated from the same source (chorionic villi). The mechanism by which maternal cell contamination overgrows ex vivo cultured term chorionic villi‐derived pMSC has yet to be delineated.
As our literature review failed to identify any common dissection, cell isolation or culture method that preferentially leads to fetal over maternal MSC 11, we set out experimentally to define the critical parameters necessary to isolate fetal MSC from the chorionic villi niche of the human term placenta. We compared fetal:maternal cell ratios obtained with both previously published and novel isolation methods using XY chromosome fluorescence in situ hybridization (XY FISH) up to six passages into the ex vivo culture period. We then used this information to develop a robust method for isolating fetal chorionic villi (fCV)‐MSC without maternal decidual (D)‐MSC contamination from the human term placenta. This method has the potential to be scaled up for clinical production of fetal CV‐MSC for clinical trials.
Materials and Methods
Sample Collection
The Human Research Ethics Committee of the Royal Brisbane and Women's Hospital and the University of Queensland approved research and collection of human placenta. Patients gave written informed consent for the use of tissue for research purposes in compliance with national guidelines. Third trimester placentas were obtained from healthy mothers during routine caesarean births after uncomplicated pregnancies. Only pregnancies with a male fetus were used.
Standard Fetoplacental MSC Culture
Third trimester placental MSCs were isolated as described previously 13, 20. A video of the dissection and isolation process, as well as a detailed written protocol, is provided in the following reference 20. A flow diagram of the procedures outlined below and summary of results obtained for each condition is provided in Supporting Information Figure 1. Placentas were used as soon as practically possible after delivery, commonly within 1–3 hours. Briefly, the umbilical cord and external membranes (amniotic sac) were positioned to expose the fetal surface of the placenta. Amniotic membrane was mechanically peeled from around the cord exposing the chorionic plate. The upper 0.5 cm of proximal tissue (the chorionic plate) was dissected off the chorionic villous tissue. Approximately 1 cm3 pieces of the chorionic villi remaining under the plate were collected (∼10 g total tissue, to the 10–15 ml mark of two 50 ml tubes), being careful to avoid ∼0.5 cm depth of the distal maternal/decidual surface underneath. Pieces with obvious residual decidual surface were discarded (a lighter red color indicates the maternal surface, see 20). Tissue portions were washed with 2–3 changes of PBS in the 50 ml tubes, and finely minced in a petri dish, then returned to a 50 ml tube and incubated with digest media. Digest media was comprised of 100 U/ml Collagenase type I (Invitrogen/ThermosFisher, https://www.thermofisher.com/au/en/home.html), 5 μg/ml, DNase I (Sigma‐Aldrich, http://www.sigmaaldrich.com/australia.html), and 2.4 U/ml dispase (Invitrogen) in serum free DMEM‐HG (4.5 g/l glucose, Invitrogen) for 1.5–2 hours, at 37°C with gentle rocking. Freshly prepared digest media was incubated in at least a 1:1 ratio with tissue (e.g., tissue filled up to the 10‐ml mark of a 50‐ml tube plus a further 10 ml digest medium).
Standard MSC medium was added to inactivate/wash out the enzymes and samples were pulse centrifuged (i.e., the centrifuge was allowed to reach 340 g, spun for 5 seconds and then the centrifuge was stopped). Centrifugation forced the large debris to collect together at the bottom of the tube, but left the cells suspended in the supernatant. The supernatant was collected, and 20 ml of fresh MSC media was added to the tissue debris to recover more cells, and the tubes were shaken vigorously for 10 seconds by hand. The samples were briefly pulse centrifuged and this step repeated a third time. All supernatants were pooled, centrifuged for 5 minutes at 340 g, and carefully decanted, leaving some solution over the fragile cell pellet. Cell pellets were resuspended in 35 ml of MSC media and filtered through 100 μm mesh. The entire filtered cell suspension (35 ml) was transferred to one T175 flask and cultured at 37°C in a humidified atmosphere with 5% CO2. Flasks were washed with PBS and media changed 48 hours after isolation.
Modifications Evaluated
To isolate fetal cells from term placenta, we tested three different methods. Unless otherwise stated, identical digestion was used for all methods and culture media consisted of DMEM‐HG (high glucose, 4.5 g/l) supplemented with nonheat inactivated MSC batch tested FCS, antibiotic/antimycotic solution and nystatin (AAN, all from Invitrogen) as above. This media was used in both isolation culture and maintenance and is referred to as standard MSC medium (DMEM+10). Alternatively, alpha MEM was used as the base medium in some experiments, with additives as per standard MSC medium (AMEM+10). EGM2 medium (Lonza, http://www.lonza.com) supplemented with 10% nonheat inactivated MSC batch‐tested FCS (referred to as EGM2 + 10) was used for comparison to standard MSC medium 21, 22.
Anatomical Dissection Approach
MSC were isolated as described but with modifications to select placental tissue based on anatomical regions of the villous tissue, specifically the chorionic plate (CP), chorionic villi (CV), or decidua (D) 20.
Chorionic Plate‐Derived (CP)‐MSC
Upon removal of the amniotic membrane from the fetal surface of the placenta, the chorionic plate from the region closest to the umbilical cord was dissected, (∼1 cm wide by 0.5 cm deep into the villi). Tissue pieces were washed in HBSS x2, finely minced with scissors and measured to fill up to the 10‐ml mark of a 50‐ml tube (∼10 g). An equal volume of digest media was added (∼10 ml), and the tube placed in a gently rocking incubator at 37°C for 1.5–2 hours with vigorous shaking for 10 seconds every 30 minutes by hand. Digested cells were filtered and plated in tissue culture flasks as per the standard method above.
Chorionic Villi‐Derived (CV)‐MSC
Approximately 3 cm2 × 0.5 cm deep into placental tissue was dissected from CV from the region closest to the cord, and ≥1 cm away from the maternal side of the placenta (decidua). Ten grams of tissue was washed, minced, digested, filtered, and plated on 1x T175 tissue culture flask as per the standard method above.
Decidua‐Derived (D)‐MSC
The remaining CV tissue of approximately 0.5 cm thickness was dissected from the maternal side of the placenta (decidua). Ten grams was washed, minced, digested, filtered, and plated on 1x T175 tissue culture flask as per the standard method above.
Explant Methods
The placenta was placed with the maternal surface facing upward (see Supporting Information Fig. 2). Care was taken to choose and isolate a single, large, round cotyledon. The decidua/basal plate was cut off the entire outer surface of the cotyledon (∼0.5–1 cm deep) and discarded 23, 24, 25, 26. The degree of internal extension of decidua from the basal plate is approximate as visual distinction by gross anatomy is imprecise, although a slight difference in fibrous texture and/or a white or lighter red color can be observed.
A 1 cm3 piece of CV tissue was removed from the cotyledonary center, similar to that described by Fukuchi et al., Igura et al., and Abumaree et al. 23, 24, 25, 26. The central tissue was transferred to a clean dish with clean tools and washed with HBSS until the blood was removed. After finely mincing tissue, either 40 mg or 3x more (120 mg) was transferred to a 1.5 ml tube with 1 ml of enzymatic digest solution (incubation for 45–60 minutes at 37°C with gentle rocking). The sample was centrifuged at 400 g for 5 minutes, the supernatant discarded and the pellet transferred to culture vessel by pipetting. Samples were not filtered, but small pieces of tissues were cultured along with mononuclear cells. Alternatively, for some experiments minced tissue was transferred to dishes without enzymatic digestion, this is indicated in the text.
Small‐Scale Explant Method
The digested tissue pieces were evenly spread in 1 well of a 6‐well plate (40 mg for the first experiment and 120 mg in subsequent experiments) were allowed to dry onto the dish by leaving it uncovered in the tissue culture hood for 20–30 minutes. Two‐milliliter media (EGM2 + 10, Amniomax‐II or DMEM+10) was added to relevant wells and incubated under standard tissue culture conditions. Media was refreshed 48 hours and 7 days after isolation. After 14 days, the large pieces of tissue were removed by gently dislodging with a pipette tip and the media replaced. At approximately 20 days post isolation, the outgrowing mononuclear cells and remaining tissue clumps were passaged and expanded. The media was changed in all cultures approximately every 7 days if not passaged.
Large‐Scale Explant Method
Approximately 1 g or the whole 1 cm3 portion from the center of a cotyledon was used for large‐scale culture (outlined in Supporting Information Fig. 2). Samples were incubated in digest media for 1 hour in 5 ml of digestion solution in a 15‐ml tube with gentle rocking at 37°C. Samples were centrifuged at 400 g/5 minutes, supernatant removed and resuspended in 10 ml of EGM2 + 10 and plated into a T75 flask. The large‐scale culture did not require drying of tissue to the flask before adding media.
Media Selection
To determine which media were necessary for optimal fetal CV‐MSC culture, samples were prepared as above and cultured into DMEM+10, AMEM+10, or EGM2 + 10 medium for a direct comparison (N = 3 donors).
Statistics
Data are shown as mean +/− SEM. Long‐term growth kinetics were calculated as described previously 27; N = 4 donors, fetal, and maternal MSC from the same placentas, first trimester fetal bone marrow‐derived MSC (fbmMSC) were from different donors. Data were analyzed using unpaired t test. Flow cytometry data were analyzed with Galios flow cytometer and Kaluza software (Beckman Coulter, https://www.beckmancoulter.com/wsrportal/wsr/index.htm), using two‐way ANOVA and Bonferoni's multiple comparison test (N = 4 donors, fetal, and maternal cells from the same placentas, PL‐EPC or fbmMSC from different donors).
Other Procedures
Methodologies are detailed in the supplementary text for isolation protocols, XY FISH, PCR mesodermal differentiation, flow cytometery, and so forth. 27, 28, 29, 30, 31. Antibodies for flow cytometry are detailed in the Supporting Information Table 1 21, 27, 28. Control fetal bone marrow MSC samples were isolated as previously described 27, 31, 32 and characterized in the Supporting Information Figure 3. PL‐EPC (N = 4 donors), and CD34+ sorted placental‐MSC were isolated, cultured in EGM2 + 10 medium and characterized as previously reported 21, 22.
Results
Anatomical Chorion Dissection Approaches Yield Maternal MSC in Culture
We first incorporated careful macroscopic dissection of the chorion into layers we hypothesized would produce purely fetal MSC. Tissues were dissected separately from the fetal surface, from chorionic plate (CP‐MSC) and the villi themselves (CV‐MSC), taking care to avoid collecting tissue with obvious maternal surface attached. The remaining portion of the placental tissue closest to the maternal surface and containing the decidual tissue was termed Decidua‐derived MSC (D‐MSC), which we expected to produce either pure maternal MSC or mixed fetal/maternal populations (chorionic villi with decidual surface).
Upon plating the mononuclear cells obtained from the three layers into MSC medium, cells attached to the culture dish within 24–48 hours. MSCs were isolated from all three layers of the chorion (n = 3–5 independent donors per layer). The D‐MSC cultures, were generally the first to become confluent, grew as slim long cells in whirlpool‐shaped populations when confluent (Fig. 1A). The CV‐ and CP‐MSC initially proliferated somewhat slower with less defined whirlpool shaped populations than the D‐MSC (P0–P1). However, by P2 CP‐ and CV‐MSC appeared indistinguishable from D‐MSC (Fig. 1A). Non‐MSCs were evident in most cultures at early passage (P0–P1), but did not proliferate under MSC culture conditions. These cells were likely red blood cells, hematopoietic mononuclear cells, endothelial cells, and/or trophoblasts as they are the predominant cell types in the placenta 33. The in vitro expansion capacity of these MSC populations was assessed to P6 (D‐MSC for 70 days) or P8 (CV‐MSC and CP‐MSC for 97 days) (Fig. 1B). The population doubling times (DT) for representative donor isolated CP‐, CV‐, and D‐MSC were 122 ± 16 hours, 143 ± 25 hours, and 158 ± 42 hours, respectively, in contrast to the positive control first trimester fbmMSC, which proliferated more rapidly in long‐term culture (fbmMSC, Fig. 1B, 50 ± 20 hours).
Figure 1.
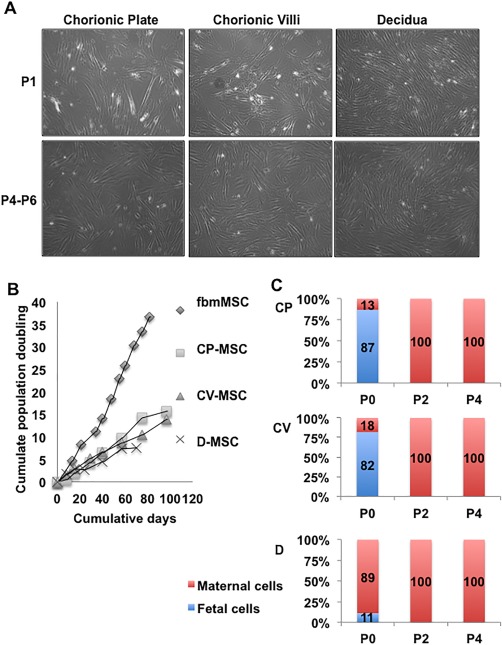
Comparison of placenta‐derived mesenchymal stem/stromal cell (pMSC) isolated from fetal and maternal components of placenta. (A): Morphology of MSC isolated by anatomical selection from the chorionic plate, chorionic villi and decidua. Upper panel—representative images from passage 1 (P1), lower panel—passages 4–6. The total magnification of images is ×40. (B): Representative long‐term growth kinetics of cells listed above, from one donor, compared to fetal bone marrow‐derived MSC. This experiment was not permitted to continue to senescence. (C): Quantitative determination of fetal (male) and maternal (female) cells during ex vivo expansion of pMSC using XY FISH. Passage numbers are indicated on the X‐axis and percentage of the population that was XY male (blue) or XX female (red) on the Y‐axis. (N = 3–5 donors, each data point from 2–5 donors). Abbreviations: CP‐MSC, chorionic plate‐MSC; CV‐MSC, chorionic villi‐derived MSC; D‐MSC, Decidua‐derived MSC; fbmMSC, fetal bone marrow‐derived MSC.
To quantitate changes in the percentage of fetal and maternal cells in culture, XY chromosome FISH was performed on cells isolated from the CP, CV, and D tissue (N = 3–5 independent donors per layer). The CP‐MSC cultures were 87% fetal in origin at P0, but 100% maternal at P2 and P4 (Fig. 1C). Similarly, the CV‐MSC cultures were 93% fetal at P0, but 100% maternal at P2 and P4 (Fig. 1C). The D‐MSC cultures contained 98% maternal cells and only 2% fetal cells at P0 with 100% maternal cells at P2 and P4 (Fig. 1C). Again, the fetal cells observed in the P0 or P1 cultures are not likely to be MSC, but rather other, nonproliferating cell types described above.
The Source of Maternal MSC Contamination—Decidual Cells Lining the Septal Space
To investigate where maternal cell contamination was originating, we next examined whole placental morphology. Figure 2A shows the maternal surface of the placenta, with septal folds reaching to within 10 mm of the chorionic plate or fetal surface in a placenta in situ (Fig. 2A ii). We characterized the origin of cells present in the septum by sectioning the placental tissue so as to keep the septal space intact (Fig. 2A iii) and then determined maternal cell presence by three methods (Fig. 2B). First, using serial formalin fixed paraffin embedded (FFPE) sections, hematoxylin and eosin (H&E) staining showed the classic dark pink stained fetal origin intermediate trophoblasts and light pink stained maternal decidual cells within fibrinoid matrix 33. Second, XY FISH on paraffin sections confirmed both female (XX) and male (XY) cells in the region of the septum. Third, vimentin positive maternal decidual cells appeared along the length of the septa and were distinct from the vimentin negative fetal basal plate intermediate trophoblasts located in the same region 33. In this particular case, decidual cells have differentiated from the maternal endometrial stromal cells and retain expression of the mesodermal marker vimentin 33. Whereas the fetal intermediate trophoblast cells have originated from the trophectoderm layer contiguous with the embryonic ectoderm and are known to be vimentin negative 33. Fetal perivascular cells surrounding the villi, a likely source of the fetal CV‐MSC, are also vimentin positive, but are in a distinctly different location to the maternal decidual cells (not shown).
Figure 2.
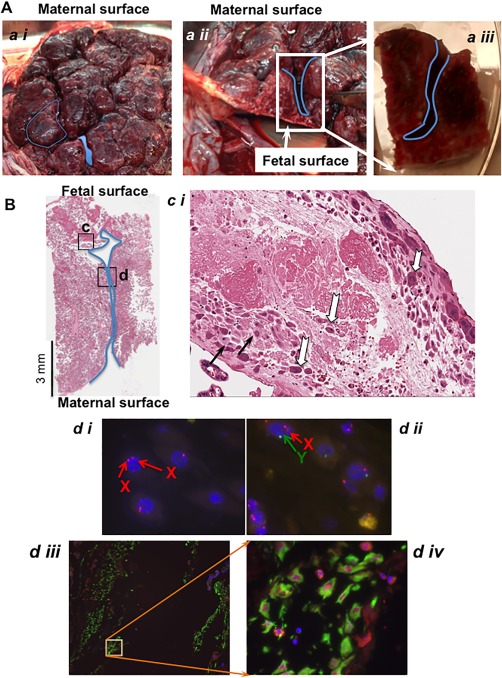
The septal source of maternal cell contamination in isolating fetal cells from the chorionic villi. (A): A representative image of term placenta from maternal surface. The demarcation created between the cotyledons and the extents to which the septa can reach near to the fetal membrane are shown (a ii), in cross section). The blue lines show the location of decidual invagination in the septal space. The specimen shown in (a iii) is an example of one used for subsequent histology and XY FISH studies. Images are captured by a standard photographic camera and not magnified. (B) Sagittal section of septum from the tissue specimen with septa shown in (a iii), stained with hematoxylin and eosin (×1 total mag.). The boxed regions c and d refer to the following more detailed pictures. Scale bar equals 3 mm. (c i) One hundred times magnification of the boxed region c showing characteristic H&E staining where the maternal decidual cells have light pink cytoplasmic staining (black arrow) and the fetal intermediate trophoblasts with dark pink staining in cytoplasm (white arrows). (d i) XY FISH on a consecutive paraffin embedded slide sectioned serially to the slide, image (d i) show maternal XX cells (two red signals) and image (d ii, ×1000 total mag.) shows a male fetal cell with one red and one green signal. Image (d iii) is an adjacent slide to that shown in B, of the region indicated by the boxed region in B labeled (d), stained with vimentin characteristic for maternal decidual cells. Image (d iv, ×400 total mag.) is inset of (d iii, ×200 total mag.).
Methodological Factors Favoring Ex Vivo Expansion of Pure Fetal CV‐MSC
Cotyledonary Core Dissection, Enzymatic Digestion, Explant Culture of Unfiltered Tissue, and EGM2 + 10 Medium Combine to Allow for Pure Fetal CV‐MSC Expansion
Methods in the literature were typically insufficiently detailed to determine precisely where and how placental tissue was isolated, but some commonalities in isolating fetal MSC were the use of small pieces of CV tissue (e.g., 40 mg of the ∼500 g placenta) and explant culture with or without enzymatic digestion. Therefore, we combined dissection methods reported in detail by Fukuchi et al., Igura et al., and Abumaree et al. 23, 25, 26 (eponymously termed the cotyledonary core approach). We tested explant culture of CV tissue, enzymatic digestion protocols, and different culture media to promote the ex vivo expansion and maximize the purity of cultured fetal CV‐MSC (Supporting Information Fig. 1).
Three culture media were tested for ability to support growth of fetal CV‐MSC from explant cultures: (i) DMEM+10% FCS (DMEM+10), standard for culturing fetal bmMSC 27, 34, (ii) Amniomax‐II complete medium as used in clinical cytogenetic laboratories to enhance the growth of fetal cells rather the maternal cells in prenatal diagnostic specimens 35, and (iii) EGM2 + 10% FCS (EGM2 + 10), as reported by us to culture placenta‐derived endothelial progenitor cells (PL‐EPC) 21. Fetal cell outgrowth with or without collagenase/dispase or trypsin digestion was assessed in each medium.
The only medium/digest combination that supported the growth of any cells from the small‐scale explant method to passage 1 was EGM2 + 10 with enzyme digest (Fig. 3A, N = 3 independent donors). Small pieces of tissue, similar to cell colonies or those named “mushroom‐like” colonies by Nazarov et al. 36, remained in the culture and were passaged with the cells, persisting over several passages. However, it took ∼27 days from isolation for each 35‐mm dish to become confluent and required scale up culture into a T75 flask. Generally, no cells proliferated in DMEM+10 and Amniomax‐II media past initial plating (passage 0) (Fig. 3A) or from undigested tissue in any media. Trypsin digest was not as successful as the collagenase/dispase cocktail in allowing cell outgrowth in EGM2 + 10 medium (data not shown).
Figure 3.
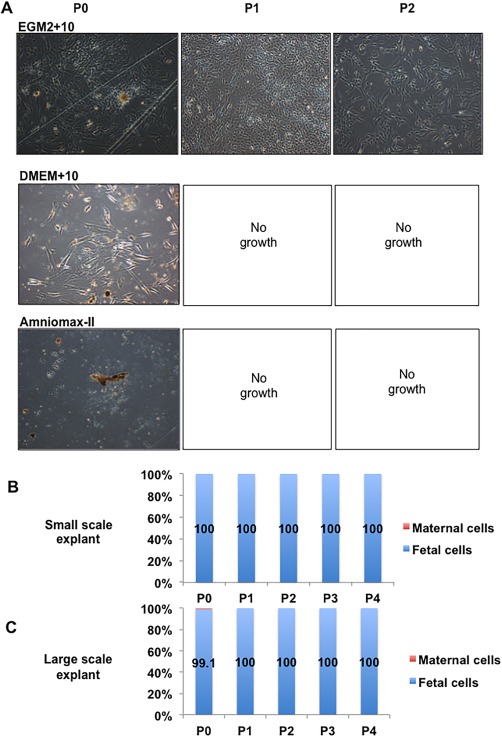
Isolation of pure fetal populations of mesenchymal stem/stromal cell from chorionic villous tissue using an explant method. (A): Representative images of light microscopic images of cells isolated from explant cultures using various media at different passages. (B): Quantitation of fetal:maternal cells isolated from the small scale explant cultures. (C): Quantitation of fetal:maternal cells isolated from the large scale explant cultures (fetal cells shown as blue in graph, maternal cells are shown as red in graph, N = 3–5). The total magnification of images is ×40.
XY FISH on cells isolated by the method above and cultured in EGM2 + 10 medium from 27‐day explant culture were 100% fetal in origin (passage 0–passage 4, N = 3, Fig. 3B). As these MSC cultures were determined to be purely fetal in origin, they were named fetal CV‐MSC (fCV‐MSC). D‐MSC isolated from the same placentas as per the anatomical dissection method and cultured in DMEM+10 were ∼100% maternal from P2 (data not shown).
All Four Steps Are Critical to Pure Fetal CV‐MSC Culture
To further confirm the necessity of all four steps in combination when whole, nonspecific regions of the placental villous tissue were enzymatically digested and treated as per the standard isolation method, including filtering to obtain mononuclear cells. When plated into EGM2 + 10, a mixed population of colonies proliferated, including placental endothelial progenitor cell (PL‐EPC, small cobblestone‐like cells) colonies, fetal MSC colonies (small fibroblastic cells), and maternal MSC colonies (large fibroblastic cells) were observed (Supporting Information Fig. 4). However, after passaging, only the maternal MSC remained in culture (Supporting Information Fig. 4).
Increased Starting Material Leads to More Rapid Expansion of Fetal CV‐MSC
Initially, the amount of starting CV tissue for the explant method was 40–120 mg per 35 mm culture dish. However, when increased to ∼1 cm3 (∼1 g) of starting tissue plated directly into a T75 flask, cells grew out of the explants as early as day 3–4 after isolation, with the flasks becoming confluent by 7–14 days. These populations were purely fetal when analyzed from P1 to P4 (Fig. 3C), and accordingly this large‐scale explant procedure was used for subsequent optimization (Supporting Information Fig. 1).
EGM2 + 10 Medium is Critical for Fetal CV‐MSC Isolation and Propagation
When DMEM+10 was substituted for EGM2 + 10 in already adherent cells, fCV‐MSC remained alive for approximately the first 7 days. However, regardless of whether fCV‐MSC were passaged after DMEM+10 preconditioning or were passaged directly into DMEM+10, they senesced (Fig. 4A). However, transferring D‐MSC into EGM2 + 10 after initial isolation in DMEM+10 made cells qualitatively smaller but retained their whirlpool‐like morphology at high confluence (Fig. 4B, 4C), but did not change surface marker expression (Fig. 4D).
Figure 4.
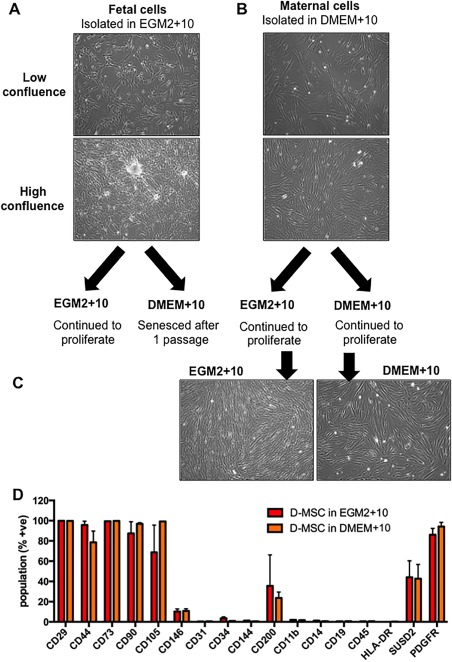
Determining the effect of transferring pure fCV‐mesenchymal stem/stromal cell (MSC) or Decidua‐derived MSC (D‐MSC) cultures into different culture media. (A): Morphology at low and high confluence of fetal CV‐MSC in EGM2 + 10 culture medium and (B) maternal D‐MSC in DMEM+10 medium. (C): Morphology of D‐MSC subsequently cultured in maternal EGM2 + 10 or DMEM+10. (D): The MSC cell surface markers were assessed by flow cytometry for D‐MSC cultured in DMEM+10 or EGM2 + 10 were (red bars are maternal D‐MSC in EGM2 + 10, orange bars are maternal D‐MSC in DMEM+10, N = 3). The total magnification of images is ×40. Statistical analysis was carried out on the mean +/− SD of N = 3 independent donors, using two‐way ANOVA and Bonferoni's multiple comparison test and comparing D‐MSC in EGM2 + 10 versus D‐MSC in DMEM+10 for each cell surface marker. p = not significantly different. Abbreviation: D‐MSC, Decidua‐derived MSC.
Using the large‐scale explant procedure, digested CV tissue was plated directly into T75 flasks and different media conditions (EGM2 + 10, DMEM+10, or AMEM+10). In all media, tissue explants attached, survived and cells proliferated to some degree, with EGM2 + 10 being the most conducive medium (Fig. 5A). All cultures were passaged simultaneously at 14 days post isolation and continued to be cultured in the same medium as for isolation. Cells in EGM2 + 10 survived and proliferated rapidly, requiring further passage at approximately weekly intervals D21, D30, and so forth. However, with cultures isolated and passaged into DMEM+10 and AMEM+10, very few cells continued to grow after passage 1; a few small colonies survived to D30 in DMEM+10 but not further. With AMEM+10, cultures proliferated slowly to confluence at D30 (Fig. 5A). However, cells in AMEM+10 had the morphology analogous to maternal cell cultures and XY FISH confirmed they were 100% maternal while cells in EGM2 + 10 retained a fetal CV‐MSC‐like morphology and were 100% fetal (Fig. 5B, 5C). The use of attachment factor (0.1% gelatin) coating on the flasks or trypsin pretreatment had little effect on the outgrowth of cells in any media (not shown). Finally, although AMEM basal medium has almost twice the number of components of DMEM (mainly vitamins, amino acids, and nucleic acids, see www.invitrogen.com), this did not facilitate fetal CV‐MSC expansion ex vivo. Therefore, the EGM2 basal media or, more likely, its proprietary concentrations of growth factors (EGF, IGF‐1, bFGF, and VEGF) contributed directly and appeared critical for fetal CV‐MSC proliferation.
Figure 5.
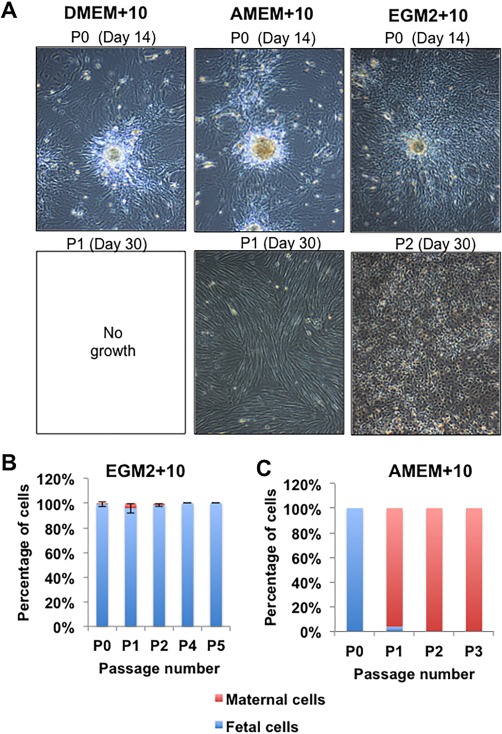
Determining the role of media in the isolation of fetal CV‐mesenchymal stem/stromal cell after large scale explant culture. (A): Representative light microscopic images of cells isolated from explant cultures using various media (N = 4 donors). Cells proliferated or migrated out from explant tissue in all medium at passage 0 (upper images). However, only some conditions allowed cells to reattach and proliferate to the flask after passage and continue to proliferate. These are AMEM+10 in some cases and EGM2 + 10 in all cases, as illustrated by the lower panel of images. The morphology of cells proliferating in the AMEM+10 medium had a distinctly different morphology than those present in the EGM2 + 10 medium. (B and C): Quantitation of fetal and maternal cells by XY FISH in the resulting cell populations from the EGM2 + 10 or AMEM+10. Cells did not proliferate in DMEM+10. The total magnification of images is ×40.
fCV‐MSC Are Not Placental Endothelial Progenitor Cells Despite Their Requirement for Endothelial Growth Medium
We directly compared the characteristics of the fetal CV‐MSC to placenta‐derived endothelial progenitor cells (PL‐EPC) isolated previously by our group 21. A number of similarities exist between the methods to isolate the fetal MSC and the fetal EPC from villous tissue, namely the tissues used, the enzymatic digestion process, and the requirement for culture in EGM2 + 10 medium. Differences in the methods were namely the requirement to use CD45 negative magnetic cell sorting depletion step and a CD34+CD31+ fluorescence activated cell sorting step, and culture on collagen coated dishes. It was of critical importance to exclude the chance that the cells we obtained were PL‐EPC and also to clarify which cells surface markers or functional characteristics were different between the fetal MSC and EPC 37 as this was not clear from the literature.
There was a clear difference in morphology of the cells, with PL‐EPC displaying a classic round, cobblestone appearance, and fCV‐MSC a more square, fibroblastic appearance (Fig. 6B vs. A). The PL‐EPC did not undergo osteogenic or adipogenic differentiation in vitro, whereas the fCV‐MSC did (Fig. 6C vs. Fig. 7D, 7E).
Figure 6.
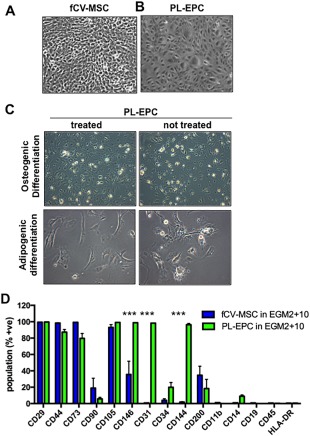
Direct comparison of fetal CV mesenchymal stem/stromal cell (MSC) and placental villi‐derived endothelial progenitor cells (PL‐EPC). (A): Morphology of fCV‐MSC and PL‐EPC (B). (C): PL‐EPC do not undergo mesodermal differentiation to osteocytes or adipocytes. (D): Cell surface markers as assessed by flow cytometry fCV‐MSC and PL‐EPC. The total magnification of images is ×100. Statistical analysis was carried out on the mean +/− SD of N = 3 independent donors, using two‐way ANOVA and Bonferoni's multiple comparison test and comparing fCV‐MSC in EGM2 + 10 versus PL‐EPC in EGM2 + 10 for each cell surface marker. ***, p > .001. Other values were not significantly different.
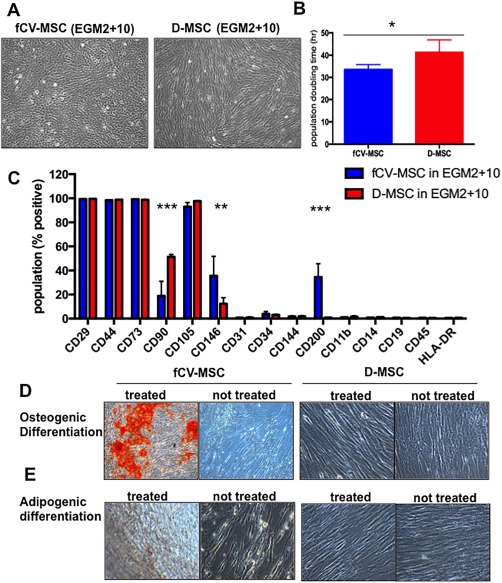
Figure 7.
Comparison of pure fetal and maternal placenta‐derived mesenchymal stem/stromal cell (MSC) (A) Morphology of fetal CV‐MSC and maternal Decidua‐derived MSC (D‐MSC) in EGM2 + 10 medium. (B): Growth kinetics of fCV‐MSC versus D‐MSC in short‐term population doubling assay (pop. doubling time in hours, N = 4 donors). (C): Immunological surface markers that define human MSC were assessed by flow cytometry (blue bars are fetal, red bars are maternal, N = 3). (D and E): Mesodermal differentiation capacity of fCV‐MSC and D‐MSC. fetal CV‐MSC (left side) and control cells D‐MSC (right side) cultured under osteogenic differentiation (D) or adipogenic differentiation conditions (E). The total magnification of images is ×40 (A), ×100 (D), and ×200 (E). Statistical analysis was carried out on the mean +/− SD of N = 3–4 independent donors, using two‐way ANOVA and Bonferoni's multiple comparison test and comparing fCV‐MSC in EGM2 + 10 vs. D‐MSC in EGM2 + 10. *, p > .05, **, p > .05, ***, p > .001. Abbreviation: D‐MSC, Decidua‐derived MSC.
The endothelial markers CD31 and CD144 were not expressed by fCV‐MSC (<2%, Fig. 6D), but were expressed by >95% of PL‐EPC (p < .0001 for each). CD146 was expressed by a moderate number of fCV‐MSC (36% +/− 16, Fig. 6D) but a high number of PL‐EPC (98% +/− 0.17, p < .0001). Markers expressed in common between the two cell types, and known MSC markers, included CD29, CD44, and CD105, with CD73 being expressed at >80% in both populations. The expression of CD34, CD90, and CD200 trended toward differential expression between the two cell types, but these values did not reach statistical significance. Both cell types were negative for hematopoietic markers (HLA‐DR, CD45, CD11b, CD14, and CD19, Fig. 6D).
Comparison Between Fetal and Maternal MSC Isolated from Term Chorionic Villi
The morphology of fCV‐MSC in EGM2 + 10 differed markedly from maternal D‐MSC from the same placenta (Fig. 7A). fCV‐MSC were small, with a short fibroblastic appearance, and cells grew as clusters at low confluence, whereas the maternal D‐MSC appeared longer and formed the classic whirlpool pattern at high confluence. To an experienced user, the microscopic morphology of a flask of cells at confluence could easily distinguish fetal CV‐MSC cultures from those of D‐MSC. The fCV‐MSC in EGM2 + 10 proliferated more rapidly than D‐MSC in EGM2 + 10 in short‐term proliferation assays (Fig. 7B, mean +/− SEM of 33 hours +/− 1.1 vs. 41 hours +/− 2.8 population doubling time, respectively, N = 4 donors).
Cell surface markers and mesodermal differentiation were also different between fetal and maternal MSC. The cell surface marker CD90 was low in fetal CV‐MSC (mean +/− SEM of 19% +/− 12 vs. 51% +/− 1.6, p < .0001, Fig. 7C). Furthermore, fCV‐MSC had more cells positive for CD200 compared to D‐MSC (35% +/− 11 vs. 1% +/− 0.06, p < .0001) and similarly for CD146 (36% +/− 16 vs. 12% +/− 5, p < .006) (Fig. 7C). However, neither CD90, CD146 nor CD200 expression were mutually exclusive in these cell populations and therefore were not suitable as markers for prospective isolation of fetal or maternal pMSC. MSC markers CD73, CD105, CD29, and CD44 were expressed by >98% of cells of both cell types, with both negative for hematopoietic (CD11b, CD14, CD19, CD34, CD45, and HLA‐DR) and endothelial markers (CD144, CD31). fCV‐MSC showed enhanced in vitro osteogenic differentiation capacity compared to D‐MSC which had very little differentiation evident by 21 days (Fig. 7D, 7E).
Discussion
After showing here how standard MSC isolation and culture methods predispose to maternal cell contamination and overgrowth, we report a method whereby pure fetal cells can be isolated and cultured from term placenta, free of maternal contamination. The three chief findings are (i) standard MSC media (DMEM+10) is selective for the ex vivo proliferation of maternal over fetal placental‐MSC (ii) dissection and culture methods are critical to exclude maternal cells, and (iii) serum plus the added growth supplements contained in EGM2 + 10 media are necessary for ex vivo proliferation of fetal CV‐MSC. Such a rational approach to the large‐scale isolation and culture of fetal CV‐MSC ex vivo has not previously been reported in the literature, nor a method that we speculate may be reproducible globally due to the commercially availability of EGM2 medium. Further, novelty lies in that we have extensively quantified of fetal and maternal MSC proliferation kinetics, from various populations isolated via different techniques. We have defined optimal approaches for culturing pure populations of either fetal or maternal pMSC. Being able to selectively culture fetal or maternal pMSC has important implications for the regenerative medicine potential of these cells, which are currently undergoing numerous clinical trials worldwide without determining the best cell type for their application 38, 39, 40.
Decidual Septa Contribute to Maternal Cell Contamination and Overgrowth
Cells isolated from anatomical locations of the villous placenta to favor selection of fetal or decidual cells were each 100% maternal in origin from P2 to P6 on XY FISH, and not surprisingly thus displayed the same morphology, growth kinetics, and differentiation propensities. Interestingly, cells derived from the chorion at P0 cells were initially mixed in origin but later maternal cells overtook in culture with no fetal cells detected beyond P2. Fetal cells were derived in all initial (P0–1) cultures of CP, CV, and D with little between donor variations. Studies in the literature support our finding of maternal cell overgrowth, although few if any quantitated the rate of change over passage as here.
As simple regional selection failed, we next applied an explant method that led to fetal CV‐MSC cultures without maternal contamination. The explant culture method also required removal of all or at least the majority of maternal cells by culturing tissue explants that were dissected from a central cotyledonary core. This specific dissection method excluded not only the decidua basalis but also the septal region as well. Septa are visible indentations evident on the maternal surface of the placenta after birth and consist of anchoring columns between the cotyledons formed by the folds in the decidua as a result of traction during the villous development 33. We confirmed using XY FISH that maternal cells exist in the septa of the placenta, but do not appear elsewhere in great numbers. Anatomically these can extend right up to the chorionic plate, so it is not surprising that the locoregional approaches failed 33. Thus, if tissue samples are taken from the fetal side dissecting between cotyledons, some decidual stromal cells can be collected which then overgrow the fetal cultures. Our findings accord with one previous study where cells isolated from the center of a cotyledon 24 that outgrew from explants were confirmed to be fetal over several passages (P0–P4) without maternal cell contamination.
Interestingly, studies that have derived fetal MSC have done so more often by dissecting the tissue from the maternal side to clearly identify the cotyledon 23, 24, 26. The cotyledonary or lobular structure is only evident from the maternal surface and not from the chorionic plate or fetal side of the placenta. This may explain why maternal cells were easily isolated with both the standard and anatomical dissection methods, where tissue was collected from the fetal surface, and must have unintentionally also collected some decidual tissue that ran close to the fetal membranes.
Core Explant Approach in EGM2 + 10 Medium Critical for Isolation of Pure Fetal CV‐MSC
After determining the relative contributions of media and tissue preparation to the specific isolation of fetal CV‐MSC, we present an optimized isolation procedure with several key steps. First there was the cotyledonary core dissection, with enzymatic digestion and then an explant culture method. Next, a methodological modification was made that utilized a greater amount of starting tissue (1 cm3) and facilitated a larger scale expansion of fetal CV‐MSC. Finally, fetal CV‐MSC only grew out of digested tissue explants when cultured in EGM2 + 10 medium. Excluding any one of these steps resulted in no cell growth or maternal cell contamination and overgrowth.
Did Digest Time or Filtering Influence Fetal Versus Maternal MSC Isolation?
Although there are subtle differences in enzymatic digestion between the anatomical and explant approaches (<1 hour and greater than 1.5 hours), these were not likely to be the reason fetal or maternal cell populations were obtained during each procedure. However, the enzymatic digestion step itself is critical, as we observed no cell proliferation from undigested explants. We decreased enzymatic digestion time as necessary for the amount of tissue used for each procedure (1 g vs. 10 g) so that the appearance of the digested tissue looked the same for each (see 20 for a visual example of the digested placental tissue). The anatomical approach is demonstrated visually in our maternal MSC protocol video 20. We also discovered during the initial experiments with the small‐scale culture that over digestion (1.5 hours) resulted in poor tissue attachment to the flask and limited cell outgrowth.
Filtering the digested mononuclear cells from the remaining tissue is another critical difference in the fetal MSC isolation process. The partially digested tissue that remains in the filter and is discarded in the anatomical approach is, in fact, the tissue pieces that attach to the flask and from which the fetal MSC proliferate out from in the explant procedure. However, the EGM2 + 10 medium contains a critical growth factors for fetal MSC proliferation that are missing from DMEM+10 medium while the specific dissection process removes the majority of decidual tissue containing the maternal cells. In conclusion, the critical points of the process are (i) specific cotyledonary dissection to remove maternal tissue, (ii) mincing and enzymatic digestion to loosen/release the cells from placental villi structures, (iii) not filtering the digested tissue, but plating tissue pieces in explant culture, and (iv) the use of EGM2 + 10 culture medium containing critical growth factors for fetal CV‐MSC proliferation.
We found that the choice of media supplemented to the explants was critical to deriving fetal CV‐MSC cultures. Fetal CV‐MSC survived and proliferated only in EGM2 + 10 media, whereas some cells grew out of the tissue but did not proliferate in DMEM (likely hematopoietic or trophoblastic cells), and did not appear at all in Amniomax‐II. Cells transferred from EGM2 + 10 media to AMEM or DMEM do not survive beyond one passage. The same holds true for cells isolated in AMEM or DMEM. Fetal cells arguably exist in a highly proliferative intrauterine environment and therefore need more/specific growth factors. EGM2 has been used to derive pericytes from a number of sources, with MSC and pericytes considered analogous by some 41. However, it is important to note that DMEM+10 can facilitate maternal D‐MSC and fetal early trimester tissue‐derived (e.g., bone marrow, liver, blood), reported in this study and others 6, 27, 28, 34 but not term fetal placenta CV‐MSC in this study. It is a critical next step to determine exactly what additives in the EGM2 + 10 medium are critical for fCV‐MSC proliferation. Understanding the growth factors required for the ex vivo proliferation of fCV‐MSC would also facilitate the move to serum‐free growth conditions. We anticipate that the commercially available EGM2 media will facilitate the reproduction of our method of fCV‐MSC culture in different laboratories around the world and also in the production of fCV‐MSC for clinical trials. However, we appreciate that the ideal culture medium composition requires further refinement and for FCS to be completely excluded for the optimal clinical production of fCV‐MSC.
Is EGM2 + 10 Medium Able to Select for Fetal Cells Over Maternal Cells?
Both fetal and maternal MSC proliferate in EGM2 as shown in Figure 7A–7C and previously described by us 21, 22. It is not that EGM2 necessarily favors fetal MSC over maternal MSC, but that DMEM+FCS is not conducive to the expansion of fetal MSC. Then the fetal MSC proliferate in the EGM2 more rapidly than the maternal MSC (Fig. 7B) and the maternal MSC get progressively reduced in number over time (as may have occurred in some cultures shown in Fig. 5B).
The selection of the precise area of tissue at the center of a cotyledon removes the source of the maternal cells, that is, the decidual tissue lining the entire cotyledonary surface. As seemingly unexpected by most stem cell researchers, maternal decidual cells can be found merely 5–10 mm from the fetal surface (Fig. 2A, 2B). However, we have shown that careful dissection will limit the number of maternal MSC from the beginning of culture. These maternal (decidual) cells can also proliferate in EGM2 + 10 medium, whether isolated in DMEM+10 or EGM2 + 10, as shown in Figure 4B‐4D and Figure 7A‐7C.
Recent Publications Detailing Methods for Isolating Fetal CV‐MSC
Our group has previously eluded to the fact that the digest of the whole placenta (i.e., via our standard method) that is plated into EGM2 + 10 gave mixed mesenchymal and endothelial cell colonies 21 (complete data are shown in Supporting Information Fig. 4). These large MSC/fibroblastic colonies were found to be maternal MSC and later isolated specifically using a flow cytometric sorting method 22. Maternal pMSC were found to be the CD34−CD31−CD45− population and could be cultured in both EGM2 + 10 and DMEM+10% FCS 22. The cells that were sorted for CD34+CD31−CD45− could grow in EGM2 + 10 but not DMEM+10. These CD34+ MSC were found to be fetal MSC. The CD34+CD31+CD45− population comprised the placental endothelial colony forming cells (PL‐EPC). Thus, the isolation of placental EPC in EGM2 + 10 21 led serendipitously to the method previously reported by our group for isolating fetal and maternal MSC from human term placental villi 22. This CD34+ method was being carried out concurrently but independently to the experiments described in the current manuscript.
However, this CD34+ sorting method is laborious, time‐consuming (∼8 hours procedure), expensive (requires MACS and FACS of samples), and not easily amenable to the large‐scale clinical production of fetal MSC (uses fluorescence activated cell sorting). Furthermore, the Patel et al. short report does not address the mechanism of where the maternal MSC are located or why they took over the fetal MSC or EPC during ex vivo culture. Therefore, for this current study we hypothesized that maternal MSC contamination and overgrowth could be specifically avoided using a physical/dissection procedure.
The Systematic Review—An Oversimplification of Published Methods?
As mentioned above, it was puzzling as to why the term placenta, a largely fetal tissue, was reported in the literature to produce MSC cultures purely of maternal origin, even in our own hands 13, 20, 38. Therefore, a systematic review was carried out in order to (i) clarify published results on the origin of MSC obtained from the human chorion, (ii) determine if common methodologies resulted in MSC of a particular origin, and (iii) summarize the techniques used determine the origin of MSC obtained. Surprisingly, the main outcome was that only 18% of studies reported MSC origin (N = 26 studies of 147) and only 10% (N = 15) reported both MSC origin and satisfied MSC minimal criteria. Next, the systematic review reported that seven studies reported pure fetal MSC cultures, seven reported maternal pure MSC cultures, one reported mixed fetal and maternal cultures, and one reported the separate isolation of pure fetal or pure maternal MSC 42. However, two studies included in the first trimester placental samples were from the same laboratory so would obviously use the same techniques 43, 44.
Igura et al. 26 was the only paper in the systematic review 11 to isolate fetal pMSC using an explant method, whereas the five other papers on term placenta used enzymatic digestion of tissue pieces or perfusion to isolate fetal pMSC. Culture media and incubation conditions were otherwise similar in all papers (DMEM, AMEM, +10%–15% FCS, no additional growth factors). However, although we found that growth media was one of the four critical methodological points for successful culture fCV‐MSC, culture medium components were not specifically assessed in the systematic review 11.
Wang et al. was the only study that claimed to isolate fetal and maternal pMSC from the same placenta 42. Interestingly they used human umbilical cord blood serum as a substitute for FCS in the culture medium. Also, the authors report that they isolated fetal MSC from the amniotic side (i.e., fetal side) of the placenta (“1cm thick” was described, but not other dimensions or tissue weight) and maternal MSC from the maternal side of the placenta (“0.5 cm thick” tissue). Clearly, this paper lacked sufficient description of the placental dissection method to repeat the procedure accurately. Further, their claim of fetal‐origin MSC was not backed up by quantitative origin testing techniques, such as XY FISH, or late passage fetal origin testing, so one could question the validity of their claims of obtaining both fetal and maternal chorionic villi‐derived MSC.
Caveats From This Study
We did not use placentas from female babies in this study due to the difficulties in quantitating fetal:maternal cell populations in a mixture of two genetically related female donors cells. Although, similar results would be expected, and could be easily assessed by microscopic observations of cell morphology differences that we observed between fetal and maternal pMSC. Gene expression studies that are currently underway may yield cell surface markers specific to fetal or maternal pMSC and could allow rapid and precise quantitation of the fetal:maternal cell ratio in a population of pMSC via flow cytometry, independent of fetal gender. We also determined that fetal CV‐MSC proliferate more rapidly and are superior at osteogenic differentiation than D‐MSC, so fetal CV‐MSC might be preferred for some applications including for skeletal regeneration in the fetus, infant, or child 45.
Several components included in EGM2 medium have been determined to enhance human MSC proliferation, including the well‐known bFGF 46, 47, and also EGF, IGF‐1, and ascorbic acid 48, 49, 50, 51, 52, 53. It would be of interest to determine in future which factors are critical for fetal CV‐MSC proliferation. We surmise that the discrepancy between our results and other papers in isolating fCV‐MSC could be due to the FCS sourced in these studies containing factors necessary for fetal CV‐MSC proliferation, and that the FCS we used did not contain the same concentrations or bioavailability of such growth factors 54. In this study, we used two different lots of Australian sourced fetal calf serum purchased from the same company and produced the same finding. These are likely to more similar than two lots of FCS from vastly different herds, collected by different companies and/or in different countries. FCS is an animal derived product of complex and unknown composition and contains serum of numerous donors pooled together. Numerous studies have proven that multiple factors (e.g., genetics, time of day, age, diet, and environment) can affect in the composition of cows milk of (e.g., concentration and type of lipids, nutrients, proteins, and antibodies) 55, 56, 57, 58, 59, 60, 61, 62, 63. Like cow milk, FCS would likely have minor differences in growth factor composition/degradation depending on the specific herd of animals, season, food source, gestation of the fetuses, and as well as differences in serum collection, storage, and preparation methods. Few, if any, of these variables standardized by FCS producers to limit lot‐to‐lot variation. The numerous components known to be present in fetal calf serum as well as the requirements for bone marrow MSC is reviewed by June et al. 54. In Australia, strict government quarantine regulations do not allow the importation of FCS produced in certain countries due to risk of disease contamination. Alternatively, methodologies for isolation were incompletely described in the original articles, differences in reagents used (e.g., enzymes, antibodies) or cells not thoroughly assessed for fetal:maternal cell ratio, are potential reasons for our findings contrasting with those of other groups. We have previously reported discrepancies in gene expression in fbmMSC isolated by similar methods to collaborators in different countries, possibly due to such uncontrollable factors 64. We acknowledge that the future of commercial large‐scale manufacture of clinical grade MSC will be challenging, if not impossible, with fetal bovine serum‐supplemented media. Animal‐free, fully defined cell culture medium is seen as a critical future development for the MSC field 54, but remains to be validated by the MSC‐research community.
Conclusion
This is the first study to detail the requirements to isolate reproducibly fetal cells from human term placental chorionic villi. We confirmed the high frequency of MCC as a genuine problem in placental MSC cultures and quantified how maternal MSC overgrowth increases rapidly with time in culture. The dissection method, culture media, and explant method are critical determinants for successful fCV‐MSC propagation and elimination of maternal MSC contamination over serial passages. An anatomical approach required the dissection of the cotyledonary core clear of decidual septa, and culture required the avoidance of standard media promoting maternal MSC growth in favor of endothelial growth media, which contained factors essential for fetal cell proliferation. Our four‐step method comprising of (i) villous tissue dissection using the cotyledonary core approach, (ii) enzymatic digestion, (iii) the use of an explant culture technique to allow fCV‐MSC to outgrow from the villous tissue pieces, and (iv) the culture in EGM2 + 10 medium to specifically promote proliferation of fCV‐MSC and suppression of maternal cell contamination. In conclusion, the methods we have detailed here allow for the culture and expansion of pure fetal CV‐MSC and may be suitable for the future large‐scale clinical production of fetal CV‐MSC.
Author Contributions
V.S.: carried out sample collection and preparation, data acquisition, data analysis and edited the manuscript; A.S.: contributed to some sample preparation, data acquisition and analysis; N.F.: developed the original concept, provided funding, intellectual input, editing and final approval of the manuscript; R.P.: developed the experimental design, supervised data acquisition, carried out sample collection, data acquisition, undertook data analysis, drafted the paper, carried out revisions and had final approval of the manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
Supporting information
Supporting Information Figure 1.
Supporting Information Figure 2.
Supporting Information Figure 3.
Supporting Information Figure 4.
Supporting Information.
Acknowledgments
R.P. was supported by a National Health and Medical Research Council (NHMRC) Postdoctoral Training Fellowship. VS was supported by a University of Queensland International Postgraduate Student scholarship. A.S. was supported by a Queensland University of Technology International Postgraduate Student scholarship. We thank Weili Wang for technical/laboratory assistance. We thank clinical and nursing staff for assisting in patient consent and sample collection.
References
- 1. Bianco P, Cao X, Frenette PS et al. The meaning, the sense and the significance: Translating the science of mesenchymal stem cells into medicine. Nat Med 2013;19:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Phinney DG, Sensebe L. Mesenchymal stromal cells: Misconceptions and evolving concepts. Cytotherapy 2013;15:140–145. [DOI] [PubMed] [Google Scholar]
- 3. Keating A. Mesenchymal stromal cells: New directions. Cell Stem Cell 2012;10:709–716. [DOI] [PubMed] [Google Scholar]
- 4. Guillot PV, De Bari C, Dell'Accio F et al. Comparative osteogenic transcription profiling of various fetal and adult mesenchymal stem cell sources. Differentiation 2008;76:946–957. [DOI] [PubMed] [Google Scholar]
- 5. Millard SM, Fisk NM. Mesenchymal stem cells for systemic therapy: Shotgun approach or magic bullets? Bioessays 2013;35:173–182. [DOI] [PubMed] [Google Scholar]
- 6. Zhang ZY, Teoh SH, Chong MS et al. Superior osteogenic capacity for bone tissue engineering of fetal compared with perinatal and adult mesenchymal stem cells. Stem Cells 2009;27:126–137. [DOI] [PubMed] [Google Scholar]
- 7. Zhang ZY, Teoh SH, Hui JH et al. The potential of human fetal mesenchymal stem cells for off‐the‐shelf bone tissue engineering application. Biomaterials 2012;33:2656–2672. [DOI] [PubMed] [Google Scholar]
- 8. Parolini O, Alviano F, Bagnara GP et al. Concise review: Isolation and characterization of cells from human term placenta: Outcome of the first international Workshop on Placenta Derived Stem Cells. Stem Cells 2008;26:300–311. [DOI] [PubMed] [Google Scholar]
- 9. Parolini O, Alviano F, Bergwerf I et al. Toward cell therapy using placenta‐derived cells: Disease mechanisms, cell biology, preclinical studies, and regulatory aspects at the round table. Stem Cells Dev 2010;19:143–154. [DOI] [PubMed] [Google Scholar]
- 10. Abdulrazzak H, Moschidou D, Jones G et al. Biological characteristics of stem cells from foetal, cord blood and extraembryonic tissues. JR Soc Interface 2010;7(suppl 6):S689–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heazlewood CF, Sherrell H, Ryan J et al. High incidence of contaminating maternal cell overgrowth in human placental mesenchymal stem/stromal cell cultures: A systematic review. Stem Cells Transl Med 2014;3:1305–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dominici M, Le Blanc K, Mueller I et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006;8:315–317. [DOI] [PubMed] [Google Scholar]
- 13. Barlow S, Brooke G, Chatterjee K et al. Comparison of human placenta‐ and bone marrow‐derived multipotent mesenchymal stem cells. Stem Cells Dev 2008;17:1095–1107. [DOI] [PubMed] [Google Scholar]
- 14. Castrechini NM, Murthi P, Qin S et al. Decidua parietalis‐derived mesenchymal stromal cells reside in a vascular niche within the choriodecidua. Reprod Sci 2012;19:1302–1314. [DOI] [PubMed] [Google Scholar]
- 15. Jaramillo‐Ferrada PA, Wolvetang EJ, Cooper‐White JJ. Differential mesengenic potential and expression of stem cell‐fate modulators in mesenchymal stromal cells from human‐term placenta and bone marrow. J Cell Physiol 2012;227:3234–3242. [DOI] [PubMed] [Google Scholar]
- 16. Li C, Zhang W, Jiang X et al. Human‐placenta‐derived mesenchymal stem cells inhibit proliferation and function of allogeneic immune cells. Cell Tissue Res 2007;330:437–446. [DOI] [PubMed] [Google Scholar]
- 17. Macias MI, Grande J, Moreno A et al. Isolation and characterization of true mesenchymal stem cells derived from human term decidua capable of multilineage differentiation into all 3 embryonic layers. Am J Obstet Gynecol 2010;203:495.e499–495.e423. [DOI] [PubMed] [Google Scholar]
- 18. Ringden O, Erkers T, Nava S et al. Fetal membrane cells for treatment of steroid‐refractory acute graft‐versus‐host disease. Stem Cells 2013;31:592–601. [DOI] [PubMed] [Google Scholar]
- 19. Semenov OV, Koestenbauer S, Riegel M et al. Multipotent mesenchymal stem cells from human placenta: Critical parameters for isolation and maintenance of stemness after isolation. Am J Obstet Gynecol 2010;202:193.e191–193.e113. [DOI] [PubMed] [Google Scholar]
- 20. Pelekanos R, Sardesai V, Futrega K et al. Isolation and expansion of mesenchymal stem/stromal cells derived from human placenta tissue. J Vis Exp 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patel J, Seppanen E, Chong MSK et al. Prospective surface marker‐based isolation and expansion of fetal endothelial colony‐forming cells from human term placenta. STEM CELLS TRANSL MED 2013;2:839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Patel J, Shafiee A, Wang W et al. Novel isolation strategy to deliver pure fetal‐origin and maternal‐origin mesenchymal stem cell (MSC) populations from human term placenta. Placenta 2014;35:969–971. [DOI] [PubMed] [Google Scholar]
- 23. Abumaree MH, Al Jumah MA, Kalionis B et al. Phenotypic and functional characterization of mesenchymal stem cells from chorionic villi of human term placenta. Stem Cell Rev 2013;9:16–31. [DOI] [PubMed] [Google Scholar]
- 24. Chen CP, Liu SH, Huang JP et al. Engraftment potential of human placenta‐derived mesenchymal stem cells after in utero transplantation in rats. Human Reprod 2009;24:154–165. [DOI] [PubMed] [Google Scholar]
- 25. Fukuchi Y, Nakajima H, Sugiyama D et al. Human placenta‐derived cells have mesenchymal stem/progenitor cell potential. Stem Cells 2004;22:649–658. [DOI] [PubMed] [Google Scholar]
- 26. Igura K, Zhang X, Takahashi K et al. Isolation and characterization of mesenchymal progenitor cells from chorionic villi of human placenta. Cytotherapy 2004;6:543–553. [DOI] [PubMed] [Google Scholar]
- 27. Chen YS, Pelekanos RA, Ellis RL et al. Small molecule mesengenic induction of human induced pluripotent stem cells to generate mesenchymal stem/stromal cells. STEM CELLS TRANSL MED 2012;1:83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pelekanos RA, Ting MJ, Sardesai VS et al. Intracellular trafficking and endocytosis of CXCR4 in fetal mesenchymal stem/stromal cells. BMC Cell Biol 2014;15:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pelekanos RA, Sardesai VS, Dekker Nitert M et al. Rapid method for growth hormone receptor exon 3 delete (GHRd3) SNP genotyping from archival human placental samples. Endocrine 2015;49:643–652. [DOI] [PubMed] [Google Scholar]
- 30. Matigian N, Brooke G, Zaibak F et al. Multipotent human stromal cells isolated from cord blood, term placenta and adult bone marrow show distinct differences in gene expression pattern. Genom Data 2015;3:70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ryan JM, Matigian N, Pelekanos RA et al. Transcriptional ontogeny of first trimester human fetal and placental mesenchymal stem cells: Gestational age versus niche. Genom Data 2014;2:382–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Campagnoli C, Roberts IA, Kumar S et al. Identification of mesenchymal stem/progenitor cells in human first‐trimester fetal blood, liver, and bone marrow. Blood 2001;98:2396–2402. [DOI] [PubMed] [Google Scholar]
- 33. Benirschke K, Burton GJ, Baergen RN. Pathology of the Human Placenta, sixth edition. Springer, 2012. [Google Scholar]
- 34. Guillot PV, Gotherstrom C, Chan J et al. Human first‐trimester fetal MSC express pluripotency markers and grow faster and have longer telomeres than adult MSC. Stem Cells 2007;25:646–654. [DOI] [PubMed] [Google Scholar]
- 35. Kuligowski S, Csirke B, Biddle W Battista P. AmnioMAX‐II Complete: A Second‐Generation AmnioMAX Medium for Amniocyte Culture. Focus 1999;21:35–37. [Google Scholar]
- 36. Nazarov I, Lee JW, Soupene E et al. Multipotent stromal stem cells from human placenta demonstrate high therapeutic potential. Stem Cells Transl Med 2012;1:359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shafiee A, Fisk NM, Hutmacher DW et al. Fetal endothelial and mesenchymal progenitors from the human term placenta: Potency and clinical potential. Stem Cells Transl Med 2015;4:419–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brooke G, Rossetti T, Pelekanos R et al. Manufacturing of human placenta‐derived mesenchymal stem cells for clinical trials. Br J Haematol 2009;144:571–579. [DOI] [PubMed] [Google Scholar]
- 39. Prather WR, Toren A, Meiron M. Placental‐derived and expanded mesenchymal stromal cells (PLX‐I) to enhance the engraftment of hematopoietic stem cells derived from umbilical cord blood. Expert Opin Biol Ther 2008;8:1241–1250. [DOI] [PubMed] [Google Scholar]
- 40. Prather WR, Toren A, Meiron M et al. The role of placental‐derived adherent stromal cell (PLX‐PAD) in the treatment of critical limb ischemia. Cytotherapy 2009;11:427–434. [DOI] [PubMed] [Google Scholar]
- 41. Crisan M, Yap S, Casteilla L et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 2008;3:301–313. [DOI] [PubMed] [Google Scholar]
- 42. Wang L, Yang Y, Zhu Y et al. Characterization of placenta‐derived mesenchymal stem cells cultured in autologous human cord blood serum. Mol Med Rep 2012;6:760–766. [DOI] [PubMed] [Google Scholar]
- 43. Poloni A, Maurizi G, Babini L et al. Human mesenchymal stem cells from chorionic villi and amniotic fluid are not susceptible to transformation after extensive in vitro expansion. Cell Transplant 2011;20:643–654. [DOI] [PubMed] [Google Scholar]
- 44. Poloni A, Rosini V, Mondini E et al. Characterization and expansion of mesenchymal progenitor cells from first‐trimester chorionic villi of human placenta. Cytotherapy 2008;10:690–697. [DOI] [PubMed] [Google Scholar]
- 45. Gotherstrom C, Westgren M, Shaw SW et al. Pre‐ and postnatal transplantation of fetal mesenchymal stem cells in osteogenesis imperfecta: A two‐center experience. Stem Cells Transl Med 2014;3:255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bianchi G, Banfi A, Mastrogiacomo M et al. Ex vivo enrichment of mesenchymal cell progenitors by fibroblast growth factor 2. Exp Cell Res 2003;287:98–105. [DOI] [PubMed] [Google Scholar]
- 47. Choi SC, Kim SJ, Choi JH et al. Fibroblast growth factor‐2 and −4 promote the proliferation of bone marrow mesenchymal stem cells by the activation of the PI3K‐Akt and ERK1/2 signaling pathways. Stem Cells Dev 2008;17:725–736. [DOI] [PubMed] [Google Scholar]
- 48. Fan VH, Tamama K, Au A et al. Tethered epidermal growth factor provides a survival advantage to mesenchymal stem cells. Stem Cells 2007;25:1241–1251. [DOI] [PubMed] [Google Scholar]
- 49. Tamama K, Kawasaki H, Wells A. Epidermal growth factor (EGF) treatment on multipotential stromal cells (MSCs). Possible enhancement of therapeutic potential of MSC. J Biomed Biotechnol 2010;2010:795385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Potdar PD, D'Souza SB. Ascorbic acid induces in vitro proliferation of human subcutaneous adipose tissue derived mesenchymal stem cells with upregulation of embryonic stem cell pluripotency markers Oct4 and SOX 2. Hum Cell 2010;23:152–155. [DOI] [PubMed] [Google Scholar]
- 51. Sun LY, Pang CY, Li DK et al. Antioxidants cause rapid expansion of human adipose‐derived mesenchymal stem cells via CDK and CDK inhibitor regulation. J Biomed Sci 2013;20:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Krampera M, Pasini A, Rigo A et al. HB‐EGF/HER‐1 signaling in bone marrow mesenchymal stem cells: Inducing cell expansion and reversibly preventing multilineage differentiation. Blood 2005;106:59–66. [DOI] [PubMed] [Google Scholar]
- 53. Huang YL, Qiu RF, Mai WY et al. Effects of insulin‐like growth factor‐1 on the properties of mesenchymal stem cells in vitro. J Zhejiang Univ Sci B 2012;13:20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jung S, Panchalingam KM, Rosenberg L et al. Ex vivo expansion of human mesenchymal stem cells in defined serum‐free media. Stem Cells Int 2012;2012:123030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tzompa‐Sosa DA, van Valenberg HJ, van Aken GA et al. Milk fat triacylglycerols and their relations with milk fatty acid composition, DGAT1 K232A polymorphism, and milk production traits. J Dairy Sci 2016;99:3624–3631. [DOI] [PubMed] [Google Scholar]
- 56. Rafiee‐Yarandi H, Ghorbani GR, Alikhani M et al. A comparison of the effect of soybeans roasted at different temperatures versus calcium salts of fatty acids on performance and milk fatty acid composition of mid‐lactation Holstein cows. J Dairy Sci 2016;99:5422–5435. [DOI] [PubMed] [Google Scholar]
- 57. Eisenberg SW, Boerhout EM, Ravesloot L et al. Diurnal differences in milk composition and its influence on in vitro growth of Staphylococcus aureus and Escherichia coli in bovine quarter milk. J Dairy Sci 2016;99:5690–5700. [DOI] [PubMed] [Google Scholar]
- 58. Boudon A, Johan M, Narcy A et al. Dietary cation‐anion difference and day length have an effect on milk calcium content and bone accretion of dairy cows. J Dairy Sci 2016;99:1527–1538. [DOI] [PubMed] [Google Scholar]
- 59. Schwendel BH, Morel PC, Wester TJ et al. Fatty acid profile differs between organic and conventionally produced cow milk independent of season or milking time. J Dairy Sci 2015;98:1411–1425. [DOI] [PubMed] [Google Scholar]
- 60. Tzompa‐Sosa DA, van Aken GA, van Hooijdonk AC et al. Influence of C16:0 and long‐chain saturated fatty acids on normal variation of bovine milk fat triacylglycerol structure. J Dairy Sci 2014;97:4542–4551. [DOI] [PubMed] [Google Scholar]
- 61. Marino VM, Schadt I, Carpino S et al. Effect of Sicilian pasture feeding management on content of alpha‐tocopherol and beta‐carotene in cow milk. J Dairy Sci 2014;97:543–551. [DOI] [PubMed] [Google Scholar]
- 62. Yayota M, Tsukamoto M, Yamada Y et al. Milk composition and flavor under different feeding systems: A survey of dairy farms. J Dairy Sci 2013;96:5174–5183. [DOI] [PubMed] [Google Scholar]
- 63. Halmemies‐Beauchet‐Filleau A, Kairenius P, Ahvenjarvi S et al. Effect of forage conservation method on plasma lipids, mammary lipogenesis, and milk fatty acid composition in lactating cows fed diets containing a 60:40 forage‐to‐concentrate ratio. J Dairy Sci 2013;96:5267–5289. [DOI] [PubMed] [Google Scholar]
- 64. Ryan JM, Pettit AR, Guillot PV et al. Unravelling the pluripotency paradox in fetal and placental mesenchymal stem cells: Oct‐4 expression and the case of the Emperor's New Clothes. Stem Cell Rev 2013;9:408–421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1.
Supporting Information Figure 2.
Supporting Information Figure 3.
Supporting Information Figure 4.
Supporting Information.


