Abstract
Salt tolerance is an important trait that is required to overcome salinity-induced reduction in plant productivity. We have reported previously the isolation of a pea DNA helicase 45 (PDH45) that exhibits striking homology with the eukaryotic translation initiation factor eIF-4A. Here, we report that PDH45 mRNA is induced in pea seedlings in response to high salt, and its overexpression driven by a constitutive cauliflower mosaic virus-35S promoter in tobacco plants confers salinity tolerance, thus suggesting a previously undescribed pathway for manipulating stress tolerance in crop plants. The T0 transgenic plants showed high levels of PDH45 protein in normal and stress conditions, as compared with WT plants. The T0 transgenics also showed tolerance to high salinity as tested by a leaf disk senescence assay. The T1 transgenics were able to grow to maturity and set normal viable seeds under continuous salinity stress without any reduction in plant yield in terms of seed weight. Measurement of Na+ ions in different parts of the plant showed higher accumulation in the old leaves and negligible accumulation in seeds of T1 transgenic lines as compared with the WT plants. The possible mechanism of salinity tolerance is discussed. Overexpression of PDH45 provides a possible example of the exploitation of DNA/RNA unwinding pathways for engineering salinity tolerance without affecting yield in crop plants.
Keywords: DEAD box protein, eIF-4A, salinity stress
Soil salinity is an increasing threat for agriculture and is a major factor in reducing plant productivity; therefore, it is necessary to obtain salinity-tolerant varieties. A typical characteristic of soil salinity is the induction of multiple stress-inducible genes (1). Some of the genes encoding osmolytes (2), ion channels (3), or enzymes (4, 5) are able to confer salinity-tolerant phenotypes when transferred to sensitive plants. Because salinity stress affects the cellular gene-expression machinery, it is evident that molecules involved in nucleic acid processing, including helicases, are likely to be affected as well. DNA helicases unwind duplex DNA and are involved in replication, repair, recombination, and transcription, whereas RNA helicases unfold the secondary structures in RNA and are involved in transcription, ribosome biogenesis, and translation initiation (6, 7). Most helicases are members of the DEAD-box protein super-family, and the eukaryotic translation initiation factor (eIF-4A) is a prototypic member of this family (7, 8). It is an essential component of the cap-binding complex that facilitates ribosome assembly on mRNA by unwinding the secondary structures in the 5′ untranslated region (9).
The involvement of RNA helicases in response to stress has been reported previously. The cyanobacterial, CrhC (10), Clostridium perfringes RNA helicase (11), and fission yeast DEAD-box protein, Ded1 (12), respond to cold, oxidative, and cellular stress, respectively. There is also evidence for the involvement of the mammalian RNA helicase, RHII/Gu, in RNA polymerase II-catalyzed transcription during stress response (13). By subtractive hybridization and differential display, salinity-regulated genes have been identified, showing the involvement of DEAD-box RNA helicases (14). An Arabidopsis RNA helicase has been reported as a regulator of the transcriptional activator, CBF, in chilling resistance (15).
We previously have isolated and characterized a functional DNA helicase from pea, pea DNA helicase 45 (PDH45), that exhibits striking homology with eIF-4A and contains ATP-dependent DNA and RNA helicase, DNA-dependent ATPase, and ATP-binding activities (16). In this paper, we report the up-regulation of PDH45 in response to abiotic stresses. Its overexpression in tobacco plants confers increased tolerance to salinity, indicating its potential for improving stress tolerance in crop plants.
Materials and Methods
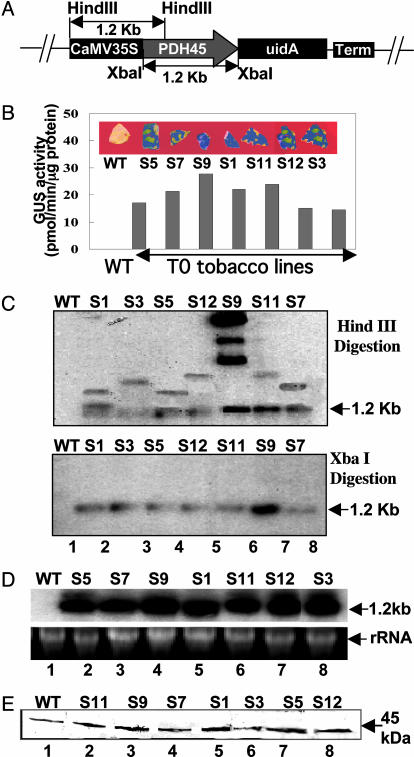
Vectors and Plant Transformation. The complete ORF of PDH45 cDNA (1.2 kb) was PCR amplified by using primers P45F1 (5′-ATGGCGACAACTTCTGTGG-3′) starting from the translation initiation site and P45R1 (5′-GAGCTCGAGTTATATAAGATCACCAATATTC-3′) designed to create an XbaIsiteatthe3′ end (underlined above) next to the translation termination codon (double underlined). The amplified fragment was cloned into pGEMTeasy vector. The EcoRI fragment from this vector was ligated into vector pBSK, and the XbaI fragment from this vector was ligated into vector pBI121 (Clontech) to create pBI-PDH45 (see Fig. 3A). This vector contains PDH45 and GUS (uidA) under a single cauliflower mosaic virus-35S promoter; however, a stop codon has been inserted between PDH45 and the reporter gene to avoid translational fusions. The pBI-PDH45 also carries the NPTII (kanamycin) gene as a selectable marker.
Fig. 3.
Analysis of transgenic plants (T0) to confirm the integration, copy number, and expression of the gene. (A) pBI-PDH45 sense construct. Shown is a schematic representation of the construct used to overexpress PDH45 cDNA in tobacco plants. (B–E) The WT and PDH45 T0 transgenic lines (S) were analyzed by the following methods. Analysis was performed by GUS assay (B) and Southern blot (C) to show the presence and copy number of the gene. (C) DNA was digested with HindIII (Upper) and XbaI(Lower). (D) Northern blot to confirm the transcript. Lower shows rRNA to confirm equal loading. (E) Western blot analysis using anti-PDH45 antibodies.
Tobacco (Nicotiana tabacum cv. Xanthi) leaf disks were transformed through a procedure (17) with Agrobacterium tumefaciens (LBA4404) containing pBI-PDH45. Putative T0 transgenic plants were regenerated from the callus in the presence of kanamycin and further screened by using PCR and β-glucuronidase (GUS) assay (18). The seeds from these plants, i.e., T0 seeds, were germinated on kanamycin-containing medium to select for all of the transgenic T1 seedlings.
GUS Assay. GUS activity was visualized in the leaf tissue and measured fluorimetrically by using 1 mM 4-methyl-umbelliferyl-β-d-glucuronide as substrate, as described by Jefferson (18).
Stress Treatments. We transferred 7-day-old pea seedlings into various concentrations of NaCl (50, 100, 200, and 300 mM), mannitol (200 mM), LiCl (100 mM), and abscisic acid (ABA; 10 μM) for different time intervals. Wounding stress was given by puncturing the leaves, whereas exposing the seedlings to 4°C provided low-temperature stress. Young leaves and roots of the stressed seedlings were harvested after 6, 12, 24, and 48 h. Seedlings grown in water for the same period were taken as a control.
To monitor stress effects on PDH45 overexpressing plants, 10-day-old WT and kanamycin-positive T1 transgenic seedlings were transferred to Murashige and Skoog medium (Sigma) supplemented with 50, 200, or 300 mM NaCl, under culture room conditions. The number of seedlings surviving under stress was scored. The 200-mM NaCl-stressed seedlings later were transferred to earthen pots and grown in a greenhouse (16 h light/8 h dark at 25 ± 2°C), and these plants were watered every 14 days with a 200 mM NaCl solution.
Leaf Disk Assay. Leaf disks of 1.0-cm diameter were excised from healthy and fully expanded tobacco leaves of similar age from transgenic and WT plants (60 days old). The disks were floated in a 6-ml solution of NaCl or water (experimental control) for 72 h (19) and then used for measuring chlorophyll a and b spectrophotometrically after extraction in 80% acetone (20). The treatment was carried out in continuous white light at 25 ± 2°C. The experiment was repeated at least three times with different transgenic lines.
Genomic DNA PCR and Southern Analysis. Genomic DNA was isolated by grinding leaf tissue followed by extraction using the CTAB (N-acetyl-N,N,N-trimethylammonium bromide) method (21). PCR was performed by using the oligonucleotides P45F1 as the forward primer and GUS-R1, 5′-TCATTGTTTGCCTCCCTGCTGC-3′, as the reverse primer. The pBI-PDH45 was used as a positive control template. For Southern analysis, 20 μg of genomic DNA from PCR-positive tobacco lines was digested with HindIII and XbaI separately, electrophoresed, and blotted on Hybond N membranes (Amersham Pharmacia). Radiolabeled 1.2-kb complete ORF of PDH45 cDNA was used as a probe. Hybridization and washing were carried out at 55°C as described in ref. 16.
Northern Analysis. Total RNA was isolated by using an extraction method described in ref. 22. About 30 μg of total RNA samples resolved on a 1.0% formaldehyde-agarose gel were transferred on Hybond N membranes (Amersham Pharmacia) and probed as described above.
Immunoblotting. Total soluble proteins from the tissue were separated by 12% SDS/PAGE, and analysis was performed by using PDH45 antibodies (1:5,000 dilution) as described in ref. 16.
Root Growth Assay. Seeds of the T0 transgenic tobacco plants were selected by germinating them on Murashige and Skoog (23) medium in the presence of kanamycin. These seedlings representing the T1 generation were tested for their sensitivity of root growth to salinity stress. Individual seedlings were selected for uniformity, and their initial root lengths were recorded. The seedlings then were transferred to the salt solution for 48 h, and their root length was monitored.
Endogenous Ion Content. Mature T1 transgenic plants grown in water or in 200 mM NaCl were used for Na+ measurements. Young and old leaves and seeds were collected from different plants of each line and thoroughly rinsed in deionized water. The fresh weight of each sample was determined. The samples were dried at 65°C for 24 h, and the dry weight of each sample was measured. The material was digested in concentrated HNO3 overnight. Samples then were picked in 2 M HCl and analyzed for Na+ content by using simultaneous inductively coupled argon-plasma emission spectrometry (ICP trace analyzer, Labtam, Braeside, Australia).
Results
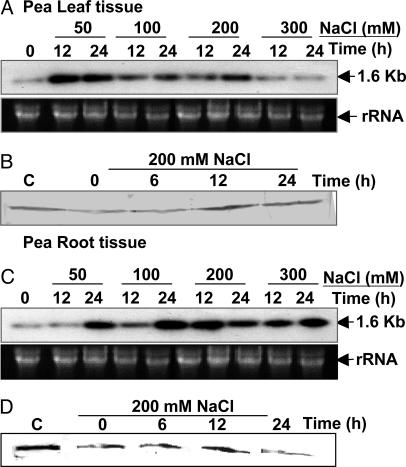
PDH45 Is Induced by Salinity Stress in Pea. To study the effect of salt stress on the expression of the PDH45 gene, 7- to 8-day-old pea seedlings were exposed to 50–300 mM NaCl, and the transcript levels were analyzed. In leaf tissue, a 2.5-fold induction of PDH45 mRNA was observed after 12-h exposure to 50 mM NaCl. The induction was less upon increasing the concentration of NaCl and decreased to the control levels at 300 mM NaCl (Fig. 1A). In the root tissue, the induction of transcript (Fig. 1C) is slow, and a 6-fold higher transcript level was seen after 24-h exposure to 100 mM and 12-h exposure to 200 mM NaCl. At 300 mM NaCl, the mRNA levels slowly decreased to a 2-fold higher level compared with the controls (Fig. 1C). The Western blot analysis of pea leaves and root tissue stressed with 200 mM NaCl also showed a time-dependent accumulation of PDH45 protein up to 12 h of stress treatment (Fig. 1 B and D). At 24 h after stress, transcript and protein levels equilibrated at a slightly lower level. To check whether the effect was specific to Na+, the seedlings were stressed with LiCl. As shown in Fig. 2A, treatment with LiCl (100 mM), in contrast to NaCl, did not result in significant induction in the transcript level.
Fig. 1.
Analysis of total RNA and PDH45 proteins in the leaves and roots of pea seedlings stressed with various concentrations of NaCl. (A and C) Northern blot of total RNA isolated from leaves (A) and roots (C) of pea seedlings stressed with various concentrations of NaCl for 12 and 24 h. The 0-h time point served as the control. (Lower) rRNA as loading control is shown. (B and D) The PDH45 protein in the leaves (B) and roots (D) of seedlings stressed with 200 mM NaCl was analyzed by immunoblotting. Lane C shows the recombinant protein as positive control.
Fig. 2.
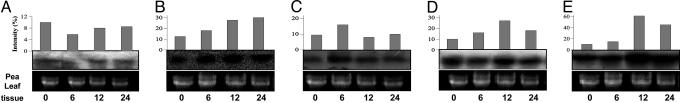
Transcript analysis of pea seedlings stressed with 100 mM LiCl (A), 200 mM mannitol (B), wounding stress (C), low temperature (4°C) (D), and 10 μM ABA (E) for 6, 12, and 24 h. The 0-h time point served as the control. Bottom shows rRNA as loading control, Middle shows the pea leaf tissue, and Top shows the quantitative data.
PDH45 Up-Regulation Under Other Abiotic Stresses in Pea. Inducibility of the PDH45 transcript was studied in dehydration, wounding, and low temperature (4°C). Up-regulation of the transcript was observed in all of the above stresses (Fig. 2 B–D). A 12-h treatment with 200 mM mannitol at 4°C showed 2- to 2.5-fold enhancement in the level of mRNA, with no increase at the 6-h time point (Fig. 2 B and D). The response to low-temperature stress was reconfirmed by RT-PCR (data not shown). Wounding stress for 6 h resulted in 1.5-fold induction in the transcript, and no further change was observed after longer exposure to stress (Fig. 2C). The stress effect may be mediated by ABA, because a 3-fold increase in the mRNA level was observed after 12 h of 10 μM ABA treatment (Fig. 2E). Thus, the PDH45 transcript is up-regulated by mannitol, low temperature, and ABA, although it exhibits maximum induction during salinity stress.
Overexpression of PDH45 in Tobacco Plants. To establish the functional significance of the PDH45 gene, the complete ORF (Fig. 3A) was overexpressed in tobacco plants by using Agrobacterium-mediated transformation. The preliminary screening of kanamycin-resistant putative tobacco transformants was performed using a GUS assay (Fig. 3B). The presence of PDH45 was confirmed in the GUS positive plants by PCR using P45F1 and GUS-R1 as primers (data not shown). We selected 30 independently transformed T0 transgenic plants and grew them to maturity; however, only 7 different plants (S1, S3, S5, S7, S9, S11, and S12) were randomly selected for subsequent analysis. The morphological and growth characteristics of T0 plants were similar to the WT plants.
Analysis of the T0 Transgenic Plants. To check the stable integration and copy number of the transgene, an equal amount of HindIII-digested genomic DNA was probed with PDH45 cDNA. The expected band pattern per insertion was one band at the 1.2-kb position (containing the entire cauliflower mosaic virus promoter and part of PDH45 cDNA) and a second band of variable size. The intensity of the hybridized 1.2-kb band and the size of the second band varied among different transgenic lines, indicating random integration events. The observed copy number was one (Fig. 3C Upper, lanes 2–5, 7, and 8) and three (Fig. 3C Upper, lane 6) copies. The integration of the gene was further confirmed by hybridization of the XbaI-digested DNA with the same probe, and a single band of 1.2 kb, corresponding to the PDH45 cDNA, was observed (Fig. 3C Lower). No band was detected in the WT plants (Fig. 3C, lane 1).
Northern blots of T0 lines (Fig. 3D) showed the presence of a single 1.2-kb transcript in all of the transgenic plants (lanes 2–9), but the corresponding band was missing in the WT plants (lane 1), possibly due to the use of a heterologous probe. The presence of a single transcript indicated that transcription initiation and termination of PDH45 mRNA occurred as expected. Immunoblot of the T0 transgenics by using PDH45 antibodies identified a 45-kDa band corresponding to the pea protein, and its intensity varied between the different transgenic lines (Fig. 3E). However, in contrast to the Southern and Northern results, a protein band of the same size was recognized in the WT plants. This characteristic could be due to the cross-reactivity of the polyclonal antibodies with native tobacco protein.
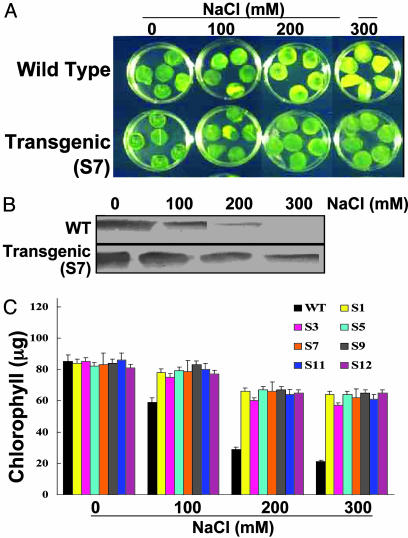
Tolerance of T0-Transgenic Plants to Excess Salinity. To test for salinity tolerance, leaf disks from T0 transgenic and WT tobacco plants were floated separately on 100, 200, and 300 mM NaCl and H2O for 72 h. Salinity-induced loss of chlorophyll was lower in PDH45-overexpressing lines compared with those from the WT plants (Fig. 4A). The damage caused by stress was reflected in the degree of bleaching observed in the leaf tissue after 72 h. It was evident that transgenic plants have a better ability to tolerate salinity stress. Moreover, in WT plants the endogenous protein decreased in response to stress, whereas in T0 transgenics the protein was evident in treatments up to 300 mM NaCl in all seven lines (Fig. 4B). This observation could be due to high expression of the gene from the cauliflower mosaic virus promoter. Fig. 4 A and B represent pictures of line S7, although experiments were performed on all of the lines. Measurement of the chlorophyll content of the leaf disks from different transgenic lines stressed with 200 mM NaCl (Fig. 4C and Table 1) provided further support for a positive relationship between the overexpression of PDH45 and tolerance of salinity stress.
Fig. 4.
Leaf disk senescence assay for salinity tolerance in transgenic tobacco plants (T0). (A and B) Representative picture to show phenotypic differences in leaf disks (A) and level of PDH45 protein in them after immunostaining with anti-PDH45 antibodies (B). Data for line S7 alone are provided here. The experiments were performed on all seven lines with similar results. (C) Chlorophyll content from leaf disks of WT and different T0 plants after incubation in 100, 200, and 300 mM NaCl solution. Disks floated in water served as the experimental control.
Table 1. Comparison of various growth parameters of the WT and PDH45 T0 and T1 transgenic plants grown in the presence of water and 200 mM NaCl, respectively.
| WT water*
|
Transgenics in 200 mM NaCl
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameter | Generation | S1 | S3 | S5 | S7 | S9 | S11 | S12 | |
| Transgene copy number | T0 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | |
| Total protein,† mg/g FW | T0 | 0.95 | 0.86 | 0.87 | 0.93 | 0.83 | 0.91 | 0.78 | 0.81 |
| Chlorophyll,† mg/g FW | T0 | 8.5 | 5.5 | 6.1 | 6.3 | 4.9 | 5.7 | 5.1 | 5.9 |
| Segregation ratio,‡Kr:Ks [n] | T1 | 3.5:1 [150] | 2.03:1 [225] | 2.77:1 [171] | 2.70:1 [163] | 4.2:1 [149] | 2.9:1 [183] | 3.16:1 [215] | |
| Root growth inhibition,§ % | T1 | 0 | 11.0 | 21.0 | 17.0 | 17.0 | 19.0 | 16.0 | 16.0 |
| Fresh weight,† g per leaf | T1 | 3.9 | 3.9 | 3.8 | 3.9 | 3.9 | 3.9 | 3.8 | 3.8 |
| Height,¶ cm | T1 | 113.0 | 106 | 109 | 104 | 113 | 101 | 109 | 107 |
| Flowering time,¶ days | T1 | 122.0 | 133 | 131 | 133 | 131 | 129 | 131 | 130 |
| Seed weight per pod‡ | T1 | 119.0 | 118 | 119 | 118 | 120.7 | 120.4 | 122 | 123.1 |
Experiments were repeated at least three times, and the values represent means of different samples. SD in each case was <3%. In all cases, n = 15 except for T0 seeds. n is mentioned for each line in brackets.
WT plants did not survive under salinity stress
Recordings made from mature leaf. FW, fresh weight
Recordings made from seed
Recordings made from seedling
Recordings made from mature plant
Analysis of the T1 Transgenic Progeny. To determine whether PDH45-imparted salinity tolerance is functionally and genetically stable, the T1 progeny were characterized. Seeds from the T0 plants, when plated onto kanamycin-containing medium, segregated in the expected 3:1 ratio of Kanr/Kans (Table 1). The seedlings from each line were further confirmed for the presence of the transgene by PCR (data not shown) and GUS assay.
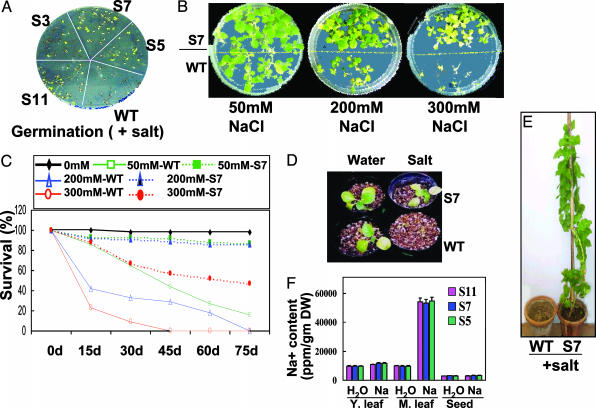
T1-Transgenic Plants Grow Normally and Set Viable Seeds Under Salinity Stress. To assess the effect of high salt on PDH45-overexpressing plants, the kanamycin-positive T1 seedlings were characterized. T1 and WT seedlings were morphologically similar when grown without NaCl. In the presence of NaCl, the WT seedlings showed growth retardation, whereas transgenic plants did not develop any sign of stress (Fig. 5A); however, the growth rate of the T1 seedlings was affected under stress in a dose-dependent manner (Fig. 5B). In the presence of 50 mM NaCl, both WT and transgenic seedlings were able to survive, but under 200 mM NaCl stress, WT seedlings showed chlorosis and stunted phenotypes and ultimately died, whereas the T1 transgenics continued to grow. A distinct difference could be observed on exposing the seedlings to 300 mM NaCl, when only the transgenic plants showed slow growth. Statistically similar results were obtained for the seven transgenic lines (Table 1 and Fig. 5 B–E, representative picture of S7 line). To follow the degree of NaCl tolerance, the percentage survival of WT and kanamycin-resistant T1 seedlings under different concentrations of NaCl was scored at 15-day intervals after transfer under control growth conditions. The increased NaCl tolerance manifested by the transgenics with respect to the WT plants was apparent at both 200 and 300 mM NaCl concentrations (Fig. 5C). For instance, at 60 days 80% of the transgenic seedlings were tolerant to 200 mM salt compared with 20% survival of the WT seedlings (Fig. 5C). Similar results were obtained for all of the transgenic lines (data not shown), indicating that the lowest expression level was sufficient to confer salinity tolerance.
Fig. 5.
Representative picture to show relative salinity tolerance of WT and kanamycin-positive T1-PDH45 tobacco transgenic plants. (A) Seedling growth of WT and T1 transgenic lines (S3, S5, S7, and S11) on medium supplemented with NaCl. (B) Comparative picture of WT and S7 seedlings growing on medium supplemented with 50, 200, and 300 mM NaCl for 25 days. (C) Percentage survival of WT and S7 seedlings grown in soil supplied with 50, 200, and 300 mM NaCl solution. (D) Phenotype of WT and S7 transgenic seedlings growing in soil pots supplied with water or 200 mM NaCl solution every 14 days. (E) WT and S7 transgenic plants in soil pots supplied with 200 mM NaCl solution. Note that the WT plant could not sustain growth under salinity stress. The experiments in B–E were performed on all seven lines with similar results. (F) Total Na+ concentration of young (Y.) and mature (M.) leaves and T1 seeds of S5, S7, and S11 transgenic lines growing in H2O and 200 mM NaCl (Na). The values represent the average of four independent samples.
Several critical growth parameters, such as plant height, fresh weight of leaves, time required for flowering, and seed weight, were scored as an indicator of salinity stress to test tolerance in T1 transgenic lines (Table 1) as compared with the WT plants grown in H2O. It should be noted here that similar data for WT plants grown under salinity could not be obtained because these plants failed to sustain growth in the presence of NaCl. No major difference in the overall performance or total seed yield of the transgenic plants grown in the presence of 200 mM NaCl was found. Although the transgenic seedlings exposed to continuous stress of 200 mM NaCl in soil showed a slightly slower growth rate initially (Fig. 5D) compared with similar plants growing without stress, they continued to grow, reached maturity, flowered, and set seeds. Conversely, growth of WT seedlings was severely affected by continuous exposure to salinity stress, and they could not survive under these conditions (Fig. 5E, representative plants shown). The flowering time in salinity-stressed plants was delayed by a week when compared with plants growing without stress, but there was no change in the flower morphology and yield (Table 1). Significantly, the pod and seed size, seed number, and seed weight were statistically similar to those from WT plants grown without stress.
To examine the basic mechanism resulting in salt-tolerant phenotypes of PDH45-overexpressing lines, we measured the endogenous Na+ levels in leaves and seeds of T1 plants growing in the presence and absence of NaCl. The Na+ concentration was lower in young leaves compared with mature leaves when grown in 200 mM NaCl. There was approximately five times more Na+ accumulation in old leaves of transgenic plants growing in the presence of 200 mM NaCl as compared with plants in water, but no significant difference was observed between the young leaves and T1 seeds (Fig. 5F).
Discussion
The response of plants to salt stress consists of a coordinated function of several biochemical pathways to regulate cellular ion homeostasis, enabling the plant to show tolerance and yield stability in saline environments. However, these are complex multigenic traits that are difficult to establish in crops. Several genes such as barley HVA1 (24), rice CDPK (25), alfalfa Alfin1 (26), tobacco NPK1 (27), and Brassica GlyI (4) have been ectopically expressed in transgenic plants to enhance their stress tolerance. These reports suggest that transfer of a single gene also can improve stress tolerance in plants (3, 25, 28). Implementation of biotechnology strategies to improve salinity tolerance requires that research efforts be focused to identify salt tolerance effectors and the regulatory components that control these during the stress episode (1). Our research provides insight into the plant tolerance to salinity by identifying a DEAD-box helicase, PDH45, as a genetic determinant of salt tolerance and yield stability.
In this study, we observed >3-fold induction of pea PDH45 mRNA in response to NaCl. The transcript showed abundant but delayed induction in roots compared with the leaves. It is possible that rapid xylem transport causes faster accumulation of Na+ in the leaves, whereas lateral transport within the root tissue is slow. This phenomenon is reflected in the PDH45 induction as well because in leaves the transcript levels show an early induction pattern with maximum induction observed at 12 h of 50 mM NaCl, and the levels decrease to the control levels upon further increasing the salt stress to 12 h of 300 mM NaCl. In contrast, in the root tissue, the induction of transcript is slow and is at maximum at 24 h of 100 mM and 12 h of 200 mM NaCl, beyond which it slowly decreases. This response was specific to Na+ stress, because treatment with Li+ did not induce the transcript. Li+ isaNa+ analogue with higher toxicity and can be used at concentrations lower than that of Na+ to reduce the osmotic effect and follow the ionic effect (29). The PDH45 protein also increased at 12 h of NaCl treatment in both leaves and roots, compared with the water control. However, a correlation could not be drawn between the protein and transcript levels at the given time point, probably because the rates of turnover of the PDH45 mRNA and protein are different. The PDH45 transcript also was up-regulated by other abiotic stresses, which suggested that the increase could be due to water stress resulting from salinity- and mannitol-induced desiccation (30). The induction of transcript was stimulated by the phytohormone, ABA, which is known to modulate gene activation and repression under multiple environmental stress conditions such as drought, cold, and salinity (31). Earlier studies on animal and microbial systems have suggested a possible up-regulation of DEAD-box helicases in response to environmental stresses (10–13). In plants the putative RNA helicase, HVD1, is induced under salinity stress in barley shoots (32).
Tobacco plants overexpressing PDH45 show increased tolerance to salinity stress as indicated by the presence of higher chlorophyll content and retention of protein in the leaf disks of salt-stressed T0 transgenics, whereas WT leaves become yellow and have less protein under stress conditions. These data suggest that tolerance in transgenics is conferred not only by overexpression under cauliflower mosaic virus promoter but also by regulating the protein turnover under salt stress. It was observed that a single copy transgenic line with low PDH45 levels (S7) also exhibits salt tolerance, clearly indicating that position of integration or copy number does not influence the expression pattern under stress. This observation also indicates that there must be a minimum threshold of overexpressed protein above the untransformed levels that is sufficient to confer tolerance. Moreover, the T1 seedlings were able to grow, flower, and set viable seeds in the presence of high salt. This result indicates that the introduced trait is functional in transgenic plants and stable. Significantly, the salt-stressed transgenics showed yield stability as measured by no loss in seed number and seed weight. Ionic measurements made in different parts of the salt-stressed T1 plants revealed that the old leaves accumulated greater amounts of Na+, whereas there was no disturbance in the ion balance of young leaves and seeds. This finding suggests that the old leaves seem to function as ion sinks, thus keeping the young leaves and seeds essentially free from the additional uptake of Na+ ions. It has been reported that Na+ is always higher in plant leaves (xylem fed) than in their fruits or seeds (phloem fed), thereby indicating that plants maintain lower Na+ concentrations in the seeds by controlling the transport of Na+ (2, 5).
The exact mechanism of PDH45-mediated tolerance of salinity stress is not understood. However, based on the properties of PDH45 that were studied previously (16) and those known for DEAD-box proteins (6, 8), we suggest that there may be two sites of action of this dual helicase: (i) it may act at the translation level to enhance or stabilize protein synthesis, or (ii) it may associate with DNA multisubunit protein complexes to alter gene expression. Regarding support for the first hypothesis, we have demonstrated previously that antibodies against PDH45 inhibited in vitro protein synthesis, which suggested its role in translation (16). It is evident that mRNA and protein synthesis are very sensitive to stress, so factors involved in transcription and translation are potential targets of salt toxicity in plants. Lubin (33) showed that in bacteria the toxic effect of Na+ is mainly in translation rather than in RNA synthesis. Recently, it was shown that in yeast, factors involved in translation initiation, eIF-4A (34) and eIF-1A (35), improve tolerance to lithium and salinity stress, respectively. The mechanisms of translation initiation are conserved among eukaryotes, and the regulation of translation occurs at the step of initiation (9). We envisage the PDH45 function, considering that it is homologous to eIF-4A and contains RNA helicase activity (16). The RNA helicase activity of PDH45 could facilitate transcription by altering the structure of nascent RNA, a process that can stimulate reinitiation and/or elongation. The second hypothesis is supported by the demonstration of the interaction of PDH45 with topoisomerase I (16). It was reported that RNA helicase A facilitates the relaxation of double-stranded nucleic acids by interacting physically and functionally with topoisomerase IIα (36). This interaction is proposed to play an important role in regulating chromatin structure during RNA polymerase II-mediated transcription. PDH45 may have an important role at the level of transcriptional regulation.
In this paper, we provide direct evidence for the involvement of a DEAD-box helicase (PDH45) in conferring salinity tolerance in transgenic tobacco plants, thus suggesting a previously undescribed pathway for manipulating stress tolerance in crop plants.
Acknowledgments
We thank Drs. R. Haselkorn (University of Chicago, Chicago), R. A. Bressan (Purdue University, West Lafayette, IN), R. Wu (Cornell University, Ithaca, NY), and R. Tuteja (International Centre for Genetic Engineering and Biotechnology) for their insightful comments. We also thank Drs. V. Rajamani and J. K. Tripathi (Jawaharlal Nehru University, New Delhi) for help with ionic measurements. This work was partially supported by grants from the Department of Science and Technology and Department of Biotechnology (India).
Author contributions: N.S.-M. and N.T. designed research; N.S.-M. and X.H.P. performed research; N.S.-M., S.K.S., and N.T. analyzed data; and N.S.-M. and N.T. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ABA, abscisic acid; eIF-4A, eukaryotic translation initiation factor; GUS, β-glucuronidase; PDH45, pea DNA helicase 45.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. Y17186).
References
- 1.Hasegawa, P. M., Bressan, R. A., Zhu, J. K. & Bonhert, H. J. (2000) Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463-499. [DOI] [PubMed] [Google Scholar]
- 2.Garg, A. K., Kim J. K., Owens, T. G., Ranwala, A. P., Choi, Y. D., Kochian, L. V. & Wu, R. J. (2002) Proc. Natl. Acad. Sci. USA 99, 15898-15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang, H. K., Hodson, J. N., Williams, J. P. & Blumwald, E. (2001) Proc. Natl. Acad. Sci. USA 98, 12832-12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veena, Reddy, V. S. & Sopory, S. K. (1999) Plant J. 17, 385-395. [DOI] [PubMed] [Google Scholar]
- 5.Singla-Pareek, S. L., Reddy, M. K. & Sopory, S. K. (2003) Proc. Natl. Acad. Sci. USA 100, 14672-14677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuteja, N. & Tuteja, R. (1996) Nat. Genet. 13, 11-12. [DOI] [PubMed] [Google Scholar]
- 7.Luking, A., Stahl, U. & Schmidt, U. (1998) Crit. Rev. Biochem. Mol. Biol. 33, 259-296. [DOI] [PubMed] [Google Scholar]
- 8.Linder, P., Lasko, P. F., Ashburner, M., Leroy, P., Nielsen, P. J., Nishi, K., Schnier, J. & Slonimski, P. P. (1989) Nature 337, 121-122. [DOI] [PubMed] [Google Scholar]
- 9.Gingras, A. C., Raught, B. & Sonenberg, N. (1999) Annu. Rev. Biochem. 68, 913-963. [DOI] [PubMed] [Google Scholar]
- 10.Yu, E. & Owttrim, G. W. (2000) Nucleic Acids Res. 28, 3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briolat, V. & Reysset, G. (2002) J. Bacteriol. 184, 2333-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, H. Y. Nefsky, B. S. & Walworth, N. C. (2002) J. Biol. Chem. 277, 2637-2643. [DOI] [PubMed] [Google Scholar]
- 13.Westermarck, J., Weiss, C., Saffrich, R., Kast, J., Musti, A. M., Wessely, M., Ansorge, W., Seraphin, B., Wilm, M., Valdez, B. C. & Bohmann, D. (2002) EMBO J. 21, 451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinnemeier, J. & Hagemann, M. (1999) Arch. Microbiol. 172, 377-386. [DOI] [PubMed] [Google Scholar]
- 15.Gong, Z., Lee, H., Xiong, L., Jagendorf, A., Stevenson, B. & Zhu, J. K. (2002) Proc. Natl. Acad. Sci USA 99, 11507-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pham, X. H., Reddy, M. K., Ehtesham, N. Z., Matta, B. & Tuteja, N. (2000) Plant J. 24, 219-229. [DOI] [PubMed] [Google Scholar]
- 17.Horsch, R. B., Fry, J. E., Hoffmann, N. L., Eichholtz, D., Rogers, S. G. & Farley, R. T. (1985) Science 227, 1229-1231. [Google Scholar]
- 18.Jefferson, R. A. (1987) Plant Mol. Biol. Rep., 5, 387-405. [Google Scholar]
- 19.Fan, L., Zheng, S. & Wang, X. (1997) Plant Cell 9, 2183-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lichtenthaler, H. K. (1987) Methods Enzymol. 148, 350-366. [Google Scholar]
- 21.Murray, M. G. & Thompson, W. F. (1980) Nucleic Acids Res. 8, 4321-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chomczynski, P. & Sacchi, N. (1987) Anal. Biochem. 162, 156-159. [DOI] [PubMed] [Google Scholar]
- 23.Murashige, T. & Skoog, F. (1962) Physiol. Plant. 15, 473-479. [Google Scholar]
- 24.Xu, D., Duan, X., Wang, B., Hong, B., Ho, T. & Wu, R. (1996) Plant Physiol. 110, 249-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saijo, Y., Hata, S., Kyozuka, J., Shimamoto, K. & Izui, K. (2000) Plant J. 23, 319-327. [DOI] [PubMed] [Google Scholar]
- 26.Winicov, I. (2000) Planta 210, 416-422. [DOI] [PubMed] [Google Scholar]
- 27.Kovtun, Y., Chiu, W. L., Tena, G. & Sheen, J. (2000) Proc. Natl. Acad. Sci. USA 97, 2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K. & Shinozaki, K. (1999) Nat. Biotechnol. 17, 287-291. [DOI] [PubMed] [Google Scholar]
- 29.Serrano, R. (1996) Int. Rev. Cytol. 165, 1-52. [DOI] [PubMed] [Google Scholar]
- 30.Bray, E. A. (1997) Trends Plant Sci. 2, 48-54. [Google Scholar]
- 31.Jonak, C., Kiegerl, S., Ligterink, W., Barker, P. J., Huskisson, N. S. & Hirt, H. (1996) Proc. Natl. Acad. Sci. USA 93, 11274-11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura, T., Muramoto, Y. & Takabe, T. (2004) Plant Sci. 167, 63-70. [Google Scholar]
- 33.Lubin, M. (1963) in The Cellular Functions of Membrane Transport, ed. Hoffman, J. F. (Prentice–Hall, Englewood Cliffs, NJ) pp. 193-211.
- 34.Montero-Lomeli, M., Morais, B. L., Figueiredo, D. L., Neto, D. C., Martins, J. R. & Masuda, C. A. (2002) J. Biol. Chem. 277, 21542-21548. [DOI] [PubMed] [Google Scholar]
- 35.Rausell, A., Kanhonou, R., Yenush, L., Serrano, R. & Ros, R. (2003) Plant J. 34, 257-267. [DOI] [PubMed] [Google Scholar]
- 36.Zhou, K., Choe, K. T., Zaidi, Z., Wang, Q., Mathews, M. B. & Lee, C. G. (2003) Nucleic Acids Res. 31, 2253-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]