Abstract
Objectives:
The increase in resistance of methicillin resistant Staphylococcus aureus (MRSA) strains to vancomycin has been perceived as a formidable threat in the therapeutic fields. The present study investigated the vancomycin resistance traits of MRSA isolates [vancomycin resistant S. aureus (VRSA)] collected from burn patients.
Materials and Methods:
Twenty-nine of 40 isolates of Staphylococcus spp. were identified as S. aureus which were further tested against 20 commercially available antibiotics to determine antibiotic susceptibility patterns.
Results:
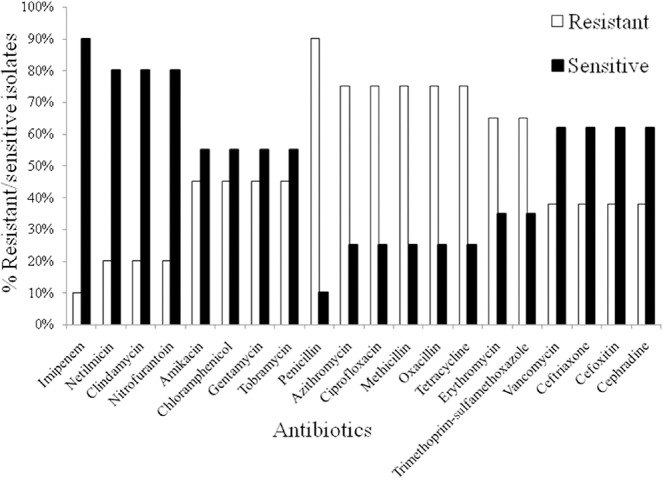
Imipenem was the most potential antibiotic resulting in 90% sensitivity, followed by netilmicin, clindamycin, and nitrofurantoin (80% sensitivity). All isolates were found to be resistant to penicillin. Approximately 75% of them were found to be resistant to methicillin, oxacillin, azithromycin, cipro-floxacin, and tetracycline. Approximately 45% isolates exhibited resistance to amikacin, chloramphenicol, gentamycin, and tobramycin. Twenty-one of the 29 strains of S. aureus were MRSA, of which 11 were resistant to vancomycin when employing the disc diffusion method. However, when the broth micro-dilution procedure was used to measure the minimum inhibitory concentration (MIC) of vancomycin, eight isolates were resistant to vancomycin, six with an MIC of 32 μg/mL and two with an MIC of 64 μg/mL.
Conclusion:
A significant fraction of VRSA was found among MRSA strains in this study, revealing the necessity for new and effective drugs against MRSA.
Keywords: Antibiotic resistance, Methicillin resistant Staphylococcus aureus (MRSA), Minimum inhibitory concentration (MIC), Vancomycin intermediate S. aureus (VISA), Vancomycin resistant S. aureus (VRSA), Vancomycin sensitive S. aureus (VSSA)
1. Introduction
Over the past few decades, there has been an alarming increase in the prevalence of antibiotic resistant pathogens and strains in serious infections [1,2,3,4,5,6,7,8,9,10,11]. The occurrence of bacterial infection had decreased with the discovery of penicillin in 1940 until Staphylococcus aureus began producing β-lactamase, which destroys the penicillin β-lactam core ring [11,12]. This increase in resistance towards penicillin drove the development of methicillin drugs, which are virtually resistant against many genetic variations of the β-lactamase enzyme. Infection by S. aureus was well controlled using methicillin until the isolation of the first strain of methicillin resistant S. aureus (MRSA) in 1961 [1,12,13]. Since then, MRSA has become endemic in hospitals and nursing homes worldwide [1,3,7,14].
Burn patients are susceptible to infection, especially skin and soft tissue infections such as burn wound impetigo, burn wound cellulitis, and invasive types infection, because of their impaired immune system. Hence hospital-associated strains of MRSA have become a great concern, mostly due to treatment failure [7]. S. aureus was noted in the skin and mucosa of up to 40% of all burn patients of which 30% had severe cases of toxic shock [4,15,16]. In one study, the frequency of S. aureus infection reached 47% in burn patients and the prevalence of MRSA was up to 45% [15]. Although vancomycin has been the most reliable therapeutic agent against infections caused by MRSA, there has been an alarming emergence of vancomycin-resistant S. aureus (VRSA), possibly due to: (1) the widespread use of vancomycin to treat infections caused by MRSA; (2) a patient's immune status; (3) surgical procedures; and (4) involvement of healthcare workers infected with MRSA [17,18,19,20,21,22,23]. With the increasing resistance of S. aureus as well as the emergence of multidrug resistant strains, the choice of medication remains one of the most challenging concerns in the burn management unit.
Although nosocomial infections are associated with remarkable morbidity and mortality both in developed and developing countries, information concerning such infections in Bangladesh in the international literature is limited [4,7]. The detection rates of MRSA in hospitals in different cities in Bangladesh were recently reported to be 32–63%, which is high compared with the United States and European countries [18]. In this context, information on the antimicrobial susceptibility patterns of MRSA could help in the selection of appropriate treatment. Based on this rationale, the current study investigated the occurrence of MRSA strains in burn patients and the susceptibility patterns of these strains against various antibiotics used to treat hospitalized patients in Bangladesh.
2. Materials and methods
2.1. Study area, sampling, and sample processing
The experiment was carried out in the Microbiology Laboratory of the Department of Microbiology, Stamford University Bangladesh, Dhaka, Bangladesh from April 23, 2012 to January 15, 2013. A total of 40 wound samples from patients with tertiary burns of partial or full thickness (deep reticular dermis) were collected aseptically with a sterile cotton swab by a clinician wearing gloves (US Safety & Supply Co.) and a mask in the burn unit of Dhaka Medical College Hospital, Dhaka, Bangladesh [7,24]. The patients were under treatment with antibiotics including trimetho-prim–sulfamethoxazole, methicillin, and ceftriaxone (Oxoid, UK). All patients were men aged between 20 years and 45 years from a lower middle-class community. The samples were inoculated on Mannitol salt agar (MSA) (HiMedia, India) plates immediately after sample collection for isolation of S. aureus and transported to the laboratory as early as possible [19,23,25].
2.2. Isolation and identification of Staphylococcus aureus
All MSA plates were incubated for 24 hours at 37°C. After incubation, isolated colonies suspected to be Staphylococcus were allowed to grow on nutrient agar plates (HiMedia, India) and then identified microscopically, biochemically, and serologically [19,20]. For microscopic observation, a pure colony was selected and subjected to Gram staining. Then the shape, arrangement, and Gram reactions of the isolates were observed under a light microscope (Max-plank-Ring 21 D-65205, Wiesbaden, Germany) (at a magnification of 100x) [25]. Required confirmatory biochemical tests including catalase and triple sugar iron agar tests were performed to identify suspected S. aureus following standard protocols [25].
2.3. Hemolytic activity and coagulase test
The hemolytic activity of S. aureus isolates was tested using blood agar plates containing 5% defibrinated sheep blood. An isolated colony from a nutrient agar (NA) plate was inoculated on blood agar and incubated at 37°C for 24 hours. The hemolytic zones were characterized as α (partial hemolysis), β (complete hemoly-sis), and γ (no hemolysis) depending on the extent of each colony [25]. A coagulase test (Becton Dickinson Microbiology Systems, USA) was performed to differentiate the hospital-acquired isolates. For this purpose, 10 μL of the antiserum was placed on the slide and a suspension of the organism was added. Agglutination was observed against light and the results were recorded [25].
2.4. Assay of antibacterial susceptibility
A standard agar-disc diffusion (Kirby–Bauer) assay using Mueller–Hinton agar (MHA) (HiMedia, India) plates was conducted to determine the susceptibility of the isolated S. aureus to different antibiotics [26,27,28]. A suspension of the test organism was prepared by adjusting the turbidity of the broth in phosphate buffer saline by comparing with that of the McFarland standard solution of 0.5 [27,28]. By means of a sterile cotton swab, a uniform lawn of bacterial growth was prepared on the MHA plates. A total of 20 antibiotic discs including amikacin (30 μg), azithromycin (15 μg), ceftriaxone (30 μg), cefoxitin (30 μg), cephradine (30 μg), chlor-amphenicol (30 μg), ciprofloxacin (5 μg), clindamycin (2 μg), erythromycin (15 μg), gentamycin (10 μg), imipenem (10 μg), methicillin (5 μg), netilmicin (30 μg), nitrofurantoin (300 μg), oxacillin (1 μg), penicillin (10 μg), tetracycline (30 μg), tobramycin (10 μg), trimethoprim/sulfamethoxazole (25 μg), and vancomycin (30 μg) were applied aseptically on the surface of the inoculated plates in an appropriate spatial arrangement using a sterile needle. The plates were incubated at 37°C for 12–18 hours and examined for zones of inhibition (mm) [26,29].
2.5. Identification of MRSA
For the detection of MRSA, oxacillin (1 μg) and methicillin (5 μg) were introduced on the MHA plates against the growth of S. aureus. For this purpose, a bacterial suspension was prepared in sterile saline by selecting colonies produced by overnight incubation on NA agar plates. After 5–7 hours of incubation, the cell turbidity was adjusted to 0.5 McFarland standards [27,28]. Subsequently, the suspensions were inoculated onto MHA plates and the antibiotic discs were then placed onto the plates [20,21]. All plates were incubated for 24 hours at 37°C to observe for oxacillin and methi-cillin resistant S. aureus.
2.6. Identification of VRSA through disc diffusion methods
MHA plates were inoculated with the bacterial suspension which was previously adjusted to 0.5 McFarland standards. Afterward, a 30 μg vancomycin disc and a blank disc as a control were aseptically placed over the surface of the MHA plates at a distance of 5 mm to observe the range of the zone diameter for the detection of strains of VRSA [20,21].
2.7. Determination of vancomycin resistance by minimum inhibitory concentration test
The minimum inhibitory concentration (MIC) of vancomycin (Oxoid, UK) was determined by the tube dilution method [7,30,31,32]. Muller–Hinton Broth was prepared with 4–512 μg/mL of vancomycin. By using a direct colony suspension method, 0.5 McFarland equivalent bacterial inoculums were prepared in normal saline after culturing for 24 hours on an agar plate. The suspension was further diluted to achieve the desired inoculum concentration. All strains were spotted onto Muller–Hinton plates containing different concentrations of vancomycin. The plates were incubated for 24 hours at 37°C and checked for any visible growth [26].
3. Results
In recent years, Staphylococcus aureus has become one of the most dangerous pathogens due to its increased resistance to β-lactam antibiotics and vancomycin [33,34,35]. Studies showed that MRSA is a causative agent of hospital-acquired infection and an incipient community pathogen in many geographical regions [13,36,37,38]. In the present study, the isolation rate of S. aureus (72.5%) from burn wound patients was high, as the microorganism was confirmed in 29 of the 40 isolated strains of Staphylococcus spp. based on cultural, biochemical, and coagulase properties (Table 1). In addition, imipenem was found to be the most effective antibiotic against the isolates with 90% of strains exhibiting sensitivity to this drug (Fig. 1). Most of the isolates (80%) were also sensitive to netilmicin, clindamycin, and nitrofurantoin. For vancomycin, 62% of isolates showed sensitivity. Almost 55% of isolates were sensitive to amikacin, chloramphenicol, gentamycin, and tobramycin. However, 26 of 29 strains of S. aureus were resistant to penicillin G and 75% of isolates were resistant to azithromycin, ciprofloxacin, methicillin, oxacillin, and tetracycline. Approximately 65% of isolates exhibited resistance to erythromycin, and trimethoprim–sulfamethoxazole (Fig. 1).
Table 1.
Biochemical determination of the presumptively selected Staphylococcus aureus.a
| Isolates ID | Catalase test | TSI | Hemolytic activity | Coagulase test | |
|---|---|---|---|---|---|
| Slan | Butt | ||||
| S-1 | + | A | A | β | + |
| S-2 | + | A | A | β | + |
| S-3 | + | A | A | β | + |
| S-4 | + | A | A | β | + |
| S-5 | + | A | A | β | + |
| S-6 | + | A | A | α | + |
| S-7 | + | A | A | β | + |
| S-8 | + | A | A | β | + |
| S-9 | + | A | A | β | + |
| S-10 | + | A | A | β | + |
| S-11 | + | A | A | Β | + |
| S-12 | + | A | A | β | + |
| S-13 | + | A | A | β | + |
| S-14 | + | A | A | β | + |
| S-15 | + | A | A | β | + |
| S-16 | + | A | A | β | + |
| S-17 | + | A | A | β | + |
| S-18 | + | A | A | β | + |
| S-19 | + | A | A | β | + |
| S-20 | + | A | A | β | + |
| S-21 | + | A | A | β | + |
| S-22 | + | A | A | β | + |
| S-23 | + | A | A | β | + |
| S-24 | + | A | A | β | + |
| S-25 | + | A | A | β | + |
| S-26 | + | A | A | β | + |
| S-27 | + | A | A | β | + |
| S-28 | + | A | A | α | + |
| S-29 | + | A | A | β | + |
| S-30 | + | A | A | β | + |
| S-31 | + | A | A | β | + |
| S-32 | + | A | A | β | + |
| S-33 | + | A | A | β | + |
A = acidic reaction; S = Staphylococcus aureus isolates; TSI = Triple Sugar Iron.
aAll the experiments were performed in triplicates and the results were reproducible.
Fig. 1.
Resistance and susceptibility patterns of Staphylococcus aureus towards commonly used antibiotics including amikacin (30 μg), azithromycin (15 μg), ceftriaxone (30 μg), cefoxitin (30 μg), cephradine (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), clindamycin (2 μg), erythromycin (15 μg), gentamicin (10 μg), imipenem (10 μg), methicillin (5 μg), netilmicin (30 μg), nitrofurantoin (300 μg), oxacillin (1 μg), penicillin (10 μg), tetracycline (30 μg), tobramycin (10 μg), trimethoprim/sulfamethoxazole (25 μg), and vancomycin (30 μg). Among 20 tested antibiotics, imipenem was found to be the potent antibiotic against all isolated S. aureus. However, 90% of strains exhibited resistance to penicillin G.
Twenty-one of the 29 S. aureus isolates studied were found to be MRSA (Table 2). The prevalence of methicillin resistance among staphylococci isolated from burn patients in our hospital has not been determined accurately to date. In this study, the prevalence of MRSA was 72% (Table 2), which varied from findings in other studies in other countries. In three separate studies in Iran, 56%, 72%, and 58% of staphylococci were identified as methicillin resistant [39,40,41]. Interestingly, a study in Korea in 2001 showed that the incidence of MRSA in burn cases could be as high as up to 98% [42]. A study in the United States in 2006 showed the rate of MRSA in a burn center was 33% [43]. Therapeutic strategies for severe MRSA infections are indeed limited to a few antibiotics including vancomycin. Thus the acquisition of high-level vancomycin resistance by MRSA is a major health concern. Genomic studies have provided information on the evolutionary history of VRSA and identified genetic features that may focus on the acquisition mechanism of vancomycin resistance genes [37,38,39,40,41,42,43,44].
Table 2.
Detection of methicillin resistant Staphylococcus aureus (MRSA).a
| No of isolates | Antibiotics | Resistant presence (%) |
|---|---|---|
| n = 21 (out of 29) | Oxacillin & methicillin | |
| S-2 | R | 72% |
| S-3 | R | |
| S-4 | R | |
| S-5 | R | |
| S-6 | R | |
| S-8 | R | |
| S-9 | R | |
| S-10 | R | |
| S-12 | R | |
| S-14 | R | |
| S-21 | R | |
| S-23 | R | |
| S-24 | R | |
| S-25 | R | |
| S-26 | R | |
| S-27 | R | |
| S-28 | R | |
| S-29 | R | |
| S-31 | R | |
| S-32 | R | |
| S-33 | R |
R = resistant.
aAll the experiments were performed in triplicates and the results were reproducible reproducible.
To determine vancomycin resistance among isolated MRSA in the current study, S. aureus strains were further tested using both agar disc diffusion and broth microdilution procedures (MIC). A total of 11 of the MRSA isolates (S-8, S-9, S-14, S-21, S-23, S-25, S-26, S-27, S-28, S-31, and S-33) were resistant to vancomycin with the disc diffusion method (Table 3). Subsequently, MIC assay showed that eight strains of S. aureus (28%) were resistant to vancomycin. Two of these strains had MIC values of 64 μg/mL and the other six strains had MIC values of 32 μg/mL, which were defined as VRSA in accordance with the laboratory breakpoints published by the Clinical and Laboratory Standards Institute [45]. Interestingly, among 29 samples 16 strains were noted to be vancomycin intermediate S. aureus (VISA), eight strains with MIC values of 8 μg/mL and another eight strains with MIC values of 16 μg/mL, and five samples were vancomycin sensitive S. aureus (VSSA) with MIC values of 5 μg/mL (Table 4). We assume the MIC values of vancomycin for these 21 MRSA isolates varied because of different levels of expression of the vanA gene in these isolates or other mechanisms [46,47].
Table 3.
Identification of vancomycin resistant Staphylococcus aureus (VRSA) among MRSA through disc diffusion method.a
| No of isolates | Antibiotic | Resistant presence (%) |
|---|---|---|
| n = 11 (out of 21) | Vancomycin | |
| S-8 | R | 52% |
| S-9 | R | |
| S-14 | R | |
| S-21 | R | |
| S-23 | R | |
| S-25 | R | |
| S-26 | R | |
| S-27 | R | |
| S-28 | R | |
| S-31 | R | |
| S-33 | R |
R = resistant.
aAll the experiments were performed in triplicates and the results were reproducible.
Table 4.
Determination of vancomycin susceptibility pattern (VRSA, VISA & VSSA) of Staphylococcus aureus through minimal inhibitory concentration (MIC).a
| S. aureus strain | Vancomycin MIC (mg/mL) | Vancomycin phenotype | Resistant presence (%) |
|---|---|---|---|
| S-21 | 32 | VRSA | 28% |
| S-23 | 32 | ||
| S-25 | 64 | ||
| S-26 | 32 | ||
| S-27 | 64 | ||
| S-28 | 32 | ||
| S-31 | 32 | ||
| S-33 | 32 | ||
| S-1 | 8 | VISA | 55% |
| S-2 | 8 | ||
| S-3 | 8 | ||
| S-4 | 16 | ||
| S-6 | 16 | ||
| S-8 | 16 | ||
| S-10 | 8 | ||
| S-11 | 8 | ||
| S-12 | 16 | ||
| S-14 | 16 | ||
| S-16 | 8 | ||
| S-19 | 8 | ||
| S-29 | 16 | ||
| S-30 | 16 | ||
| S-32 | 16 | ||
| S-33 | 8 | ||
| S-5 | 5 | VSSA | 17% |
| S-9 | 5 | ||
| S-18 | 5 | ||
| S-20 | 5 | ||
| S-22 | 5 |
MIC = minimal inhibitory concentration, VISA = vancomycin intermediate Staphylococcus aureus, VRSA = vancomycin resistant Staphylococcus aureus, VSSA = vancomycin sensitive Staphylococcus aureus.
aAll the experiments were performed in triplicates and the results were reproducible.
In the past few years, several antibiotics have been noted to be less effective in the context of disease mitigation worldwide, as an array of pathogenic microorganisms are gradually becoming resistant to these therapeutic agents [48]. This raises the possibility of greatly increased mortality from simple infections and treatment-mediated failures. Along with multi-drug resistant (MDR) and extensively-drug resistant Mycobacterium tuberculosis, the MRSA strains, VRSA strains, coagulase-negative staphylococci, glycopeptide intermediate sensitive S. aureus, vancomycin-resistant Enterococcus species, penicillin-resistant Streptococcus pneumoniae, and the extended-spectrum β-lactamase producing bacteria are highly prominent [48]. In Bangladesh, recent studies of burn patients revealed huge growth in aerobic viable bacteria including Pseudomonas spp., S. aureus, and Klebsiella spp, Enter-obacter spp. and Escherichia coli of which most were found to be MDR [7,14]. The current findings on the prevalence of VRSA strains among MRSA isolates further demonstrates the necessity for research emphasis on the microbiology of burn injuries, which in turn, could enhance the overall treatment of burns [7,14].
The major drawback of this study was the lack of molecular characterization of the isolates and detection of virulent genes, which could be investigated in future research. Such study may help physicians generate new treatment policies as well as to develop new drugs against the resistant properties of isolates.
The current study revealed a high percentage of VISA isolates compared with VRSA and VSSA in isolated MRSA, highlighting the necessity for local or country-based investigations to characterize and monitor MRSA and to develop strategies that will accelerate MRSA management and control. In our study, imipenem was found to be the most effective drug against MRSA. In addition, the application of antibiotic combination therapy against VISA and VRSA, and maintenance of proper hygiene by hospitalized patients and staff could effectively reduce the rate and dissemination of such cases. Further molecular studies are required to identify resistance-conferring genes.
Acknowledgments
We thank the Microbiology Laboratory, Stamford University Bangladesh for the technical facilities.
Footnotes
Conflict of interests: none.
References
- [1].Brown D, Edwards D, Hawkey P, Morrison D, Ridqway G, Towner K, et al. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA) Antimicrob Agents Chemother. 2005;65:1000–8. doi: 10.1093/jac/dki372. [DOI] [PubMed] [Google Scholar]
- [2].Chessa D, Ganau G, Mazzarello V. An overview of Staphylococcus epidermidis and Staphylococcus aureus with a focus on developing countries. J Infect Dev Ctries. 2015;9:547–50. doi: 10.3855/jidc.6923. [DOI] [PubMed] [Google Scholar]
- [3].Jones RN. Key considerations in the treatment of complicated Staphylococcal infections. Clin Microbiol Infect. 2008;14:3–9. doi: 10.1111/j.1469-0691.2008.01923.x. [DOI] [PubMed] [Google Scholar]
- [4].Dutta S, Hassan MR, Rahman F, Jilani MFA, Noor R. Study of antimicrobial susceptibility of clinically significant microorganisms isolated from selected areas of Dhaka, Bangladesh. Bang J Med Sci. 2013;12:34–42. [Google Scholar]
- [5].Noor R, Hossain A, Munshi SK, Rahman F, Kamal SM. Slide drug susceptibility test for the detection of multi-drug resistant tuberculosis in Bangladesh. J Infect Chemother. 2013;19:818–24. doi: 10.1007/s10156-013-0566-0. [DOI] [PubMed] [Google Scholar]
- [6].Noor R, Akhter S, Rahman F, Munshi SK, Kamal SMM, Feroz F. Frequency of extensively drug resistant tuberculosis (XDR-TB) among retreatment cases in NIDCH, Dhaka, Bangladesh. J Infect Chemother. 2013;19:243–8. doi: 10.1007/s10156-012-0490-8. [DOI] [PubMed] [Google Scholar]
- [7].Alam SMS, Kalam MA, Munna MS, Munshi SK, Noor R. Isolation of pathogenic microorganisms from burn patients admitted in Dhaka Medical Collage and Hospital and demonastration of their drug-resistant traits. Asian Pac J Trop Dis. 2014;4:402–7. [Google Scholar]
- [8].Aurin TH, Munshi SK, Kamal SM, Rahman MM, Hossain MS, Marma T, et al. Molecular approaches for detection of the multi-drug resistant tuberculosis (MDR-TB) in Bangladesh. PLoS ONE. 2014;9:e99810. doi: 10.1371/journal.pone.0099810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Molton JS, Tambyah PA, Ang BSP, Ling ML, Fisher DA. The global spread of healthcare-associated multidrug-resistant bacteria: a perspective from Asia. Clinical Infec Dis. 2013;6:1310–8. doi: 10.1093/cid/cit020. [DOI] [PubMed] [Google Scholar]
- [10].Tenover FC. Mechanisms of antimicrobial resistance in bacteria. Am J Med. 2006;119:3–10. doi: 10.1016/j.amjmed.2006.03.011. [DOI] [PubMed] [Google Scholar]
- [11].Khan SA, Feroz F, Noor R. Study of extended spectrum β-lactamase producing bacteria from urinary tract infection in Dhaka city, Bangladesh. Tzu Chi Med J. 2013;25:39–42. [Google Scholar]
- [12].Peacock S. Staphylococcus aureus. In: Gillespie S, Hawkey P, editors. Principle and practice of clinical bacteriology. 2nd ed. England: John Wiley and Sons Ltd; 2006. p. 620. [Google Scholar]
- [13].Lowy F. Staphylococcal infections. In: Fauci A, Braunwarld E, Kasper D, Hauser S, Longo D, Jameson J, editors. Harrison's principles of internal medicine. 16th ed. New York: MacGraw-Hill Companies; 2005. p. 2958. [Google Scholar]
- [14].Noor R, Munna MS. Emerging diseases in Bangladesh: current microbiological research. Tzu Chi Med J. 2015;27:49–53. doi: 10.1016/j.tcmj.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bagdonas R, Tamelis A, Rimdeika R. Staphylococcus aureus infection in the surgery of burns. Medicina. 2003;39:1078–81. [PubMed] [Google Scholar]
- [16].Matsushima A, Kuroki Y, Nakajima S, Sakai T, Kojima H, Ueyama M. Low level of TSST-1 antibody in burn patients with toxic shock syndrome caused by methicillin-resistant Staphylococcus aureus. J Burn Care Res. 2015;36:e120–4. doi: 10.1097/BCR.0000000000000128. [DOI] [PubMed] [Google Scholar]
- [17].Montazeri EA, Khosravi AD, Jolodar A, Ghaderpanah M, Azarpira S. Identification of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from burn patients by multiplex PCR Burns. 2015;41:590–4. doi: 10.1016/j.burns.2014.08.018. [DOI] [PubMed] [Google Scholar]
- [18].Haq JA, Rahman MM, Haque Asna SM, Hossain MA, Ahmed I, Haq T, et al. Methicillin-resistant Staphylococcus aureus in Bangladesh–a multi centre study. Int J Antimicro Agents. 2005;25:276–7. doi: 10.1016/j.ijantimicag.2005.01.004. [DOI] [PubMed] [Google Scholar]
- [19].Abdel Rahman AT, Hafez SF, Abdelhakam SM, Ali-Eldin ZA, Esmat IM, Elsayed MS, et al. Antimicrobial resistant bacteria among healthcare workers in intensive care units at Ain Shams University Hospitals. J Egypt Soc Parasitol. 2010;40:71–83. [PubMed] [Google Scholar]
- [20].Kaiser ML, Thompson DJ, Malinoski D, Lane C, Cinat ME. Epidemiology and risk factors for hospital-acquired methicillin-resistant Staphylococcus aureus among burn patients. J Burn Care Res. 2011;32:429–34. doi: 10.1097/BCR.0b013e318217f92d. [DOI] [PubMed] [Google Scholar]
- [21].Schweizer M, Ward M, Cobb S, McDanel J, Leder L, Wibbenmeyer L, et al. The epidemiology of methicillin-resistant Staphylococcus aureus on a burn trauma unit. Infect Control Hosp Epidemiol. 2012;33:1118–25. doi: 10.1086/668032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen X, Yang HH, Huangfu YC, Wang WK, Liu Y, Ni YX, et al. Molecular epidemiologic analysis of Staphylococcus aureus isolated from four burn centers. Burns. 2012;38:738–42. doi: 10.1016/j.burns.2011.12.023. [DOI] [PubMed] [Google Scholar]
- [23].Wibbenmeyer L, Williams I, Ward M, Xiao X, Light T, Latenser B, et al. Risk factors for acquiring vancomycin-resistant Enterococcus and methicillin-resistant Staphylococcus aureus on a burn surgery step-down unit. J Burn Care Res. 2010;31:269–79. doi: 10.1097/BCR.0b013e3181d0f479. [DOI] [PubMed] [Google Scholar]
- [24].American Public Health Association. Standard Methods for the Examination of Water and Wastewater. Washington DC: American Public Health Association; 1998. [Google Scholar]
- [25].Cappuccino JG, Sherman N. Microbiology -A laboratory manual. 5th ed. Menlo Park, (CA): The Benjamin/Cummings Publishing Co., Inc; 1996. [Google Scholar]
- [26].Sharmin M, Nur IT, Acharjee M, Munshi SK, Noor R. Microbiological profiling and the demonstration of in vitro antibacterial traits of the major oral herbal medicines used in Dhaka Metropolis. Springer Plus. 2014;3:739. doi: 10.1186/2193-1801-3-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ferraro MJ, Craig WA, Dudley MN, Eliopoulos GM, Hecht DW, Hindler J, et al. Approved Standard M2-A7. 7th ed. Wayne (PA): National Committee for Clinical Laboratory Standards; 2000. Performance standards for antimicrobial disk susceptibility tests. [Google Scholar]
- [28].Bauer AW, Kirby WMM, Sherris JC, Tierch M. Antibiotic susceptibility testing by a standardized single disc method. Am J Clin Pathol. 1968;45:493–6. [PubMed] [Google Scholar]
- [29].Munshi SK, Rahman MM, Noor R. Detection of virulence potential of diar-rheagenic Escherichia coli isolated from surface water of rivers surrounding Dhaka City. J Bang Acad Sci. 2012;36:109–22. [Google Scholar]
- [30].Clinical and Laboratory Standards Institute (CLSI). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: Approved Standard. 7th Edition. Wayne (PA): CLSI; 2006. [Google Scholar]
- [31].Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing;19th informational supplement. CLSI M100–S19 Clinical and Laboratory Standards Institute. Wayne (PA): CLSI; 2009. [Google Scholar]
- [32].National Committee for Clinical Laboratory Standards (NCCLS). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7–A5. 5th ed. Wayne (PA): NCCLS; 2000. [Google Scholar]
- [33].Kuehnert MJ. Staphylococcal disease burden. J Infect Dis. 2006;193:172–9. doi: 10.1086/499632. [DOI] [PubMed] [Google Scholar]
- [34].Gardete S, Tomasz A. Mechanisms of vancomycin resistance in Staphylococcus aureus. J Clin Invest. 2014;124:2836–40. doi: 10.1172/JCI68834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Friaes A, Resina C, Manuel V, Lito L, Ramirez M, Melo-Cristino J. Epidemio-logical survey of the first case of vancomycin-resistant Staphylococcus aureus infection in Europe. Epidemiol Infect. 2015;143:745–8. doi: 10.1017/S0950268814001423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Treatment of bacteraemia: meticillin-resistant Staphylococcus aureus (MRSA) to vancomycin-resistant S. aureus (VRSA). Gould IM. Int J Antimicrob Agents. 2013;42(Suppl):S17–21. doi: 10.1016/j.ijantimicag.2013.04.006. [DOI] [PubMed] [Google Scholar]
- [37].Kobayashi SD, Musser JM, DeLeo FR. Genomic analysis of the emergence of vancomycin-resistant Staphylococcus aureus. MBio. 2012;3:e00170–12. doi: 10.1128/mBio.00170-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cesur S, Irmak H, Simsek H, Coplu N, Μις H, Arslan U, et al. Evaluation of antibiotic susceptibilities and VISA-VRSA rates among MRSA strains isolated from hospitalized patients in intensive care units of hospitals in seven provinces of Turkey. Mikrobiyol Bul. 2012;46:352–8. [PubMed] [Google Scholar]
- [39].Japoni A, Alborzi AV, Rasouli M, Pourabbas B. Modified DNA extraction for rapid PCR detection of methicillin resistant Staphylococci. Iranian Biomed J. 2004;8:61–5. [Google Scholar]
- [40].Mehdinejad M, Frajzade A, Jolodar A. Study of methicillin resistance in Staphylococcus aureus and species of coagulase negative staphylococci isolated from various clinical specimens. Pak J Med Sci. 2008;24:115–7. [Google Scholar]
- [41].Ekrami A, Kalantar E. Bacterial infections in burn patients at a burn hospital in Iran. Indian J Med Res. 2007;126:541–4. [PubMed] [Google Scholar]
- [42].Song W, Lee K, Kang H, Hoon Shin D. Microbiologic aspects of predominant bacteria isolated from the burn patients in Korea. Burns. 2001;27:136–9. doi: 10.1016/s0305-4179(00)00086-3. [DOI] [PubMed] [Google Scholar]
- [43].Hodle AE, Richter KP, Thompson RM. Infection control practices in US burn units. Mol Biol Rep. 2006;27:142–51. doi: 10.1097/01.BCR.0000203493.31642.79. [DOI] [PubMed] [Google Scholar]
- [44].Kos VN, Desjardins CA, Griggs A, Cerqueira G, Van Tonder A, Holden MT, et al. Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with methicillin-resistant S. aureus hospital-acquired infection in the United States. MBio. 2012;3:e00112–12. doi: 10.1128/mBio.00112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Clinical and Laboratory Standards Institute (CLSI). Standards for antimicrobial susceptibility testing performance;16th informational supplement. CLSI M100–S16 Clinical and Laboratory Standards Institute (formerly NCCLS) Wayne (PA): CLSI; 2006. [Google Scholar]
- [46].Qureshi NK, Yin S, Boyle-Vavra S. The role of the staphylococcal VraTSR regulatory system on vancomycin resistance and vanA operon expression in vancomycin-resistant Staphylococcus aureus. PLoS One. 2014;9:e85873. doi: 10.1371/journal.pone.0085873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Apfalter P. MRSA/MRSE-VISA/GISA/VRSA-PRP-VRE: current Gram-positive problem bacteria and mechanism of resistance, prevalence, and clinical consequences. Wien Med Wochenschr. 2003;153:144–7. doi: 10.1046/j.1563-258x.2003.03014.x. [DOI] [PubMed] [Google Scholar]
- [48].Noor R, Zerin N, Das KK. Microbiological quality of pharmaceutical products in Bangladesh: current research perspective. Asian Pac J Trop Dis. 2015;5:264–70. [Google Scholar]