Abstract
The C-terminal domains of the two α-subunits (αCTD) in Escherichia coli RNA polymerase (RNAP) recognize specific sequences called UP elements in some promoters. These interactions can increase transcription dramatically. Previously, effects of upstream DNA–αCTD interactions on transcription were quantified relative to control promoters with nonspecific DNA sequences substituted for UP elements. However, contributions of nonspecific upstream DNA–αCTD interactions to promoter activity have not been evaluated extensively. Here, we examine effects of removal of αCTD, upstream promoter DNA, or both on the rate of open-complex formation with promoters that lack UP elements. Deletion of αCTD decreased the composite second-order association rate constant, ka, of RNAP for the lacUV5 promoter by ≈10-fold. Much of this effect was attributable to a decrease in the isomerization rate constant, k2. Removal of promoter DNA upstream of the -35 element also decreased both ka and k2 ≈10-fold. Upstream DNA extending approximately to base pair -100 was sufficient for maximal association rates of wild-type RNAP with lacUV5 promoter fragments. The αCTD and upstream DNA did not affect dissociation rates from the open complex. We suggest that sequence-independent upstream DNA interactions with αCTD are major contributors to initiation at many (or all) promoters (not merely promoters containing UP elements) and that these interactions facilitate isomerization events occurring well downstream of the α-binding sites. In addition to highlighting the functional importance of nonspecific protein–DNA interactions, these results suggest also that UP element–αCTD interactions play an even larger role in transcription initiation than appreciated previously.
Keywords: α-subunit, nonspecific interactions, transcription initiation, isomerization
RNA polymerase (RNAP) interacts with 70–80 bp of DNA in open complexes formed at Escherichia coli promoters, spanning a region from ≈50–60 bp upstream to 20 bp downstream of the transcription-initiation site (1–2). Each of the four principal RNAP subunits (σ, α, β, and β′) contacts promoter DNA. The specificity subunit (σ) contacts at least three promoter regions: the -10 hexamer, extended -10 region, and -35 hexamer (1). The β- and β′-subunits form the catalytic center of the enyzme and contact DNA in the vicinity of and downstream from the transcription-start site (3–5).
All previously characterized contacts of RNAP with “upstream DNA” (defined as DNA located upstream of the -35 hexamer) are mediated by the C-terminal domains of the two α-subunits (5–7). The αCTDs bind in a sequence-specific manner at two preferred positions in the A+T-rich upstream DNA sequences referred to as UP elements, the “proximal” (centered at approximately -41) and “distal” (centered at approximately -52) UP-element subsites (8). RNAP lacking the αCTD can be reconstituted in vitro and retains the ability to recognize promoters and transcribe RNA, but it is defective in responding to UP elements and many activators (6, 9, 10).
UP elements have been characterized in several bacterial species and can increase promoter activity dramatically (>50-fold in vivo relative to the same core promoter with other upstream sequences) (6, 8, 11–16). An UP-element consensus sequence (Fig. 1) that increased transcription >100-fold was identified by using in vitro selection, followed by a screen for function in vivo (8, 10, 15).
Fig. 1.
Sequences upstream of the -35 element in the lacUV5 and λPR promoters, compared with the in vitro-selected full-consensus UP element (8). The -35 hexamers and consensus UP element are in shown in bold. Positions of the proximal and distal UP-element subsites are indicated. EcoRI sites used in cloning the promoters into plasmid vectors are underlined. Sequences upstream of -60 in the promoter fragments were obtained from plasmids pRLG770 or pSL6 (34–35) and are shown in Fig. 7. Numbering is with respect to the transcription start site.
The ≈8-kDa αCTD contains two helix–hairpin–helix motifs that bind to the DNA minor groove (17–20). Genetic studies (18) and interference and protection footprints (21) are in agreement with a high-resolution αCTD–DNA x-ray structure (19) concerning the mechanism of αCTD–DNA recognition. DNA recognition involves a number of residues in αCTD, most notably R265, whose side chain contacts the DNA backbone and makes water-mediated interactions with bases in the minor groove. The orientation of an αCTD with respect to the core promoter can vary and can be influenced by stabilizing interactions with σ region 4.2 and/or DNA-bound transcription factors (22–25).
The general mechanism of RNAP binding to a promoter consists of initial binding to form a closed complex, followed by conformational changes to form one or more additional intermediates before formation of the open complex (RPO) in which the DNA strands are unpaired to expose the transcription-initiation site (1, 26–28). The UP element in the rRNA promoter rrnB P1 increases the composite second-order association-rate constant, ka, ≈30-fold relative to the same core promoter with other DNA sequences substituted for the UP element (12). Most of this increase results from stimulation of initial closed-complex formation, although the rrnB P1 UP element might also affect isomerization (12). The rrnB P1 UP element also has an ≈10-fold effect on the isomerization rate constant of the λPRM promoter in a chimeric construct (29–30).
In addition to interacting specifically with UP elements, αCTD also interacts nonspecifically with upstream DNA in promoters that lack UP elements. These include the well characterized lacUV5 and λPR promoters, in which upstream sequences do not closely match the UP-element consensus (Fig. 1) and do not function in a sequence-specific manner (31). Replacement of these upstream sequences with other non-UP-element sequences had little (<2-fold) if any effect on promoter activities in vitro or in vivo (12, 29, 31). Nevertheless, αCTD-dependent interactions with upstream DNA were identified in RNAP footprints of lacUV5 and λPR (7, 31) and in crosslinking experiments with lacUV5 (5). At two other promoters that lack UP elements, galP1 and λPRM, reduced levels of open (KMnO4-reactive) complexes were observed with RNAP lacking αCTD relative to wild-type RNAP (29, 32).
Here, we report a kinetic analysis of the role of RNAP interactions with nonspecific upstream promoter DNA in vitro, using wild-type and αCTD-mutant RNAPs and promoter fragments of varying length. Our results suggest that upstream DNA interactions with αCTD play a role in the rate of RNAP association at all promoters, both with and without UP elements. Furthermore, because the quantitative effects of UP elements on association of RNAP with promoters were determined previously relative to effects of nonspecific DNA (rather than to effects of removal of αCTD), our results suggest that UP element–αCTD interactions can have an even larger effect on transcription initiation than appreciated previously.
Materials and Methods
RNAP Preparations. Wild-type native RNAP holoenzyme, purified by standard procedures (33), was a generous gift from R. Landick (University of Wisconsin, Madison). Wild-type, αΔ235, and αR265A RNAPs containing N-terminally histidine-tagged α-subunits were reconstituted in vitro from overexpressed subunits as described (18, 21). Active RNAP concentrations were determined by promoter fragment titration (26) as follows: 2 nM 32P-end-labeled lacUV5 promoter fragment (-100 to +40 or -130 to +40) was incubated with RNAP (1–20 nM total protein) at 37°C for 40 min in 10 mM Tris·Cl, pH 8.0/30 mM KCl/10 mM MgCl2/1m MDTT/100 μg/ml BSA. Heparin (Sigma) was added to 10 μg/ml, and after 30 s, reactions were filtered. Filters (0.45 μm; NC45, Schleicher & Schuell) were then washed, dried, and quantified by Cerenkov counting. Wild-type native RNAP was ≈70% active in this assay, and reconstituted RNAPs were 20–30% active. Three reconstituted preparations of each RNAP were tested, and similar results were obtained. All calculations are based on active concentrations of RNAP, and comparisons are always between wild-type and mutant RNAPs purified by the same method.
Promoter Fragments. Fragments were prepared from restriction digests of pRLG4264 or DNA amplified from pRLG593 by PCR using vector-specific primers (21). pRLG4264 contains lacUV5 sequence from -59 to +38, inserted as an EcoRI-HindIII fragment into pSL6 (34). pRLG593 contains the same lacUV5 fragment inserted into pRLG770 (6, 35). Vector sequences upstream of -59 are shown in Fig. 7, which is published as supporting information on the PNAS web site. lacUV5 fragments were 3′-end-labeled in the nontemplate strand at the HindIII site (+40) with Sequenase (United States Biochemical); [α-32P]dATP; and unlabeled dGTP, dCTP, and TTP to produce a blunt end. Upstream end points were derived from the PCR primer for the -100 to +40 fragment used in Fig. 8 (which is published as supporting information on the PNAS web site) or from the following restriction digests of pRLG4264 (for fragments shown in Figs. 2 and 4, 5, 6): -130, EcoRV; -100, SspI; -63, EcoRI; -45, NlaIV; and -42, BsaJI (Figs. 1 and 7). The 5′ overhanging ends were filled in with unlabeled dNTPs and Sequenase to produce blunt ends. The λPR fragments (-110 to +80; λPR sequences from -60 to +20 embedded in vector sequence) were prepared from BssHII–SmaI digests of pBR81 and 3′-end-labeled at the BssHII site with [α-32P]dCTP and Sequenase (36, 37). Fragments were purified from acrylamide gels (21).
Fig. 2.
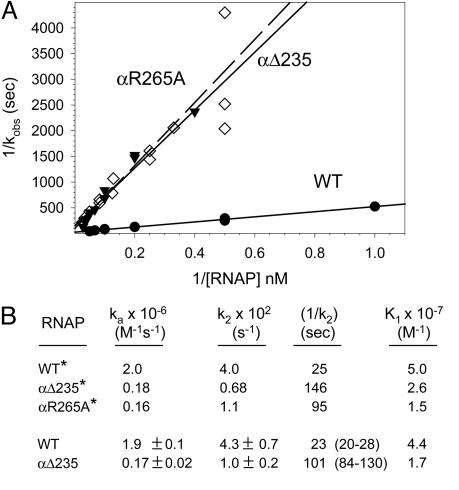
Effects of αΔ235 or αR265A on association of RNAP with the lacUV5 promoter. (A) Double reciprocal (τ) plots (see text) from reactions at a range of RNAP concentrations with the -130 to +40 fragment at 30°C. Wild-type reconstituted RNAP (•), αΔ235 RNAP (⋄), and αR265A RNAP (▾) are indicated. (B) Kinetic constants derived from the data shown in A (asterisks) or from nonlinear analysis of the same data (no asterisks; see also Fig. 9). Data from αR265A RNAP could not be fit for nonlinear analysis.
Fig. 4.
Effects of truncation of upstream DNA (to -42) on association of wild-type RNAP or αΔ235 RNAP with the lacUV5 promoter (30°C, 110 mM KCl buffer). (A) Data are displayed as a τ plot. Wild-type reconstituted RNAP and -130 to +40 lacUV5 promoter fragment (•, from Fig. 2), wild-type reconstituted RNAP and -42 to +40 lacUV5 fragment (○), and αΔ235 RNAP and -42 to +40 lacUV5 fragment (♦) are indicated. (B) Kinetic constants derived from nonlinear analysis of the data (see also Fig. 9).
Fig. 5.
Effects of upstream DNA length on association of wild-type native RNAP with the lacUV5 promoter (30°C, 110 mM KCl buffer). (A) Promoter fragments were truncated at -130 (•), -100 (▿), -63 (○), or -45 (♦). The downstream end point for each promoter fragment was +40. Data are displayed as τ plots. (B) Kinetic constants derived by nonlinear analysis of the data (see also Fig. 10).
Fig. 6.
Effects of extent of upstream DNA and αCTD on dissociation rates from lacUV5. The fraction of remaining open complexes is plotted as a function of time after addition of heparin. (A) Dissociation of native wild-type RNAP from lacUV5 fragments with upstream end points -130 (•), -100 (○), -63 (▿), or -45 (♦). Downstream end points were all +40. (B) Dissociation of reconstituted wild-type (•), αR265A (▿), and αΔ235 (♦) RNAPs from lacUV5 (-130 to +40). (C) kd values were derived from data shown and from a lacUV5 promoter fragment (-42 to +40) (data not shown), as described in Materials and Methods.
Association Kinetics. Rates of formation of heparin-resistant complexes were determined by using a filter binding assay (26, 28). RNAP (always in ≥3-fold molar excess over promoter DNA) and promoter fragment (<1 nM) were incubated at 25°C (Figs. 3 and 8) or 30°C (all other figures) in the buffer described above, except containing either 40 mM KCl (Fig. 8) or 110 mM KCl (all other figures). Aliquots were removed at time intervals to tubes containing heparin (final 10 μg/ml), filtered, and counted as described above. Counts were corrected for background retained in the absence of RNAP. The fraction of heparin-resistant complexes was determined as a fraction of input counts and plotted vs. time. Observed first-order rate constants, kobs, were determined from fitting to a single exponential [cpmobs = (cpmplateau)(1 - e-kobst)], where cpmplateau is the fraction of counts retained at equilibrium (23, 28). Maximum plateau values were ≈65% for lacUV5 and 80% for λPR. Under conditions of reversible association, observed rate constants were corrected for the equilibrium fraction of heparin-resistant complexes (28).
Fig. 3.
Effects of αR265A on association of RNAP with the λPR promoter (-110 to +80). (A) Shown are τ plots (see text) from reactions at 25°C. Wild-type reconstituted (•) and αR265A (▾) RNAP are indicated. (B) ka was derived from the linear analysis in shown in A.
Data were interpreted by using the previously characterized mechanism for RNAP association with the lacUV5 promoter under these conditions (27), and they are shown as τ plots (1/kobs vs. 1/[RNAP]), where RNAP refers to total active RNAP (Figs. 2, 3, 4, 5 and 8), or as nonlinear plots (kobs vs. [RNAP]; Figs. 9 and 10, which are published as supporting information on the PNAS web site). ka (composite second-order association rate contant) and kf (first order isomerization rate constant) were determined from τ plots by using the equation 1/kobs = (1/ka[RNAP]) + 1/kf), or they were determined from nonlinear plots by using the equation kobs = ka kf [RNAP]/(ka[RNAP] + kf). K1 (≅KB) was determined from ka and kf (K1 = ka/kf). Data were fit by using sigmaplot 8.0, and errors were determined from a weighted nonlinear analysis (28). The rate constant kf ≅ k2 under these conditions (30°C) and reflects the rate-limiting step in the formation of RPO, the isomerization of RPC to RPi (27). The nomenclature K1 (≅KB) and k2 (≅kf) is used here.
Dissociation Kinetics. RNAP (10 nM) was incubated with promoter fragments for 15 min at 30°C in the buffer described above (with 110 mM KCl). After heparin addition to 10 μg/ml, aliquots were removed at intervals, filtered, and counted. Half-lives of open complexes were determined from plots of ln(fraction remaining) vs. time. Dissociation rate constants, kd, were determined from the first-order decay equation, cpmretained = (cpmmax)e-kdt. Equivalent results were obtained with heparin at 10 μg/ml or 25 μg/ml and with 200 nM double-stranded DNA (a 60-bp “full-consensus” promoter fragment) (38) as competitor (data not shown).
Results
Deletion of αCTD Reduces the RNAP Association Rate. To determine the contribution of αCTD to open-complex formation at promoters that lack UP elements, the rates of formation of heparin-resistant complexes at the lacUV5 promoter were determined as a function of concentration of reconstituted wild-type RNAP or RNAP lacking the αCTD (αΔ235 RNAP) by using a filter binding assay (26) (see Materials and Methods). We used lacUV5 for these studies because it lacks a sequence-specific UP element (31), and its kinetics (27) and interactions with the RNAP α-subunit (5, 7) have been characterized extensively.
First-order rate constants for binding to a lacUV5 promoter fragment (-130 to +40) were determined in RNAP excess at several concentrations of RNAP by using solution conditions in which rates could be measured for both enzymes (30°C, 110 mM KCl). The data are presented as a double-reciprocal (τ) plot (39) (Fig. 2A) in which the slope is the reciprocal of the composite second order rate constant ka (1/ka = 1/K1 k2), and the y-intercept is the reciprocal of the isomerization rate constant k2 (1/k2) (see Materials and Methods). This linear plot facilitates visualization of differences in the rates of formation of the complexes, but whenever possible, the kinetic constants are provided from nonlinear analysis of the data (28) (Figs. 2B and 9), which provides a more accurate estimate of k2 and experimental error.
The overall association rate for αΔ235 RNAP and the lacUV5 promoter was ≈10-fold slower than for wild-type RNAP (ka = 1.7 × 105 M-1 s-1 for αΔ235 RNAP vs. 1.9 × 106 M-1 s-1 for wild-type RNAP). k2 for αΔ235 RNAP was ≈4-fold slower than for wild-type RNAP [τ-intercept values (1/k2) = 101 s for αΔ235 RNAP and 23 s for wild-type RNAP]. The kinetic constants for wild-type reconstituted RNAP were similar to those for wild-type native RNAP (see below) and to those obtained previously by investigators using different methods but similar solution conditions (27).
The overall association rate was also measured for a mutant RNAP containing a single amino acid substitution in αCTD (R265A) that eliminates DNA binding to the same degree as deletion of the entire C-terminal domain (6, 18). ka for αR265A RNAP was very similar to that for αΔ235 RNAP (Fig. 2). This result is consistent with the conclusion that DNA binding by αCTD is required for the effect of αCTD on the association rate, although additional interactions between αCTD and some other part of RNAP cannot be excluded.
Similar conclusions were reached from experiments with a lacUV5 fragment containing different vector-derived sequences in the upstream region, a slightly different upstream DNA end point (-100), different temperature (25°C) and salt (40 mM KCl) conditions, and different reconstituted RNAP preparations (Fig. 8). Again, the overall ka values for αΔ235 RNAP and αR265A RNAP were 10-fold lower than for wild-type RNAP and isomerization rates were slower for mutant than for wild-type RNAP. The fold difference in k2 cannot be determined with certainty in this experiment because 1/k2 observed for wild-type RNAP under these conditions is indistinguishable from zero.
Similar results were obtained also with another promoter lacking an UP element, λPR (Fig. 3). In this case, ka for αR265A RNAP was reduced ≈14-fold compared with wild-type RNAP. ka for wild-type reconstituted RNAP (2.1 × 106 M-1 s-1) was consistent with that determined previously for wild-type native RNAP and λPR under similar conditions (7.2 × 105 M-1 s-1) (26, 28). Although the τ plot suggests that a reduction in k2 is the primary effect of removal of αCTD (Fig. 3), experimental uncertainty in the data does not allow a definitive conclusion. However, other results (36) strongly support the inference that αCTD binding to upstream DNA in λPR increases k2.
Elimination of Upstream DNA Reduces the Overall Association and Isomerization Rate Constants for lacUV5. If the results given above reflect loss of sequence-independent upstream DNA interactions with αCTD, then removal of upstream DNA sequences should have effects similar to elimination of αCTD. Consistent with this prediction, the rate of association of wild-type reconstituted RNAP and a lacUV5 fragment extending only to -42 was ≈10-fold slower than for the fragment extending to -130 (ka = 1.3 × 105 vs. 1.9 × 106 M-1 s-1) (Figs. 4 and 9), similar to the effect of deleting αCTD (Fig. 2). Most of the effect of removal of upstream DNA resulted from a reduction in the isomerization rate constant (k2 = 4.3 × 10-2 s-1 vs. 0.3 × 10-2 s-1), consistent with the relatively large effect of deletion of αCTD on k2 (Fig. 2). Wild-type RNAP associated slightly faster with a fragment extending to -45 than with the -42 fragment (data not shown), consistent with the presence of an αCTD-binding site centered at -41 (5, 7).
In contrast to the large effects of removal of upstream DNA on the kinetics of association with wild-type RNAP, removal of upstream DNA had smaller effects on ka and k2 with αΔ235 RNAP (ka = 1.7 × 105 vs. 1.1 × 105 M-1 s-1, and k2 = 1.0 × 10-2 vs. 0.4 × 10-2 s-1 for the -130 and -42 fragments, respectively) (Fig. 4B). The kinetic constants for the wild-type and αΔ235 RNAPs on a promoter fragment lacking upstream DNA (-42 to +40) also differed very little (ka = 1.1 × 105 vs. 1.3 × 105 M-1 s-1, and k2 = 0.3 × 10-2 vs. 0.4 × 10-2 s-1, respectively) (Figs. 4 and 9).
Upstream DNA Length Required for Stimulation of RNAP Binding to the lacUV5 Promoter. To determine the length of upstream DNA required for maximal rates of association with RNAP, experiments were carried out by using lacUV5 fragments with upstream end points of -45, -63, -100, or -130 and wild-type native RNAP (Fig. 5). The kinetic constants ka, K1, and k2 for the two longer fragments (-100 to +40, -130 to +40) were very similar (Fig. 5) and in good agreement with those determined previously for a 211-bp lacUV5 fragment (-140 to +63) by using similar solution and temperature conditions and an abortive initiation assay (27).
Both ka and k2 for binding of RNAP to the promoter fragment extending to -45 were greatly decreased compared with the two longest fragments (Fig. 5), consistent with the effects observed with the -42 fragment shown in Fig. 4. For the intermediate-length fragment (-63), k2 was also significantly decreased relative to the longer fragments, although the reduction in ka was not as great as that observed with the -45 fragment as a consequence of an apparent increase in K1 (see Discussion). Together, the data indicate that sequences extending to -100 are sufficient for maximal rates of association, and that sequences at, or upstream of, -63 play a large role in determining the isomerization rate k2 (see Discussion).
To determine whether open complexes were formed with the short fragments, heparin-resistant complexes formed on fragments extending to -130, -63, and -45 were compared in KMnO4 footprints. The same KMnO4-reactive positions were observed on the template and nontemplate strands of each of the three promoter fragments (Fig. 7 and Fig. 11, which is published as supporting information on the PNAS web site); no differences in the identities or relative reactivities of signals were observed as a function of upstream length (see Discussion).
Dissociation Rates of lacUV5 Complexes Are Unaffected by Upstream DNA or αCTD. Dissociation rates of heparin-resistant lacUV5 promoter complexes were determined as a function of αCTD and upstream DNA length by using the filter binding assay. Complexes formed with wild-type native RNAP and lacUV5 fragments extending to -45, -63, -100, or -130 were long-lived and did not differ significantly from each other (110- to 130-min half-lives). kd values (≈1 × 10-4 s-1; Fig. 6) were very similar to that reported previously for experiments that used similar solution conditions (27). Complexes formed with reconstituted αΔ235, αR265A, or wild-type RNAPs and lacUV5 fragments extending to -130 or -42 were long-lived and had similar dissociation constants (Fig. 6 B and C, and data not shown). These results indicate that the rate-limiting step(s) in decay of the lacUV5 open complex to a competitor-sensitive intermediate are not affected by the nonspecific upstream DNA–αCTD interactions that have such profound effects on the association kinetics.
Discussion
Significance of αCTD-Nonspecific DNA Interactions for Association of RNAP. Because the αCTD is essential for viability (18) and upstream DNA cannot be removed from a promoter in vivo, we used in vitro kinetic approaches with known concentrations of active enzyme to quantify sequence-independent effects of αCTD–upstream DNA interactions. We found that removal of αCTD, upstream DNA, or both reduced the overall ka for RNAP with lacUV5 ≈10-fold. A major part of this effect was on isomerization of a heparin-sensitive intermediate (RPc) to a heparin-resistant complex (RPi) that is in rapid equilibrium with the open complex (RPO) under these conditions (27), without a concomitant decrease in the dissociation rate. Loss of αCTD–DNA interactions also resulted in a 14-fold reduction in ka for λPR, another promoter lacking an UP element. Together, these findings suggest that αCTD–upstream DNA interactions are likely to play a significant role in RNAP binding at many or all promoters, not merely promoters that have UP elements or that use transcription factors. This conclusion is consistent with previous observations that αCTD mutant RNAPs formed lower levels of KMnO4-reactive complexes than wild-type RNAP at other promoters without UP elements (29, 32). The importance of the αCTD–DNA interaction for RNAP association at many or all promoters, in addition to its importance for transcription initiation and elongation factor function (10, 40), is likely to contribute to the requirement for αCTD for cell viability (18).
DNA-binding proteins exhibit sequence-nonspecific as well as sequence-specific interactions (e.g., lac repressor) (41–42), and although nonspecific interactions are generally electrostatic and weaker, they can be crucial for regulation in vitro and in vivo. Nonspecific binding can influence the effective concentration of a protein and facilitate binding-site location by sliding, intersegment transfer, and/or hopping (43). Because the αCTDs are covalently tethered to RNAP, the locations of specific binding sites in promoters with UP elements and of nonspecific binding sites in promoters like lacUV5 are essentially the same (as indicated by hydroxyl radical footprints) (7, 8). The affinity of α for UP element DNA was estimated to be at least 15-fold greater than for nonspecific DNA (18), and we suggest that the larger effects of UP elements than nonspecific αCTD-binding sites reflect larger increases in K1, as observed at rrnB P1 (12).
Implications for UP-Element Function. The magnitude of UP-element effects on transcription or on RNAP association rates with promoters was determined previously by comparing the effects of UP element sequences to those of nonspecific sequences in the context of the same core promoter (for e.g., see refs. 6, 12, 15, 16, and 31). The results presented here suggest that because nonspecific interactions with αCTD increase the rate of RNAP association, it is likely that the previously determined effects of UP elements underestimated the overall contribution of αCTD–UP element interactions to promoter function: the specific interaction amplifies the effect of the nonspecific interaction. For example, as much as a 1,000-fold increase in ka would be predicted for the consensus UP-element sequence fused to the rrnB P1 core promoter relative to the same promoter truncated upstream of the -35 element or transcribed by RNAP lacking αCTD.
Length of Upstream DNA Required for Effects on RNAP Association. Deletion of DNA upstream of the -35 element had a large effect on the association of RNAP with lacUV5, affecting both ka and k2. Surprisingly, most of the large and unexpected effect on k2 derived from sequence between -63 and -100. The precise location of the critical sequence in this region has not been defined further.
The requirement for sequences upstream of the -35 hexamer reflects interactions with αCTD. Crosslinks of αCTD to upstream DNA occur at periodic intervals from approximately -41 to approximately -95 (5), and αCTD-dependent footprint protections extend upstream to at least -70 (7, 44). Hydroxyl radical footprints of RNAP complexes with the -130 lacUV5 fragment (Fig. 12, which is published as supporting information on the PNAS web site) indicate partial occupancy of three principal sites upstream of -35 (at -41, -52, and -63), consistent with previous reports (7).
Although sites corresponding to the UP-element proximal and distal subsites (at -41 and -52) (8) were occupied on the fragment truncated at -63 (Fig. 12), the isomerization rate for this fragment was much slower than for the full-length fragment. Sequences upstream of -63 provide additional potential binding sites for αCTD. The third principal αCTD-binding site, centered at -63, is truncated on this fragment, suggesting that this particular site might be important for lacUV5 function. Consistent with this possibility, the distal subsite of the rrnB P1 UP element (-52) was fully functional when displaced upstream to -63, although not when moved further upstream (45).
Although most of the effect of upstream sequences can be attributed to interactions with αCTD, we cannot exclude the possibility that DNA upstream of the αCTD sites could interact with another surface on RNAP. Crosslinks to an additional RNAP surface were not observed in lacUV5 open complexes with wild-type RNAP (5) (although potential transient interactions with an early transcription intermediate would not have been detected in these experiments). Deletion of upstream DNA had a small effect even with αCTD-deleted RNAP (Fig. 4) (although this effect could be attributable to a DNA-binding surface not accessible on wild-type RNAP). Potential contacts between DNA several turns upstream of the -35 element and another surface on RNAP could depend on αCTD–DNA interactions. It has been suggested that transcription factors could play this role at some promoters (46, 47).
Effects of Upstream DNA on Individual Kinetic Constants. Elimination of αCTD reduced the equilibrium constant for the initial step, K1 (≈2- to 3-fold) (Figs. 2 and 4), consistent with the stimulatory effects of UP elements on K1 observed previously at rRNA promoters (12). However, K1 was not reduced by deletion of upstream DNA: with reconstituted wild-type RNAP, there was no effect of DNA removal on K1 (Fig. 4), whereas with native RNAP, there was an increase in K1 (Fig. 5). It is possible that this difference in the effects on K1 with the two wild-type RNAPs reflects the presence or absence of other factor(s) in the enzyme preparations. For example, the ω-subunit, present in native but not reconstituted RNAP, has been proposed to facilitate RNAP assembly (48) and could result in subtle differences in the RNAPs.
The mechanism that is responsible for the effect of upstream DNA deletion on K1 with wild-type native RNAP is unclear. An increase in K1 was observed also with native RNAP upon truncation of a λPR promoter fragment, where it was attributed to reductions in the energetic costs of establishing contacts with core promoter DNA (36). Although less likely, effects are also possible from potential end-binding of RNAP to fragments whose upstream end points are close to the core promoter. However, we emphasize that the overall consequences of deleting upstream DNA correlate with the effects of removal of the αCTD; large reductions in ka and k2 occur in each case.
It is likely that the isomerization step, k2, involves conformational changes that occur far downstream of the DNA regions directly contacted by αCTD. We have observed differences in the extent of protection by RNAP in the extended -10 region (as well as in the upstream region) when comparing rrnB P1 footprints with αCTD mutant vs. wild-type RNAP (ref. 49 and W.R. and R.L.G., unpublished data). The accompanying article (36) reports differences in the extended -10 region as a function of DNA upstream of λPR in KMnO4 footprints. However, we did not detect such differences in lacUV5 as a function of upstream DNA length in footprints with wild-type RNAP using hydroxyl radicals or KMnO4 as probes (Figs. 11 and 12). Differences in the behavior of λPR and lacUV5 could reflect the structures of the complexes formed by the two promoters or the locations of the residues potentially reactive with KMnO4.
Although UP elements have been implicated in isomerization previously (12, 29, 30) and some activator proteins that bind far from the transcription-start site [e.g., λcI (50, 51) and CRP (52)] also increase k2, the mechanism by which αCTD–DNA interactions or upstream-bound activator proteins increase isomerization step(s) remains a major unresolved issue in the mechanism of gene expression. The companion article (36) to this article suggests that the entry of downstream DNA into the “jaws” of the enzyme is affected by upstream DNA interactions with αCTD at λPR (36), although the molecular basis for this effect is not yet understood. In any case, the data reported here indicate that αCTD–DNA interactions play a major role in the rate of association of RNAP, and we suggest that this role, although amplified by activators and UP elements, may be a general property of the initiation process at all promoters.
Supplementary Material
Acknowledgments
We thank R. Saecker, T. Record, and C. Davis for providing many helpful discussions, sharing results before publication, and giving comments on the manuscript; T. Gaal for suggestions; and C. Vrentas for performing one of the KMnO4 experiments. This work was supported by National Institutes of Health Grant R01 GM37048 (to R.L.G.).
Author contributions: W.R. and R.G. designed research; W.R. performed research and analyzed data; and W.R. and R.G. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: RNAP, RNA polymerase.
References
- 1.Record, M. T., Jr., Reznikoff, W. S., Craig, M. L., McQuade, K. L. & Schlax, P. J. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology, ed. Neidhardt, F.C. (Am. Soc. Microbiol., Washington, D.C.), Vol. 1, pp. 792-820. [Google Scholar]
- 2.Ozoline, O. N. & Tsyganov, M. A. (1995) Nucleic Acids Res. 23, 4533-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korzheva, N. & Mustaev, A. (2001) Curr. Opin. Microbiol. 4, 119-125. [DOI] [PubMed] [Google Scholar]
- 4.Murakami, K. & Darst, S. (2003) Curr. Opin. Struct. Biol. 1, 31-39. [DOI] [PubMed] [Google Scholar]
- 5.Naryshkin, N., Revyakin, A., Kim, Y., Mekler, V. & Ebright, R. H. (2000) Cell 101, 601-611. [DOI] [PubMed] [Google Scholar]
- 6.Ross, W., Gosink, K. K., Salomon, J., Igarashi, K., Zou, C., Ishihama, A., Severinov, K. & Gourse, R. L. (1993) Science 262, 1407-1413. [DOI] [PubMed] [Google Scholar]
- 7.Kolb, A., Igarashi, K., Ishihama, A., Lavigne, M., Buckle, M. & Buc, H. (1993) Nucleic Acids Res. 21, 319-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estrem, S. T., Ross, W., Gaal, T., Chen, Z. W., Niu, W., Ebright, R. H. & Gourse, R. L. (1999) Genes Dev. 13, 2134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Igarashi, K. & Ishihama, A. (1991) Cell 65, 1015-1022. [DOI] [PubMed] [Google Scholar]
- 10.Gourse, R. L., Ross, W. & Gaal, T. (2000) Mol. Microbiol. 37, 687-695. [DOI] [PubMed] [Google Scholar]
- 11.Banner, C. D., Moran, C. P., Jr., & Losick, R. (1983) J. Mol. Biol. 168, 351-365. [DOI] [PubMed] [Google Scholar]
- 12.Rao, L., Ross, W., Appleman, J. A., Gaal, T., Leirmo, S., Schlax, P. J., Record, M. T., Jr., & Gourse, R. L. (1994) J. Mol. Biol. 235, 1421-1435. [DOI] [PubMed] [Google Scholar]
- 13.Fredrick, K., Caramori, T., Chen, Y. F., Galizzi, A. & Helmann, J. D. (1995) Proc. Natl. Acad. Sci. USA 92, 2582-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helmann, J. D. (1995) Nucleic Acids Res. 23, 2351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estrem, S. T., Gaal, T., Ross, W. & Gourse, R. L. (1998) Proc. Natl. Acad. Sci. USA 95, 9761-9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirvonen, C. A., Ross, W., Wozniak, C. E., Marasco, E., Anthony, J. R., Aiyar, S. E., Newburn, V. & Gourse, R. L. (2001) J. Bacteriol. 183, 6305-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeon, Y. H., Negishi, T., Shirakawa, M., Yamazaki, T., Fujita, N., Ishihama, A. & Kyogoku, Y. (1995) Science 270, 1495-1497. [DOI] [PubMed] [Google Scholar]
- 18.Gaal, T., Ross, W., Blatter, E. E., Tang, H., Jia, X., Krishnan, V. V., Assa-Munt, N., Ebright, R. H. & Gourse, R. L. (1996) Genes Dev. 10, 16-26. [DOI] [PubMed] [Google Scholar]
- 19.Benoff, B, Yang, H., Lawson C. L., Parkinson, G., Liu, J., Blatter, E., Ebright, Y. W., Berman, H. M. & Ebright, R. H. (2002) Science 297, 1562-1566. [DOI] [PubMed] [Google Scholar]
- 20.Shao, X. & Grishin, N. V. (2000) Nucleic Acids Res. 28, 2643-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross, W., Ernst, A. & Gourse, R. L. (2001) Genes Dev. 15, 491-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, D. J., Busby, S. J. & Lloyd, G. S. (2003) J. Biol. Chem. 278, 52944-52952. [DOI] [PubMed] [Google Scholar]
- 23.Ross, W., Schneider, D. A., Paul, B. J., Mertens, A. & Gourse, R. L. (2003) Genes Dev. 17, 1293-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen, H., Tang, H. & Ebright, R. H. (2003) Mol. Cell 11, 1621-1633. [DOI] [PubMed] [Google Scholar]
- 25.Paul, B. J., Ross, W., Gaal, T. & Gourse, R. L. (2004) Annu. Rev. Genet. 38, 749-770. [DOI] [PubMed] [Google Scholar]
- 26.Roe, J. H., Burgess, R. R. & Record, M. T., Jr. (1984) J. Mol. Biol. 176, 495-522. [DOI] [PubMed] [Google Scholar]
- 27.Buc, H. & McClure, W. R. (1985) Biochemistry 24, 2712-2723. [DOI] [PubMed] [Google Scholar]
- 28.Saecker, R. M., Tsodikov, O., McQuade, K. L., Schlax, P. E., Capp, M. W. & Record, M. T., Jr. (2002) J. Mol. Biol. 319, 649-671. [DOI] [PubMed] [Google Scholar]
- 29.Tang, Y., Murakami, K., Ishihama, A. & deHaseth, P. L. (1996) J. Bacteriol. 178, 6945-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strainic, M. G., Jr., Sullivan, J. J., Velevis, A. & deHaseth, P. L. (1998) Biochemistry 37, 18074-18080. [DOI] [PubMed] [Google Scholar]
- 31.Ross, W., Aiyar, S. E., Salomon, J. & Gourse, R. L. (1998) J. Bacteriol. 180, 5375-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burns, H. D., Ishihama, A. & Minchin, S. D. (1999) Nucleic Acids Res. 27, 2051-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgess, R. R. & Jendrisak, J. J. (1975) Biochemistry 14, 4634-4638. [DOI] [PubMed] [Google Scholar]
- 34.Gosink, K. K., Ross, W., Leirmo, S., Osuna, R., Finkel, S. E., Johnson, R. C. & Gourse, R. L. (1993) J. Bacteriol. 175, 1580-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross, W., Thompson, J. F., Newlands, J. T. & Gourse, R. L. (1990) EMBO J. 9, 3733-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis, C. A., Capp, M. W., Record, M. T., Jr., & Saecker, R. M. (2005) Proc. Natl. Acad. Sci. USA 102, 285-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Craig, M. L., Suh, W. C. & Record, M. T., Jr. (1995) Biochemistry 34, 15624-15632. [DOI] [PubMed] [Google Scholar]
- 38.Gaal, T., Ross, W., Estrem, S. T., Nguyen, L. H., Burgess, R. R. & Gourse, R. L. (2001) Mol. Microbiol. 42, 939-954. [DOI] [PubMed] [Google Scholar]
- 39.McClure, W. R. (1980) Proc. Natl. Acad. Sci. USA 77, 5634-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Busby, S. J. & Ebright, R. H. (1999) J. Mol. Biol. 293, 199-213. [DOI] [PubMed] [Google Scholar]
- 41.von Hippel, P. H., Revzin, A, Gross, C. A., & Wang, A. C. (1974) Proc. Natl. Acad. Sci. USA 71, 4808-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Record, M. T., Jr., Lohman, T. M. & deHaseth, P. L. (1976) J. Mol. Biol. 107, 145-158. [DOI] [PubMed] [Google Scholar]
- 43.von Hippel, P. H. (2004) Science 305, 350-352. [DOI] [PubMed] [Google Scholar]
- 44.Buckle, M., Buc, H. & Travers, A. A. (1992) EMBO J. 11, 2619-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newlands, J. T., Josaitis, C. A., Ross, W. & Gourse, R. L. (1992) Nucleic Acids Res. 20, 719-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eichenberger, P., Dethiollaz, S., Buc, H. & Geiselmann, J. (1997) Proc. Natl. Acad. Sci. USA 94, 9022-9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Susanna, K. A., van der Werff, A., den Hengst, C. D., Calles, B., Salas, M., Venema, G., Hamoen, L. W. & Kuipers, O. P. (2004) J. Bacteriol. 186, 1120-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minakhin, L, Bhagat, S., Brunning, A., Campbell, E. A., Darst, S. A., Ebright, R. H. & Severinov, K. (2001) Proc. Natl. Acad. Sci. USA 98, 892-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aiyar, S. E., Gourse, R. L. & Ross, W. (1998) Proc. Natl. Acad. Sci. USA 95, 14652-14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dove, S. L, Huang, F. W. & Hochschild, A. (2000) Proc. Natl. Acad. Sci. USA 97, 13215-13220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nickels, B. E., Dove, S. L., Murakami, K. S., Darst, S. A. & Hochschild, A. (2002) J. Mol. Biol. 324, 17-34. [DOI] [PubMed] [Google Scholar]
- 52.Niu, W., Kim, Y., Tau, G., Heyduk, T. & Ebright, R. H. (1996) Cell 87, 1123-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.