Abstract
Background
Approximately 15% of primary colorectal cancers have DNA mismatch repair deficiency, causing a complex genome with thousands of small mutations—the microsatellite instability (MSI) phenotype. We investigated molecular heterogeneity and tumor immunogenicity in relation to clinical endpoints within this distinct subtype of colorectal cancers.
Methods
A total of 333 primary MSI+ colorectal tumors from multiple cohorts were analyzed by multilevel genomics and computational modeling—including mutation profiling, clonality modeling, and neoantigen prediction in a subset of the tumors, as well as gene expression profiling for consensus molecular subtypes (CMS) and immune cell infiltration.
Results
Novel, frequent frameshift mutations in four cancer-critical genes were identified by deep exome sequencing, including in CRTC1, BCL9, JAK1, and PTCH1. JAK1 loss-of-function mutations were validated with an overall frequency of 20% in Norwegian and British patients, and mutated tumors had up-regulation of transcriptional signatures associated with resistance to anti-PD-1 treatment. Clonality analyses revealed a high level of intra-tumor heterogeneity; however, this was not associated with disease progression. Among the MSI+ tumors, the total mutation load correlated with the number of predicted neoantigens (P = 4 × 10−5), but not with immune cell infiltration—this was dependent on the CMS class; MSI+ tumors in CMS1 were highly immunogenic compared to MSI+ tumors in CMS2-4. Both JAK1 mutations and CMS1 were favorable prognostic factors (hazard ratios 0.2 [0.05–0.9] and 0.4 [0.2–0.9], respectively, P = 0.03 and 0.02).
Conclusions
Multilevel genomic analyses of MSI+ colorectal cancer revealed molecular heterogeneity with clinical relevance, including tumor immunogenicity and a favorable patient outcome associated with JAK1 mutations and the transcriptomic subgroup CMS1, emphasizing the potential for prognostic stratification of this clinically important subtype.
See related research highlight by Samstein and Chan 10.1186/s13073-017-0438-9
Electronic supplementary material
The online version of this article (doi:10.1186/s13073-017-0434-0) contains supplementary material, which is available to authorized users.
Keywords: Colorectal cancer, Consensus molecular subtypes, Immunogenicity, Immunotherapy resistance, JAK1, Microsatellite instability, Mutation, Neoantigen, Prognosis
Background
Colorectal cancers (CRCs) with the microsatellite instability (MSI) phenotype are defined by high rates of insertions and deletions (indels) of nucleotides in short, repetitive sequences [1–3]. This is caused by a defective DNA mismatch repair (MMR) machinery, either by epigenetic silencing, primarily of MLH1 [4–6], or through somatic or germ line mutations [7–11]. MSI occurs in a wide range of cancer types but is prevalent in only a few [12–15]. In CRC, the MSI+ subgroup accounts for approximately 15% of cases and is characterized by a low level of DNA copy number aberrations but a higher frequency of small mutations (single nucleotide variants (SNVs), and indels) than most other types of cancer [16, 17]. The mutation profiles of MSI+ CRCs differ from those of microsatellite stable (MSS) CRCs, for example, by a strong enrichment for BRAF mutations [18]. Furthermore, low complexity sequences with short tandem repeats are prone to indel mutations and are found in the coding regions of several cancer-critical genes [19–21], including TGFBR2, which was the first MSI target gene to be discovered [22–24]. However, large-scale analysis of indel mutations has been challenging [25, 26], and a recent paper describing frameshift mutations in RNF43 in 80% of MSI+ CRCs clearly demonstrates that even highly prevalent indel mutations have gone unnoticed [27].
CRC has a high world-wide incidence and mortality rate [28], but compared with the MSS subgroup, patients with MSI+ tumors have a favorable prognosis in the primary setting [3, 29, 30]. This is possibly explained by tumor immunogenicity and the high level of lymphocyte infiltration [31, 32]. Immunogenicity in MSI+ tumors may be a result of the high mutation load and subsequent expression of mutated, tumor-specific peptides as neoantigens [33]. Neoantigens are presented on the tumor cell surface by class I HLA molecules and have the potential to activate cytotoxic T cells. Expression of neoantigens, computationally predicted based on mutation profiling, has been shown to be associated with a favorable patient outcome across tumor types [34]. In CRC, there are strong indications that the number of predicted neoantigens is also prognostic [35–37], but the subgroup of MSI+ tumors has not been specifically analyzed in this context. In the metastatic setting, MSI+ tumors are associated with a poor prognosis [38], but patients generally respond well to immunotherapy by anti-PD-1 immune checkpoint inhibition [39]. However, disease control is not obtained in all cases, and prediction of treatment response is currently a research area of high interest [40, 41].
At the gene expression level, an international consortium has recently described four consensus molecular subtypes (CMS) of CRC [42]. The majority of MSI+ tumors are found in CMS1, which is a subtype characterized by a high mutation load and infiltration of immune cells in the tumor microenvironment [42, 43]. CMS classification has clinical value independent of cancer stage, and patients with mesenchymal CMS4 tumors have a poor outcome, while CMS1 is associated with a poor patient survival after relapse. The heterogeneity and clinical value of CMS classification specifically within the subgroup of MSI+ tumors remains unknown.
In the present study, we have performed deep exome sequencing of MSI+ CRCs, integrated with DNA copy number and gene expression analyses, to (i) identify potential novel mutations and (ii) analyze the mutation-associated immunogenicity of the tumors, in relation to both CMS and clinical endpoints.
Methods
Patient samples
A total of 333 primary MSI+ CRCs from five patient series were analyzed for mutations, DNA copy number, and/or gene expression (Fig. 1a). This includes tumors from two Norwegian series (n = 78 and 31) and British patients recruited to the VICTOR adjuvant randomized trial (n = 83, 81 with clinical annotation available) [44], as well as publicly available data from The Cancer Genome Atlas (TCGA; n = 66) [16] and a French multicenter cohort (n = 75; Gene Expression Omnibus (GEO), accession number GSE39582) [45].
Fig. 1.
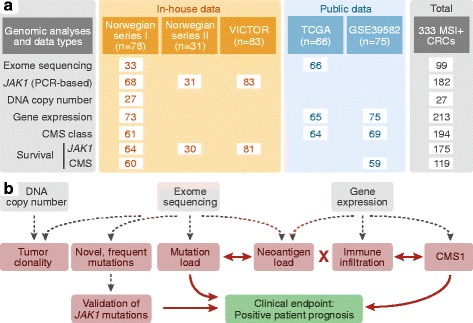
Patient samples and data analyses. a A total of 333 MSI+ CRCs were analyzed, including 192 in-house tumor samples and publicly available data from 141 patients; 248 were analyzed for gene mutations and 213 for gene expression. Among the 68 samples in Norwegian series I analyzed for JAK1 mutations by PCR-based analysis, 33 were exome sequenced, 27 were analyzed for DNA copy number aberrations, and 63 were analyzed for gene expression. b Molecular result parameters are shown in red, clinical endpoints in green, and the data types used for analysis in grey. Significant associations are indicated by red arrows and the red cross indicates no association
Fresh-frozen tumor specimens and matched normal colonic mucosa have consecutively been collected from patients treated for primary CRC at Oslo University Hospital, Norway since 2005. This patient series is referred to as Norwegian series I and all 78 patients treated for MSI+ CRC before May 2013 were included. For validation of JAK1 mutation prevalence, another 31 MSI+ tumors from an anonymized Norwegian CRC patient series treated surgically at hospitals in the Oslo region from 1987 to 1989 were analyzed (Norwegian series II; Table 1; Fig. 1a).
Table 1.
Clinicopathological data for MSI+ CRCs analyzed in-lab
| Clinicopathological parameters | Norwegian series I (n = 78) | Norwegian series II (n = 31) | VICTOR (n = 81) | |
|---|---|---|---|---|
| All patients | Exome sequenced (n = 33) | |||
| Patient age at diagnosis (years) | ||||
| Mean | 75 | 74 | 66 | 65 |
| Min | 37 | 37 | 26 | 38 |
| Max | 93 | 93 | 92 | 89 |
| Patient gender | ||||
| Female | 55 | 23 | 18 | 45 |
| Male | 23 | 10 | 13 | 36 |
| Tumor localization | ||||
| Right | 68 | 31 | 20 | 66 |
| Left | 6 | 1 | 7 | 13 |
| Rectum | 4 | 1 | 4 | - |
| Unknown | - | - | - | 2 |
| Tumor stage | ||||
| I | 16 | 1 | 3 | - |
| II | 42 | 23 | 17 | 54 |
| III | 15 | 9 | 8 | 27 |
| IV | 5 | - | 3 | - |
| 5-year survival rates | ||||
| Overall | 71% | 85% | 83% | 82% |
| Relapse-free | 70% | 82% | 72% | NA |
NA not available
DNA was extracted using a standard phenol/chloroform protocol or a magnetic bead approach according to the manufacturer’s recommendations (Maxwell 16 DNA Purification Kit; Promega, Madison, WI, USA). Microsatellite instability status of the tumors was determined using the consensus markers provided by the National Cancer Institute (Bethesda marker panel), as previously described [46]. The DNA promoter methylation status of MLH1 was analyzed by quantitative methylation-specific PCR, as previously described [47]. Primers and probe (Additional file 1: Table S1) were designed using the Methyl Primer Express 1.0 and the Primer Express 3.0 software (Thermo Fisher Scientific Inc., Waltham, MA, USA). RNA was extracted using the Qiagen Allprep DNA/RNA/miRNA Universal kit, according to the manufacturer’s instructions (Qiagen, GmBH, Hilden, Germany).
Exome sequencing and mutation analysis
Matched tumor and normal colonic mucosa samples from 33 patients in Norwegian series I were exome sequenced (Table 1; Fig. 1a). Exome libraries were generated from 1 μg of genomic DNA using the Agilent SureSelectXT Human All Exon V5 kit (Agilent, Santa Clara, CA, USA), and 2 × 100-base-pair paired-end sequencing was performed using the Illumina HiSeq 2500 system and sequencing by synthesis chemistry (Illumina, San Diego, CA, USA) at the Oslo University Hospital Genomics Core Facility (The Norwegian Radium Hospital, Oslo, Norway). We received the sequencing reads and performed sequence alignment using BWA version 0.7.8 [48]. Sequence Alignment Map (SAM) files were converted to Binary Alignment Map (BAM) files using Picard version 1.102 (http://broadinstitute.github.io/picard/). SAMtools version 0.1.19 (http://samtools.sourceforge.net/) was used for sorting and indexing. The Picard toolbox was applied to remove duplicates. Subsequent refinements of the BAM files were done according to The Genome Analysis Toolkit Best Practices guidelines (https://www.broadinstitute.org/gatk/guide/best-practices). Potential indel regions were realigned using IndelRealigner and the quality of the alignment was recalibrated using BaseRecalibrator and AnalyzeCovariates. All SNVs were called using MuTect v.1.1.7 [49] and indels were scored using the program Strelka v.1.0.14 [50]. The sequence variants were annotated using ANNOVAR [51] and the Ensembl Variant Effect Predictor v.79. The coverage threshold for each candidate somatic mutation was set to minimum 10× for normal tissue and 15× for tumor samples, and loci where <5% of the reads supported a mutation were filtered out. A single mutated read from the normal tissue was accepted to allow for minute sample contamination or the presence of circulating tumor cells. All analyses were done using human genome build hg19.
Significance of candidate mutations was evaluated using MutSigCV v.1.4 [52], and genes with a false discovery rate (q value) below 0.05 were considered significantly mutated above the background mutation rate. For comparison, the Cancer Gene Census (CGC) v.73 was downloaded from the COSMIC database (http://cancer.sanger.ac.uk/census), and the 571 listed genes are referred to as cancer-critical genes.
Candidate genes with novel frequent indels were additionally screened for hotspot indels in homopolymeric regions using the SAMtools (v.0.1.19) mpileup command. For in silico validation, aligned exome sequencing data from 78 MSI+ CRCs in TCGA were downloaded from the Cancer Genomics Hub. Using a threshold of a minimum of seven reads at each homopolymer with a mutation hotspot, 66 tumors were retained for mutation analyses using the SAMtools mpileup command. The mutation load (total number of mutations) was determined for 60 of the tumors using the same analysis pipeline as for the in-house exome sequencing data.
Estimation of MSI level
The level of MSI per sample was estimated from the exome sequencing data using the algorithm MSIsensor [53] with default settings. For a catalogue of microsatellites in the reference genome (homopolymers of a minimum of 5 base pairs and microsatellites with maximum repeat unit length 5), the number of loci with sufficient sequencing coverage to be scored per sample (minimum 20 reads) was detected by mapping sequencing reads from BAM files. Unstable microsatellites were detected by counting mapped reads with non-reference repeat lengths, and somatic events were detected as sites with significantly different mutant read count distributions between paired tumor–normal samples. An MSI score was calculated per tumor, representing the percentage of unstable microsatellites (microsatellites with indels). The MSI score was not associated with the number of microsatellites scored per sample (Spearman correlation −0.03, P = 0.8).
Mutation signatures
The mutation signatures designated in COSMIC [54] were analyzed per tumor based on SNVs and their sequence context, considering the immediately flanking 5′ and 3′ nucleotides using the R package SomaticSignatures [55] with default settings.
PCR-based validation of JAK1 indels in independent patient series
Frameshift mutations in four hotspots in homopolymeric regions of JAK1 were analyzed by PCR-based fragment analyses in a total of 182 MSI+ CRCs (Fig. 1a). Fluorescently labeled primers were designed using Primer3 [56] (Additional file 1: Table S1). The four JAK1 fragments were amplified in multiplex PCR using the Qiagen Multiplex PCR Kit according to the manufacturer’s recommendations. One primer per fragment was labeled with a fluorescent dye from the G5 dye set, and GS500 LIZ was used as a size standard (Thermo Fisher Scientific Inc.). All fragments were run in duplicate on an ABI 3730 DNA Analyzer, with default Microsatellite Analysis settings, and analyzed by the GeneMapper software v.3.7 (Thermo Fisher Scientific Inc.). DNA from disease-free individuals and water served as normal and negative controls, respectively. Electropherograms were scored independently by two researchers.
High resolution DNA copy number analysis
DNA copy number data were generated for 27 of the 33 exome sequenced MSI+ tumors in Norwegian series I using Affymetrix Genome-Wide Human SNP Arrays 6.0 (Affymetrix Inc., Santa Clara, CA; Fig. 1a). DNA (1 μg) was prepared according to the Affymetrix SNP6.0 Cytogenetics Copy Number Assay (revision 3) and hybridized onto microarrays according to the Affymetrix Genome-Wide Human SNP Nsp/Sty 6.0 User Guide (revision 4). The raw copy number data were preprocessed according to the PennCNV protocol [57] adapted for Affymetrix genotyping arrays (http://www.openbioinformatics.org/penncnv/penncnv_tutorial_affy_gw6.html) using HapMap samples [58] as reference. Single-sample segmentation and calculation of allele-specific and absolute DNA copy numbers were performed using the algorithm ASCAT (v.2.3) [59], with the penalty constant for segmentation by the Allele-Specific Piecewise Constant Fitting algorithm set to 50. The sex chromosomes were excluded from further analyses.
For comparison of DNA copy number loss and gain across samples with different segment breakpoint positions, an artificial splitting of the data into smaller segments including all breakpoint positions across the sample set was performed. For each of these smallest regions of overlap, the copy number value of the original, larger segment was kept. Copy numbers per segment were adjusted according to the overall ploidy of each tumor. Regions with loss of heterozygosity (LOH) were identified as segments with DNA copy number equal to zero for one allele and higher than one for the other allele. Copy number neutral LOH was defined as DNA copy number equal to zero for one allele and equal to the overall ploidy of the sample for the other allele. The proportion of the genome affected by copy number aberrations (CNAs) was estimated for all samples as the percentage of base pairs with a total copy number different from the overall ploidy of the sample.
Modeling of tumor clonality
Tumor clonality was modeled for the 27 MSI+ tumors in Norwegian series I analyzed by both exome sequencing and DNA copy number variation using the R package SciClone with default settings (except the threshold for the minimum total read count set to 50) [60]. The variant allele frequency (VAF) of each SNV and indel, defined as the proportion (percentage) of reads of the variant allele compared with the total number of reads at the respective locus, was used as input, and SciClone identified sample-wise mutation clusters (representing genetically distinct cellular populations/subclones) based on SNVs and indels unaffected by CNAs and LOH using a variational Bayesian mixture model. The mutation cluster with a mean VAF of approximately 50% of the aberrant cell fraction (estimated using ASCAT) corresponds to an early clone with pervasive heterozygous mutations, and mutations in this clone were scored as truncal. Mutations in clones with lower and higher mean VAFs were scored as subclonal and homozygous, respectively. The number of clones per tumor was summarized from distinct mutation clusters of truncal and subclonal mutations.
For comparison, tumor clonality was also modeled using the R package EXPANDS [61], with maxScore set to 1.5 and precision to 0.02. Predicted subclones containing less than 5% of the mutations per tumor were not considered. EXPANDS models the cellular prevalence of each SNV as a probability distribution based on its VAF, adjusted for the DNA copy number at the mutated locus.
Gene expression analyses
Genome-wide exon-level expression analysis was done for a total of 73 MSI+ tumors in Norwegian series I, including 63 of the 68 tumors analyzed for JAK1 mutations, using Affymetrix GeneChip Human Exon 1.0 ST and Human Transcriptome 2.0 Arrays (Affymetrix Inc., Santa Clara, CA, USA). These data have partly previously been published (n = 39 samples; GSE24550, GSE29638, and GSE69182), and the remaining samples (n = 34) have been deposited to the NCBI’s GEO with accession number GSE79959. Raw intensity data were background corrected, quantile normalized, and summarized at the gene-level according to the robust multi-array average (RMA) approach implemented in the Affymetrix Expression Console 1.1 software. For samples analyzed on the Affymetrix Human Transcriptome 2.0 Array, the modified Signal Space Transformation algorithm of RMA was used. Gene expression data generated from the two different platforms were matched by HUGO gene symbols and merged by batch correction using ComBat [62] implemented in the R package SVA.
Classification of the tumors according to CMS was performed using the random forest predictor implemented in the R library CMSclassifier [42]. A default posterior probability of 0.5 was used as a threshold for sample classification, assigning a CMS class to 61 (84%) of the 73 tumors.
In addition, gene expression data and CMS assignments for 65 and 75 MSI+ tumors in TCGA and GSE39582 [45], analyzed by RNA sequencing and Affymetrix HG U133 Plus 2.0 arrays, respectively, were downloaded from the Colorectal Cancer Subtyping Consortium web site at SAGE Synapse (https://www.synapse.org/#!Synapse:syn2623706/wiki/67246). Of these tumors, 64 and 69 were assigned to a CMS class, respectively. For GSE39582, clinical data (reporting relapse-free survival (RFS) and overall survival (OS) for 67 of the CMS classified patients) were obtained via the GEO accession number (Additional file 1: Table S2). Patients (n = 8 with CMS classification) reported to have an event for OS and simultaneously censored for RFS were excluded from survival analyses, due to non-compliance with the definition of end-points used in this study [63].
Gene set expression enrichment analyses were performed using the R package GSA [64] and a customized collection of 51 gene sets relevant to CRC (Additional file 1: Table S11). Sample-wise gene set expression enrichment scores were calculated using the R package GSVA [65].
Tumor immunogenicity, evaluated based on gene expression, was analyzed by the “immunophenoscore”, using the published R code [41]. For marker genes with missing expression values (7 of the 162 genes on Human Exon 1.0 arrays), imputation was done based on the overall mean expression per sample, using the impute.knn function in the R package impute. The level of infiltration of immune cells in the tumors was also evaluated based on gene expression using the R packages ESTIMATE [66] and MCPcounter v.1.1 [43]. For comparison with the MSI+ tumors, gene expression data from 160 MSS tumors from Norwegian series I, included in the previously published GEO records, were also analyzed for immune cell infiltration.
Neoantigen prediction
Mutation-associated immunogenicity was analyzed from exome sequencing and gene expression data by computational prediction of neopeptides able to bind to patient-specific HLA molecules. Genotyping of the individual patients for the HLA class I genes HLA-A, B, and C at four-digit resolution (Additional file 1: Table S3) was done from merged sequencing BAM files of paired tumor–normal samples for each patient using the algorithm Polysolver with default settings (previously reported mean overall accuracy 97%) [67]. Prior to neopeptide prediction, the list of mutations (Additional file 1: Table S4) was filtered sample-wise based on gene expression, and only mutations in genes with an expression level above the median level per tumor were kept, retaining a median of 47% of the mutations per tumor (range 42 to 54%). All possible peptides of length 8–11 amino acids containing these mutations were predicted using Topiary 0.0.14 (https://pypi.python.org/pypi/topiary), with prediction of binding affinities for the patient-specific HLA alleles using NetMHCpan 2.8 [68]. Only peptides with IC50 below 500 and a percentile rank of predicted IC50 values across interactions between all peptides and each HLA allele per tumor below 2 were retained. The resulting predicted neoantigens were further filtered to exclude peptides encoded from other, non-mutated loci in the human genome by searching for perfect matches of the peptide sequences in UniProtKB release 2015_11 using the web application Peptide Match service for UniProt Knowledgebase [69]. Lastly, to include only peptides predicted to be generated by cleavage in proteasomes, the list of neoantigens was filtered to include only peptides with predicted C-terminal cleavage sites using the stand-alone software package of NetChop 3.1 [70]. When summarizing the number of neoantigens per tumor, peptides predicted to bind to several HLA alleles were counted only once.
Survival analyses
Univariable and multivariable survival analyses were conducted with Cox’s proportional hazards regression, with calculation of P values from Wald’s tests for predictive potential using the SPSS software v.21 (IBM Corporation, Armonk, NY, USA). Kaplan–Meier survival curves were compared with the log-rank test. Five-year RFS (considering relapse or death from any cause as events, and censoring in the case of no event within five years) and OS (time to death from any cause) were used as endpoints, according to the definition by Punt et al. [63]. Time to event or censoring was calculated from the time of surgery.
Results
Molecular overview of MMR deficiency in MSI+ CRC
A total of 333 MSI+ CRCs were analyzed in the study; an overview of the genomic analyses and results is shown in Fig. 1 (parts a and b, respectively).
Tissue samples from 33 stage I–III MSI+ CRCs and matched normal colonic mucosa in Norwegian series I (Table 1) were subjected to exome sequencing with an average depth of 273× and 109×, respectively. A total of 83,706 somatic small exonic mutations, comprising 65,880 SNVs and 17,826 indels, passed the quality filtering (Additional file 1: Tables S4 and S5). The number of amino acid-affecting mutations per tumor ranged from 957 to 4614 (median 1676; Fig. 2a), and the ratio of amino acid-affecting mutations to synonymous SNVs and in-frame indels was relatively consistent among the tumors (average 2.94, standard deviation 0.25; Additional file 1: Table S5). An MSI score representing the percentage of somatically unstable microsatellites per tumor was calculated using the algorithm MSIsensor (Fig. 2b; Additional file 1: Table S5). The MSI score was strongly correlated with the number of indels per tumor, as expected, but only weakly associated with the number of SNVs (Spearman correlations 0.7 and 0.4, respectively, P = 5 × 10−6 and 0.01; Additional file 2: Figure S1a).
Fig. 2.
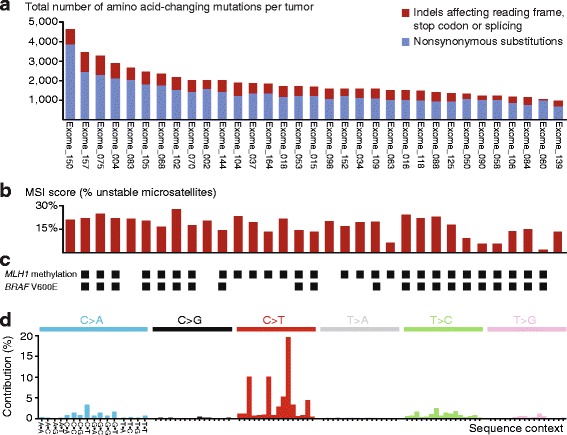
Somatic exonic mutation profiles of 33 MSI+ CRCs. a The total number of amino acid-changing mutations per tumor ranged from 957 to 4614 (median 1676). The tumors are sorted by mutation load. b The MSI score, calculated as the percentage of somatically unstable microsatellites in the exome of each tumor (microsatellites with indels), was more strongly associated with the number of indels than substitutions (Spearman correlations 0.7 and 0.4, respectively). c MLH1 promoter hypermethylation and BRAF V600E mutations were found in 28 (85%) and 21 (64%) of the tumors, respectively. There was no difference in the MSI score between tumors with and without MLH1 methylation. d Collection across all 33 tumors of substitutions in each of six categories (indicated in the top panel), classified according to sequence context (flanking nucleotides are indicated on the horizontal axis). The mutation distribution corresponds to mutation signature 6, as designated in COSMIC, which is associated with defective MMR
Consistent with their MSI phenotype, 28 (85%) of the tumors had methylation of the MLH1 promoter (Fig. 2c). Compared to tumors without methylation (n = 5), this was associated with a significant down-regulation of MLH1 gene expression (P = 0.02 by Welch’s t-test; Additional file 2: Figure S1b). The five tumors without MLH1 promoter methylation had down-regulated expression of seven other known MMR genes (Additional file 2: Figure S1c), and the sample-wise gene set enrichment score for these seven genes (calculated using the R package GSVA) was significantly lower in the unmethylated compared to the methylated samples (mean difference −0.5, P = 0.05). There was no difference in the MSI score between tumors with and without MLH1 promoter methylation (mean MSI scores 17 and 19%, respectively, P = 0.2); however, lack of methylation was associated with a younger age at diagnosis (mean age 55 years compared to 77 for patients with methylation, P = 0.01). Accordingly, familial predisposition cannot be excluded, in particular for the two youngest patients (aged 37 and 48 years).
The most common single nucleotide substitution type was C > T/G > A transitions. Analyzing all substitutions and their sequence context (the two flanking nucleotides) across all 33 MSI+ tumors, the distribution of mutations was found to correspond to mutation signature 6, as designated in COSMIC [54] (Fig. 2d). This signature is associated with hypermutation caused by MMR deficiency and was the predominant mutation signature also in all the tumors individually (Additional file 2: Figure S1d). Although ten (30%) of the tumors had amino acid-changing mutations in POLE (Additional file 1: Table S4), none of these were in known pathogenic hotspots [71], and none of the tumors had a mutation signature indicative of POLE-associated hypermutation.
In contrast to SNVs and indels, the tumors displayed few CNAs and the median proportion of the genome affected by CNAs was 2% (Additional file 2: Figures S1e and f; Additional file 1: Table S6).
Novel frequent frameshift indels in cancer-critical genes
In the 33 exome sequenced tumors, a total of 228 genes were identified as significantly mutated by MutSigCV (q values <0.05; Additional file 1: Table S7), including PHACTR4, RPL22, TFAM, TMBIM4, TTK, VPS37B, and WASF3 (ranked by q values). Compared to the whole exome, there was a clear enrichment for indels over SNVs in the significantly mutated genes; the average proportion of mutations that were indels was 80% among the tumors, significantly larger than the average of 27% for the full exome (P = 8 × 10−33, paired t-test; Additional file 2: Figure S1g). Also among the 571 genes included in the CGC, a modest increase in the sample-wise ratio of indels to SNVs was observed (the average ratio across the tumors was 32%), compared to the whole exome (P = 1 × 10−6).
Among genes in the CGC, frequent frameshift indels were detected in several genes not previously reported to be highly mutated in CRC, including CRTC1 (mutation frequency 42%), BCL9 (30%), and PTCH1 (24%) (Fig. 3a). In addition, 15 of the tumors (45%) were found to have frameshift mutations in members of the JAK-STAT pathway (Additional file 1: Table S8). The most commonly mutated gene was JAK1, in which 24% of the tumors (8 of 33) had one or more single base pair frameshift indels (Fig. 3a). For CRTC1 and BCL9, tumors with mutations had significantly higher MSI scores than the respective wild-type tumors (on average 4% more unstable microsatellites; P < 0.04 for both genes by Welch’s t-test); however, there was no significant difference in the MSI score between tumors mutated and wild type for PTCH1 or JAK1 (P > 0.3), indicating that these mutations are not associated with the level of tumor hypermutation. None of the mutations were associated with MLH1 promoter methylation (by Fisher’s exact test). For validation, we searched for the novel frequent frameshift mutations in publicly available exome sequencing data from 66 additional MSI+ CRCs from TCGA. The mutation frequencies of CRTC1, BCL9, JAK1, and PTCH1 were 27, 42, 18, and 12%, respectively (Fig. 3b; Additional file 1: Table S8).
Fig. 3.
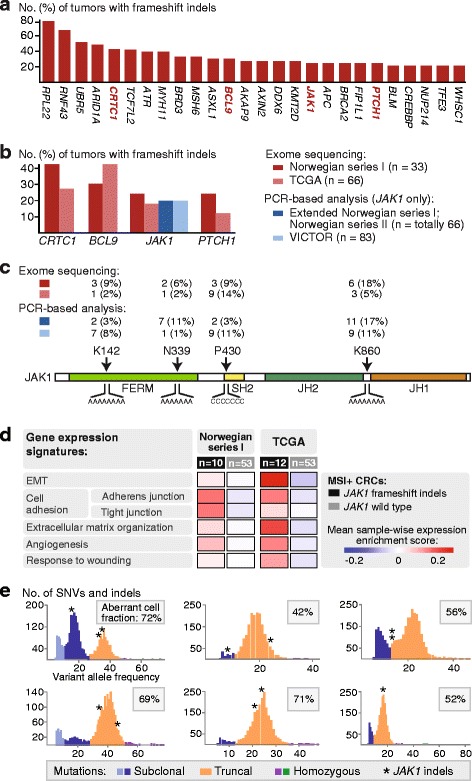
Cancer-critical genes with frequent frameshift mutations. a Mutation frequency of cancer-critical genes with most frequent indels affecting the reading frame, splice sites, or stop codons. CRTC1 (mutation frequency 42%), BCL9 (30%), JAK1 (24%), and PTCH1 (24%) were identified as novel frequently mutated genes. b Mutation validation analyses in MSI+ CRCs in independent patient series. c The JAK1 frameshift indels were found in four hotspot amino acid positions (GenBank accession NP_002218; 1154 amino acids). The number of mutations detected in each patient series and mutation hotspot is shown, confirming frequent mutations upstream of the JAK1 kinase domain (JH1). The patient series analyzed are indicated (top left) using the same color code as in b. All four homopolymers of length ≥6 in JAK1 are indicated below the protein. Protein domains are indicated by color; SH2 Src Homology 2, FERM (F, 4.1 protein; E, ezrin; R, radixin; M, moesin), JH1 kinase domain, JH2 pseudokinase domain. d Gene signatures of five processes previously found to be associated with innate resistance to anti-PD-1 treatment in melanomas were over-expressed in MSI+ CRCs with versus without JAK1 mutations in both Norwegian series I and TCGA. e Six of the tumors in Norwegian series I with JAK1 indels were available for clonality analyses. All these tumors had at least one JAK1 indel (asterisk) scored as truncal, and all JAK1 mutations were heterozygous. Each plot represents one sample and separate tumor clones, identified as mutation clusters with different variant allele frequencies, are indicated with different colors
Validation and transcriptional consequences of JAK1 loss-of-function mutations
Because of the recently suggested predictive value of JAK1 mutations indicating resistance to anti-PD-1 treatment [72, 73], this gene was selected for mutation validation analyses by PCR-based fragment analyses. Frameshift mutations in the four hotspots identified by exome sequencing were verified in the 33 tumors. These hotspots were all in homopolymers upstream of the JAK1 kinase domain (Fig. 3c), and the mutations have previously been described as truncating, loss-of-function mutations [74]. For an additional 149 MSI+ CRCs, JAK1 mutation frequency was 20%, both in independent Norwegian patients (mutations in 13 of 66 tumors) and British patients from the VICTOR trial (17 of 83) (Fig. 3b; Additional file 1: Table S8). In summary, the mutation frequency of JAK1 across all 248 primary MSI+ CRCs analyzed was 20%.
JAK1 gene expression was down-regulated in MSI+ tumors in Norwegian series I with versus without JAK1 mutations (Additional file 2: Figure S2a). Gene set expression enrichment analysis also revealed a modest depletion of JAK-STAT signaling in the mutated tumors (Additional file 1: Table S9). Resistance to anti-PD-1 treatment in hypermutated tumors with JAK1 loss-of-function mutations has been proposed to be linked to reduced expression of the PD-L1 ligand, mediated by reduced signaling in the interferon gamma (IFN-γ) receptor pathway [73]. In Norwegian series I, the sample-wise enrichment scores of a six-gene IFN-γ expression signature [75] (calculated using the R package GSVA) was significantly lower in JAK1 mutated compared to wild-type tumors (Additional file 2: Figure S2b). Also, both PD-1 signaling and gene expression of the PD-L1 ligand (CD274) was significantly down-regulated in the mutated tumors (Additional file 2: Figure S2c, d). To further explore a potential association with immune evasion, tumors were analyzed for the gene expression-based immunophenoscore [41]. Tumors with JAK1 mutations had a lower score than wild-type tumors specifically for the antigen processing component (MHC score, mean difference −0.3, P = 0.01 by Welch’s t-test), indicating immune evasion, similarly to what has been observed in endometrial cancer [76]. The total immunophenoscore (IPS score) was also lower in the mutated tumors (mean difference −0.8, P = 0.08), predicting a poor response to immune checkpoint inhibition [41]. In concordance, tumors with mutations were also found to be positively enriched for transcriptional signatures of biological processes described to be up-regulated in melanomas with innate resistance to anti-PD-1 treatment [40]. These processes include epithelial-to-mesenchymal transition (EMT), cell adhesion, extracellular matrix organization, angiogenesis, and response to wounding; sample-wise enrichment scores for gene signatures of all these five processes were higher in tumors with JAK1 mutations in both Norwegian series I and TCGA (Fig. 3d; Additional file 1: Table S9).
Clonality of MSI+ CRCs and JAK1 mutations
Resistance to anti-PD-1 treatment is found in tumors with homozygous, truncating JAK1 mutations (mutation accompanied by LOH at the JAK1 locus), resulting in complete loss of protein function [73]. To analyze the zygosity of mutations, we did computational modeling of tumor clonality based on the VAFs of exonic SNVs and indels unaffected by CNAs in the 27 tumors with both data types available, using the algorithm SciClone [60] (Additional file 2: Figure S3a, b; Additional file 1: Table S10). All tumors except three (89%) revealed intra-tumor heterogeneity with more than one subclone, and the number of clones per tumor was independent of the number of mutations (Additional file 2: Figure S3c). The pattern of clonality was similar among the tumors and the majority of mutations were found to be heterozygous and truncal (i.e., pervasive in all cancer cells in the tumor) in all tumors except one (96% of tumors; Additional file 2: Figure S3d). The proportion of truncal mutations in the full exome and genes in the CGC was proportional across the tumors, with a median of 85 and 82%, respectively (Additional file 2: Figure S2e). For comparison, tumor clonality was also modeled based on the algorithm EXPANDS [61], supporting the prediction of multiple subclones in the majority of tumors (all except one; details in Additional file 2: Supplementary text).
All truncating JAK1 indels were heterozygous and unaffected by LOH (Fig. 3e), but all mutated tumors had at least one truncal JAK1 mutation. Considering the other genes with novel frequent frameshift mutations, BCL9 and PTCH1 mutations were also mostly heterozygous and truncal, although one tumor had a homozygous BCL9 mutation and both genes were found to have a subclonal mutation in one tumor each. In contrast, mutations in CRTC1 were frequently subclonal and considering both indels and SNVs, CRTC1 was indeed the cancer-critical gene with most frequent subclonal mutations (in 30% of the 27 tumors).
The mutation load of MSI+ CRCs is correlated with the predicted neoantigen load, but not with immune cell infiltration
To analyze the association between genetic changes and infiltration of immune cells, the latter was evaluated based on gene expression using the algorithm MCPcounter [43] and the immune-score ESTIMATE [66]. In agreement with their high mutation load, the MSI+ tumors in Norwegian series I (n = 73) had a higher level of infiltration of cytotoxic lymphocytes and immune-score compared to the MSS tumors (n = 160; Additional file 1: Figure S4a). In the exome sequenced MSI+ tumors, neoantigens with high binding affinities for the patient-specific class I HLA molecules were predicted from small exonic mutations in highly expressed genes (expression level above the median across all genes per tumor). The number of predicted neoantigens per tumor (median 1169; range 376 to 4245) was correlated to the number of exonic mutations (also including synonymous; Spearman’s correlation 0.7, P = 4 × 10−5; Fig. 4a). However, this relationship was not one-to-one, indicating generation of multiple neoantigens from individual mutations (Fig. 4b). As expected, all mutations creating the largest numbers of neoantigens were frameshift indels (Additional file 2: Figure S4b), and the mean ratios between the number of neoantigens and mutations per tumor were 1.3 and 0.5 for frameshift mutations and SNVs, respectively (P = 4 × 10−10 by paired samples t-test). Still, the numbers of mutations were correlated to the corresponding numbers of neoantigens for both frameshift mutations and nonsynonymous SNVs separately (Spearman’s correlation 0.6; P = 2 × 10−4 and P = 1 × 10−4, respectively; Additional file 2: Figure S4c). Considering also the clonality of the mutations, a strong correlation (Spearman 0.9; P = 7 × 10−12) between the number of neoantigens predicted from truncal mutations and the total number of mutations per tumor was found.
Fig. 4.
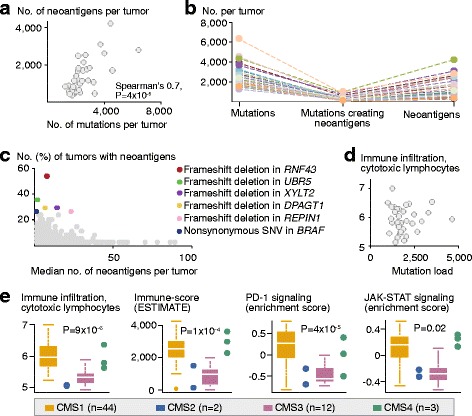
Predicted neoantigen load correlates with mutation load, while immune infiltration is highest in CMS1. a Among the 33 exome-sequenced tumors, there was a strong correlation between the number of exonic mutations (including synonymous) and predicted neoantigens. b There was not a one-to-one correspondence among the number of mutations (left), mutations predicted to create neoantigens (middle), and predicted neoantigens (right) per tumor (each tumor is indicated with a separate color), showing that individual mutations generate several neoantigens per tumor. c The individual mutations (represented by a dot) predicted to create most neoantigens per tumor (horizontal axis) were typically not recurrent in a large proportion of the tumors (vertical axis). However, several mutations (indicated by colors) created neoantigens in several tumors. d The number of amino acid changing mutations per tumor (horizontal axis) was not associated with the level of infiltration of cytotoxic lymphocytes (Spearman’s correlation −0.06, P = 0.8). e In contrast, among all MSI+ tumors in Norwegian series I, infiltration of cytotoxic lymphocytes and the ESTIMATE immune-score, as well as PD-1 and JAK-STAT signaling (based on gene expression) were significantly higher in CMS1 compared to CMS2-4 (Welch’s t-test; although high also in CMS4)
Mutations predicted to be highly immunogenic were mostly non-recurrent among the tumors (Fig. 4c). Altogether, only 0.5 and 14.7% of the SNVs and frameshift mutations, respectively, were predicted to create neoantigens in more than one tumor. Mutations in RNF43, UBR5, XYLT2, DPAGT1, REPIN1, and BRAF (V600E) were most frequently immunogenic (in 55, 36, 30, 30, 27, and 27% of the 33 tumors, respectively).
Surprisingly, there were no associations between the mutation load (or predicted neoantigen load) and the level of immune cell infiltration or the immunophenoscore among the MSI+ tumors in neither Norwegian series I (Fig. 4d; Additional file 2: Figure S4d) nor TCGA. Similarly, no indication of increased immune evasion, analyzed as PD-1 signaling, was detected in tumors with a high mutation load (Additional file 2: Figure S4e).
There was no difference in the mutation load (or predicted neoantigen load) between tumors with and without JAK1 frameshift indels (P = 0.5; Additional file 2: Figure S4f). However, in concordance with the observed down-regulation of PD-1 signaling associated with JAK1 mutations, mutated tumors also had a modestly lower level of infiltration of cytotoxic lymphocytes (analyzed based on gene expression; P = 0.01; Additional file 2: Figure S4g).
Immune cell infiltration is strongly associated with CMS1, independent of JAK1 mutation status
To further explore potential correlates to immune cell infiltration, tumors were classified according to the gene expression-based CMS classes of CRC [42]. In Norwegian series I, 44 (60%), 2 (3%), 12 (16%), and 3 (4%) of the MSI+ tumors were classified as CMS1, CMS2, CMS3, and CMS4, respectively (Additional file 2: Figure S5a). Among the exome-sequenced tumors, the mean number of mutations was higher in CMS1 than the other three CMS classes (1964 versus 1537 mutations, respectively; P = 0.08 by Welch’s t-test; Additional file 2: Figure S5b). This was also found among MSI+ tumors in TCGA (mean mutation load 1499 versus 1097, respectively; P = 0.03). There was no enrichment for JAK1 mutations in any of the CMS classes (P = 0.7 by Fisher’s exact test); mutations were found in 11% of CMS1 tumors and 18% of CMS3 tumors in Norwegian series I (the remaining mutations were found in unclassified tumors).
Gene set expression analyses revealed that the distinct biological properties of CMS1 were recapitulated also among MSI+ tumors only (Additional file 2: Figure S5c; Additional file 1: Table S11). In comparison with CMS2–4, tumors in CMS1 had a significantly higher level of infiltration of cytotoxic lymphocytes and a higher immune-score (ESTIMATE; Fig. 4e; Additional file 2: Figure S5d). This was independent of JAK1 mutation status, and analyzing tumors wild type for JAK1 only, CMS1 tumors still had a high level of immune cell infiltration (Additional file 2: Figure S5f). Furthermore, CMS1 tumors had strong PD-1 signaling and JAK-STAT signaling (Fig. 4e), independent of JAK1 mutation status (Additional file 2: Figure S5g).
JAK1 loss-of-function mutations and CMS1 are positive prognostic factors in MSI+ CRC
Considering the substantial molecular heterogeneity and associations with immunogenicity detected among MSI+ tumors, the distinct genomic features were investigated for associations with patient outcome.
Among tumors analyzed for JAK1 mutations, clinical follow-up data reporting OS were available for 175 of the 182 Norwegian and British patients (Table 1; Fig. 1a). Patients with a JAK1 frameshift indel (n = 36) had an OS rate of 94%, significantly higher than the 75% OS rate of patients wild type for JAK1 (n = 139; hazard ratio (HR) from Cox’s regression 0.2 [95% confidence interval 0.05–0.9], P value from Wald’s test of predictive potential 0.03; Fig. 5a). The positive prognostic association of JAK1 mutations was seen in all the three patient series individually (although not statistically significant; Additional file 2: Figure S6a). In multivariable analysis, the prognostic association was independent of patient age (above versus below median), patient gender, and tumor localization (right versus left and rectum; multivariable HR = 0.2 [0.06–1.0], P = 0.05). Furthermore, the majority of the patients (89%; 155 of 175) were diagnosed with stage II or III MSI+ CRC, and the prognostic association of JAK1 frameshift indels was independent of cancer stage in these patients (multivariable HR = 0.1 [0.02–0.8], P = 0.03; Additional file 2: Figure S6b).
Fig. 5.
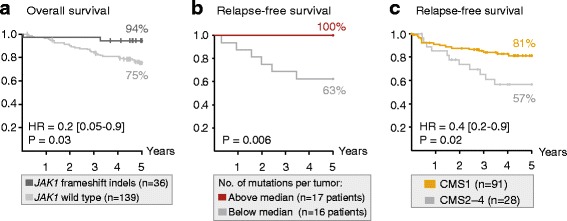
JAK1 mutations, mutation load, and CMS1 are associated with a good patient outcome. a Among 175 patients with MSI+ CRC from Norwegian series I and II and the VICTOR trial, tumors with JAK1 frameshift indels were associated with a better 5-year overall survival rate (94%) than wild-type tumors (75%; P value from Wald’s test). b Among the 33 exome-sequenced tumors in Norwegian series I, tumors with a mutation load above the median number of mutations were associated with a better 5-year relapse-free survival rate (100%) than tumors with a low mutation load (63%; P value from the log-rank test). c Among 119 patients in Norwegian series I and a publicly available dataset (GEO accession number GSE39582), patients in CMS1 had a significantly better 5-year relapse-free survival rate (81%) than patients in CMS2-4 (57%, P value from Wald’s test)
Although there was no difference in the mutation load between tumors with and without JAK1 frameshift indels (n = 33 patients in Norwegian series I), a large burden of small exonic mutations (above the median; 1676 mutations) was also associated with a favorable patient outcome (Fig. 5b), independent of cancer stage (Additional file 2: Figure S6c). In contrast, neither the MSI score nor the clonal composition of the tumors (analyzed as the number of subclones predicted by either SciClone or EXPANDS) were associated with patient survival.
The strong association between CMS1 and immune cell infiltration suggests that CMS1 is also a favorable prognostic factor in MSI+ CRC. Indeed, among 119 patients (with known CMS class and clinical information) from Norwegian series I and a publicly available dataset (GSE39582) [45], patients with CMS1 tumors (n = 91) had a 5-year RFS rate of 81%, significantly higher than the survival rate of 57% for patients with CMS2–4 tumors (n = 28) (HR = 0.4 [0.2–0.9], P = 0.02; Fig. 5c). A similar prognostic association was found for 5-year OS (HR = 0.5 [0.2–1.0], P = 0.06; Additional file 2: Figure S6d). CMS1 was associated with a better outcome than each of the subtypes CMS2, CMS3, and CMS4 separately (Additional file 2: Figure S6e). In a multivariable model including patient age, gender, and cancer stage, CMS1 (versus CMS2–4) was an independent prognostic factor (multivariable HR = 0.4 [0.2–0.9], P = 0.02; Additional file 1: Table S12).
Combined survival analysis of CMS1 and JAK1 mutations was possible for 51 patients in Norwegian series I. Here, JAK1 mutations provided additional prognostic information to CMS1. Of note, the sample numbers in the individual subgroups were low (four and two JAK1 mutated tumors in CMS1 and CMS2–4, respectively; Additional file 2: Figure S6f). However, in a multivariable model, the prognostic value of CMS1 (versus CMS2–4) was statistically independent of JAK1 mutations (multivariable HR = 0.2 [0.09–0.7] and 0.3 [0.1–0.9] for 5-year RFS and OS, respectively; P = 0.007 and 0.03).
Discussion
Accurate genome-wide analysis of MSI-type indel mutations has been challenging. Even in the most comprehensive mutational characterization studies of MSI+ CRCs published to date, scoring of indels has been limited either to known homopolymers in selected genes [16, 20, 21] or targeted sequence alignments of a reference set of microsatellite repeats [77], or mutation calling was limited to SNVs [78]. By multilevel genomics and computational analyses, we have identified heterogeneity with clinical relevance within this distinct subtype of colorectal tumors. By exome sequencing, we identified novel, frequent frameshift mutations in four cancer-critical genes (CRTC1, BCL9, JAK1, and PTCH1). The majority of mutations in CRTC1 were subclonal, possibly explaining why these have previously gone unnoticed, and highlighting the importance of high sequencing coverage. Truncating indels in JAK1, leading to loss of the entire kinase domain, were found in 20% of a total of 248 tumors. In contrast, JAK1 indels have previously been reported either as infrequent events in CRC [74] or as specific to MSI+ tumors from the endometrium [77]. We detected frameshift mutations in 18% of MSI+ CRCs from the dataset analyzed in the latter study (TCGA), illustrating the challenge of large-scale analysis of MSI-type mutations.
Consistent with a high level of intra-tumor heterogeneity and the “Big Bang model” of CRC development [79], clonality modeling predicted several subclones in most of the MSI+ tumors. There was no association between tumor clonality and patient survival, which is in contrast to reports from several other cancer types [80, 81]. On the other hand, a favorable outcome was found among patients with a high mutation load. As expected, the mutation load was strongly correlated to the predicted neoantigen load, and the level of immune cell infiltration was significantly higher in MSI+ compared to MSS tumors, consistent with recent results [37]. However, within the subgroup of MSI+ tumors, we found no association between the mutation load, or predicted neoantigen load, and the level of immune cell infiltration. This may be explained by technical issues, including the low sample number and insufficient statistical power, lower fidelity of mutation detection among hypermutated tumors, and lower sensitivity of immune cell infiltration detection based on gene expression compared to immunohistochemistry. However, similar and seemingly disparate results have recently been described in melanoma. Here, the mutation load of pre-treatment biopsies was associated with improved patient survival, but not with response to anti-PD-1 treatment, which in turn is associated with intra-tumor T-cell infiltration [40], suggesting additional determinants of tumor immunogenicity. We found immune cell infiltration to be highly dependent on the CMS class of the tumors. CMS1 is an immunogenic subtype of CRC, associated with a high prevalence of MSI [42, 43], and this study is, to our knowledge, the first to describe CMS1 as a particularly immunogenic subtype also specifically among MSI+ tumors. Clinical relevance was reinforced by a strong association between CMS1 and patient survival. CMS1 was originally described as a poor prognostic factor after relapse [42]; however, among patients with MSI+ CRC, we identify CMS1 as a favorable prognostic factor, independent of important clinicopathological parameters.
Positive prognostic associations were observed also for truncating JAK1 mutations, which is in contrast to recent results describing a poor outcome in patients with skin subcutaneous melanoma, breast invasive carcinoma, and prostate adenocarcinoma (no prognostic associations were detected in CRC in this study) [73]. These are cancer types with a low prevalence of MSI [15], suggesting heterogeneous prognostic associations according to the mutation phenotype (MSI status). JAK-STAT signaling regulates diverse cellular processes and may have oncogenic potential [82]. Preclinical data suggest that inhibition of JAK1 can block colorectal tumor growth [83], supporting a potential positive prognostic value of inactivating mutations, but additional analyses are required to explain the role of JAK1 mutations in the progression of MSI+ CRC. Furthermore, multivariable analyses indicate that JAK1 mutations and CMS1 have independent prognostic value, but combined analyses in larger patient series are needed. Still, the promising results observed for JAK1 mutations and CMS1 in altogether four independent patient series reinforce the potential for molecular prognostic stratification of MSI+ CRC.
Although hypermutated tumors generally respond well to anti-PD-1 treatment, resistance is commonly observed in metastatic MSI+ CRC. Homozygous JAK1 loss-of-function mutations, resulting in an inability to signal in the IFN-γ response pathway and with subsequent loss of PD-L1 expression, has been identified as one potential resistance mechanism [73]. At the gene expression-level, we observed similar associations with JAK1 mutations in our series of primary MSI+ CRCs, including up-regulation of transcriptional signatures associated with innate resistance to anti-PD-1 treatment in melanoma [40]. However, complete loss of JAK1 function is likely required to confer treatment resistance [73], and clonality modeling suggested that the mutations were heterozygous. We speculate that primary tumors with heterozygous, truncating mutations are more prone to biallelic inactivation and to development of resistance in the metastatic setting. However, the need for a second, inactivating hit suggests that JAK1 mutations do not account for the majority of cases with anti-PD-1 resistance. This is supported by the favorable outcome among patients with mutated tumors, predicting a low mutation frequency in metastatic disease. Consequently, there is still a need to uncover additional mechanisms of resistance to anti-PD-1 treatment in metastatic MSI+ CRC.
Conclusions
We have identified novel frequent frameshift mutations and molecular heterogeneity with clinical relevance in primary MSI+ CRC. A high mutation load was found to correlate with the predicted neoantigen load, but immune cell infiltration was highest in the transcriptomic CMS1 subgroup, and CMS1 was a predictor of a good patient outcome. Of particular interest, considering the potential for prediction of resistance to anti-PD-1 treatment, was the validation of a high prevalence of truncating JAK1 indels in independent patient series. However, the mutations were associated with a favorable patient prognosis, suggesting a low mutation frequency in metastatic disease, and reinforcing the need to discover additional mechanisms of resistance to immunotherapy.
Additional files
Supplementary Tables. An Excel file containing Supplementary Tables S1-S12. (XLSX 8497 kb)
Supplementary Text and Figures. A Word file containing Supplementary Text and Figures S1-S6. (DOCX 1054 kb)
Acknowledgements
We acknowledge granted storage space for computer files and data processing power from NorStore and TSD 2.0.
Funding
This study was supported by the Southern and Eastern Norway Regional Health Authority (Research Grant “Genome Medicine of Colorectal Cancer”, project number 2011024), the Norwegian Cancer Society (project numbers 6824048-2016 and 72190-PR-2006-0442), the foundation Stiftelsen Kristian Gerhard Jebsen, and the Research Council of Norway (grants to the Norwegian Cancer Genomics Consortium and through the Centres of Excellence funding scheme, project number 179571).
Availability of data and materials
The microarray gene expression data generated during the current study are available in the NCBI’s GEO repository (http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE79959. A complete list of mutations detected by exome sequencing during the current study is included within the article and its additional files. Additional, previously published data analyzed during the current study are available in GEO with accession numbers GSE24550, GSE29638, GSE69182, and GSE39582 (microarray gene expression data) and in the Cancer Genomics Hub (CGHub: exome sequencing data from TCGA).
Authors’ contributions
Conception and design: RAL, AS, and RIS. Acquisition of data: AS, BJ, TT, SAD, IAE, GEL, KCGB, EL, LAMZ, ED, OM, DK, IT, and AN. Analyses and interpretation of data: AS, BJ, TT, RIS, and RAL. Writing of manuscript: AS drafted the manuscript and all authors were involved in revision of the manuscript and have read and approved the final version. Study supervision: RAL.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The project is approved by the Regional Committee for Medical and Health Research Ethics, South Eastern Norway (REC number 1.2005.1629) and the Norwegian Data Inspectorate. Written consent for genome analyses is obtained from all patients in Norwegian series I (approved REC amendment 2010/1805). The research complies with the Declaration of Helsinki.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- BAM
Binary Alignment Map
- CGC
Cancer Gene Census
- CMS
Consensus molecular subtype
- CNA
Copy number aberration
- CRC
Colorectal cancer
- EMT
Epithelial-to-mesenchymal transition
- GEO
Gene Expression Omnibus
- HR
Hazard ratio
- indel
Insertions and/or deletions
- LOH
Loss of heterozygosity
- MMR
DNA mismatch repair
- MSI
Microsatellite instability
- MSS
Microsatellite stability
- OS
Overall survival
- RFS
Relapse-free survival
- RMA
Robust multi-array average
- SAM
Sequence Alignment Map
- SNV
Single nucleotide variant
- TCGA
The Cancer Genome Atlas
- VAF
Variant allele frequency.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s13073-017-0434-0) contains supplementary material, which is available to authorized users.
Contributor Information
Anita Sveen, Email: anita.sveen@rr-research.no.
Bjarne Johannessen, Email: bjarne.johannessen@rr-research.no.
Torstein Tengs, Email: torstein.tengs@vetinst.no.
Stine A. Danielsen, Email: stine.aske.danielsen@rr-research.no
Ina A. Eilertsen, Email: ina.andrassy.eilertsen@rr-research.no
Guro E. Lind, Email: guro.elisabeth.lind@rr-research.no
Kaja C. G. Berg, Email: kaja.christine.graue.berg@rr-research.no
Edward Leithe, Email: edward.leithe@rr-research.no.
Leonardo A. Meza-Zepeda, Email: leonardo.a.meza-zepeda@rr-research.no
Enric Domingo, Email: enric@well.ox.ac.uk.
Ola Myklebost, Email: ola.myklebost@rr-research.no.
David Kerr, Email: david.kerr@ndcls.ox.ac.uk.
Ian Tomlinson, Email: iant@well.ox.ac.uk.
Arild Nesbakken, Email: arild.nesbakken@medisin.uio.no.
Rolf I. Skotheim, Email: rolf.i.skotheim@rr-research.no
Ragnhild A. Lothe, Email: rlothe@rr-research.no
References
- 1.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–61. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 2.Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, et al. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–6. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 3.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–9. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 4.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–11. [PubMed] [Google Scholar]
- 5.Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, et al. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58:3455–60. [PubMed] [Google Scholar]
- 6.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–5. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–38. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 8.Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–25. doi: 10.1016/0092-8674(93)90330-S. [DOI] [PubMed] [Google Scholar]
- 9.Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–61. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 10.Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–9. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 11.Børresen AL, Lothe RA, Meling GI, Lystad S, Morrison P, Lipford J, et al. Somatic mutations in the hMSH2 gene in microsatellite unstable colorectal carcinomas. Hum Mol Genet. 1995;4:2065–72. doi: 10.1093/hmg/4.11.2065. [DOI] [PubMed] [Google Scholar]
- 12.Peltomaki P, Lothe RA, Aaltonen LA, Pylkkanen L, Nystrom-Lahti M, Seruca R, et al. Microsatellite instability is associated with tumors that characterize the hereditary non-polyposis colorectal carcinoma syndrome. Cancer Res. 1993;53:5853–5. [PubMed] [Google Scholar]
- 13.Wooster R, Cleton-Jansen AM, Collins N, Mangion J, Cornelis RS, Cooper CS, et al. Instability of short tandem repeats (microsatellites) in human cancers. Nat Genet. 1994;6:152–6. doi: 10.1038/ng0294-152. [DOI] [PubMed] [Google Scholar]
- 14.Lothe RA. Microsatellite instability in human solid tumors. Mol Med Today. 1997;3:61–8. doi: 10.1016/S1357-4310(96)10055-1. [DOI] [PubMed] [Google Scholar]
- 15.Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med. 2016;22:1342–50. doi: 10.1038/nm.4191. [DOI] [PubMed] [Google Scholar]
- 16.The Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 19.Duval A, Hamelin R. Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer Res. 2002;62:2447–54. [PubMed] [Google Scholar]
- 20.Røyrvik EC, Ahlquist T, Rognes T, Lothe RA. Slip slidin’ away: a duodecennial review of targeted genes in mismatch repair deficient colorectal cancer. Crit Rev Oncog. 2007;13:229–57. doi: 10.1615/CritRevOncog.v13.i3.20. [DOI] [PubMed] [Google Scholar]
- 21.Bruun J, Kolberg M, Ahlquist TC, Royrvik EC, Nome T, Leithe E, et al. Regulator of chromosome condensation 2 identifies high-risk patients within both major phenotypes of colorectal cancer. Clin Cancer Res. 2015;21:3759–70. doi: 10.1158/1078-0432.CCR-14-3294. [DOI] [PubMed] [Google Scholar]
- 22.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–8. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 23.Myeroff LL, Parsons R, Kim SJ, Hedrick L, Cho KR, Orth K, et al. A transforming growth factor beta receptor type II gene mutation common in colon and gastric but rare in endometrial cancers with microsatellite instability. Cancer Res. 1995;55:5545–7. [PubMed] [Google Scholar]
- 24.Parsons R, Myeroff LL, Liu B, Willson JK, Markowitz SD, Kinzler KW, et al. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res. 1995;55:5548–50. [PubMed] [Google Scholar]
- 25.Mullaney JM, Mills RE, Pittard WS, Devine SE. Small insertions and deletions (INDELs) in human genomes. Hum Mol Genet. 2010;19:R131–6. doi: 10.1093/hmg/ddq400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghoneim DH, Myers JR, Tuttle E, Paciorkowski AR. Comparison of insertion/deletion calling algorithms on human next-generation sequencing data. BMC Res Notes. 2014;7:864. doi: 10.1186/1756-0500-7-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giannakis M, Hodis E, Jasmine MX, Yamauchi M, Rosenbluh J, Cibulskis K, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet. 2014;46:1264–6. doi: 10.1038/ng.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 29.Lothe RA, Peltomaki P, Meling GI, Aaltonen LA, Nystrom-Lahti M, Pylkkanen L, et al. Genomic instability in colorectal cancer: relationship to clinicopathological variables and family history. Cancer Res. 1993;53:5849–52. [PubMed] [Google Scholar]
- 30.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–18. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 31.Guidoboni M, Gafa R, Viel A, Doglioni C, Russo A, Santini A, et al. Microsatellite instability and high content of activated cytotoxic lymphocytes identify colon cancer patients with a favorable prognosis. Am J Pathol. 2001;159:297–304. doi: 10.1016/S0002-9440(10)61695-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity. 2016;44:698–711. doi: 10.1016/j.immuni.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 33.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 34.Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, Nelson BH, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24:743–50. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angelova M, Charoentong P, Hackl H, Fischer ML, Snajder R, Krogsdam AM, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015;16:64. doi: 10.1186/s13059-015-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanz-Pamplona R, Gil-Hoyos R, Lopez-Doriga A, Alonso MH, Ausso S, Mollevi DG, et al. Mutanome and expression of immune response genes in microsatelite stable colon cancer. Oncotarget. 2016;7:17711–25. doi: 10.18632/oncotarget.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016;15:857–65. doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim CG, Ahn JB, Jung M, Beom SH, Kim C, Kim JH, et al. Effects of microsatellite instability on recurrence patterns and outcomes in colorectal cancers. Br J Cancer. 2016;115:25–33. doi: 10.1038/bjc.2016.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–62. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 42.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becht E, de Reynies A, Giraldo NA, Pilati C, Buttard B, Lacroix L, et al. Immune and stromal classification of colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clin Cancer Res. 2016;22:4057–66. doi: 10.1158/1078-0432.CCR-15-2879. [DOI] [PubMed] [Google Scholar]
- 44.Kerr DJ, Dunn JA, Langman MJ, Smith JL, Midgley RS, Stanley A, et al. Rofecoxib and cardiovascular adverse events in adjuvant treatment of colorectal cancer. N Engl J Med. 2007;357:360–9. doi: 10.1056/NEJMoa071841. [DOI] [PubMed] [Google Scholar]
- 45.Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berg M, Danielsen SA, Ahlquist T, Merok MA, Ågesen TH, Vatn MH, et al. DNA sequence profiles of the colorectal cancer critical gene set KRAS-BRAF-PIK3CA-PTEN-TP53 related to age at disease onset. PLoS One. 2010;5:e13978. doi: 10.1371/journal.pone.0013978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andresen K, Boberg KM, Vedeld HM, Honne H, Jebsen P, Hektoen M, et al. Four DNA methylation biomarkers in biliary brush samples accurately identify the presence of cholangiocarcinoma. Hepatology. 2015;61:1651–9. doi: 10.1002/hep.27707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–7. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 51.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30:1015–6. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gehring JS, Fischer B, Lawrence M, Huber W. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics. 2015;31:3673–5. doi: 10.1093/bioinformatics/btv408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–74. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 59.Van Loo P, Nordgard SH, Lingjærde OC, Russnes HG, Rye IH, Sun W, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107:16910–5. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller CA, White BS, Dees ND, Griffith M, Welch JS, Griffith OL, et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol. 2014;10:e1003665. doi: 10.1371/journal.pcbi.1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andor N, Harness JV, Muller S, Mewes HW, Petritsch C. EXPANDS: expanding ploidy and allele frequency on nested subpopulations. Bioinformatics. 2014;30:50–60. doi: 10.1093/bioinformatics/btt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 63.Punt CJ, Buyse M, Kohne CH, Hohenberger P, Labianca R, Schmoll HJ, et al. Endpoints in adjuvant treatment trials: a systematic review of the literature in colon cancer and proposed definitions for future trials. J Natl Cancer Inst. 2007;99:998–1003. doi: 10.1093/jnci/djm024. [DOI] [PubMed] [Google Scholar]
- 64.Efron B, Tibshirani R. On testing the significance of sets of genes. Ann Appl Stat. 2007;1:107–29. doi: 10.1214/07-AOAS101. [DOI] [Google Scholar]
- 65.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shukla SA, Rooney MS, Rajasagi M, Tiao G, Dixon PM, Lawrence MS, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol. 2015;33:1152–8. doi: 10.1038/nbt.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen C, Li Z, Huang H, Suzek BE, Wu CH. A fast Peptide Match service for UniProt Knowledgebase. Bioinformatics. 2013;29:2808–9. doi: 10.1093/bioinformatics/btt484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nielsen M, Lundegaard C, Lund O, Kesmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57:33–41. doi: 10.1007/s00251-005-0781-7. [DOI] [PubMed] [Google Scholar]
- 71.Rayner E, van Gool IC, Palles C, Kearsey SE, Bosse T, Tomlinson I, et al. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer. 2016;16:71–81. doi: 10.1038/nrc.2015.12. [DOI] [PubMed] [Google Scholar]
- 72.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–29. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov. 2017;7:188–201. doi: 10.1158/2159-8290.CD-16-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ren Y, Zhang Y, Liu RZ, Fenstermacher DA, Wright KL, Teer JK, et al. JAK1 truncating mutations in gynecologic cancer define new role of cancer-associated protein tyrosine kinase aberrations. Sci Rep. 2013;3:3042. doi: 10.1038/srep03042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chow LQM, Mehra R, Haddad RI, Mahipal A, Weiss J, Berger R, et al. Biomarkers and response to pembrolizumab in recurrent/metastatic head and neck squamous cell carcinoma. J Clin Oncol. 2016;34(suppl):abstr 6010. doi: 10.1200/JCO.2016.68.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stelloo E, Versluis MA, Nijman HW, de Bruyn M, Plat A, Osse EM, et al. Microsatellite instability derived JAK1 frameshift mutations are associated with tumor immune evasion in endometrioid endometrial cancer. Oncotarget. 2016;7:39885–93. doi: 10.18632/oncotarget.9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim TM, Laird PW, Park PJ. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell. 2013;155:858–68. doi: 10.1016/j.cell.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Supek F, Lehner B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. 2015;521:81–4. doi: 10.1038/nature14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47:209–16. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22:105–13. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morris LG, Riaz N, Desrichard A, Senbabaoglu Y, Hakimi AA, Makarov V, et al. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget. 2016;7:10051–63. doi: 10.18632/oncotarget.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sansone P, Bromberg J. Targeting the interleukin-6/Jak/stat pathway in human malignancies. J Clin Oncol. 2012;30:1005–14. doi: 10.1200/JCO.2010.31.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Buchert M, Burns CJ, Ernst M. Targeting JAK kinase in solid tumors: emerging opportunities and challenges. Oncogene. 2016;35:939–51. doi: 10.1038/onc.2015.150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables. An Excel file containing Supplementary Tables S1-S12. (XLSX 8497 kb)
Supplementary Text and Figures. A Word file containing Supplementary Text and Figures S1-S6. (DOCX 1054 kb)
Data Availability Statement
The microarray gene expression data generated during the current study are available in the NCBI’s GEO repository (http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE79959. A complete list of mutations detected by exome sequencing during the current study is included within the article and its additional files. Additional, previously published data analyzed during the current study are available in GEO with accession numbers GSE24550, GSE29638, GSE69182, and GSE39582 (microarray gene expression data) and in the Cancer Genomics Hub (CGHub: exome sequencing data from TCGA).


