Abstract
TP53 is the most frequently mutated tumor suppressor gene in human cancer, with nearly 50% of all tumors exhibiting a loss-of-function mutation. To further elucidate the genetic pathways involving TP53 and cancer, we have exploited the zebrafish, a powerful vertebrate model system that is amenable to whole-genome forward-genetic analysis and synthetic-lethal screens. Zebrafish lines harboring missense mutations in the tp53 DNA-binding domain were identified by using a target-selected mutagenesis strategy. Homozygous mutant fish from two of these lines were viable and exhibited mutations similar to those found in human cancers (tp53N168K and tp53M214K). Although homozygous tp53N168K mutants were temperature-sensitive and suppressed radiation-induced apoptosis only at 37°C, cells in the tp53M214K embryos failed to undergo apoptosis in response to γ radiation at both 28 and 37°C. Unlike wild-type control embryos, irradiated tp53M214K embryos also failed to up-regulate p21 and did not arrest at the G1/S checkpoint. Beginning at 8.5 months of age, 28% of tp53M214K mutant fish developed malignant peripheral nerve sheath tumors. In addition to providing a model for studying the molecular pathogenic pathways of malignant peripheral nerve sheath tumors, these mutant zebrafish lines provide a unique platform for modifier screens to identify genetic mutations or small molecules that affect tp53-related pathways, including apoptosis, cell-cycle delay, and tumor suppression.
In multicellular organisms, the tumor suppressor gene TP53 plays a major role in maintaining the integrity of the genome by responding to various types of cellular stresses and inducing cell-cycle arrest or apoptosis (1-3). Events including DNA damage, hypoxia, or oncogene activation trigger the posttranslational stabilization of the TP53 protein (4, 5). Mechanistically, TP53 causes an arrest of the cell cycle by transactivating key downstream effector genes, such as the p21 cyclin-dependent kinase inhibitor, and allowing time to repair damaged DNA (6). Furthermore, augmented levels of TP53 protein can activate apoptotic pathways through both transcription-dependent and independent mechanisms (7). Thus, the inactivation of the TP53 gene through a direct mutational event can lead to a reduced ability to activate apoptosis in cells that are compromised in DNA damage repair, which can lead to further accumulation of mutations during oncogenesis.
The pivotal role of TP53 is underscored by the fact that most human tumors have mutations that directly inactivate its function by missense mutations. Mutations in the TP53 gene are found in more than half of all human tumors, making TP53 the most frequently mutated tumor suppressor gene in human cancers (8, 9). Li-Fraumeni syndrome, an inherited disorder caused by mutations in the human TP53 gene, predisposes patients to a 50-fold higher risk of developing cancer, with >50% of Li-Fraumeni patients developing a variety of sarcomas, leukemia, breast cancer, and other forms of cancer by the age of 30 (10, 11). The TP53-DNA damage response pathway can also be altered by up-regulation of MDM2, providing additional evidence for deregulation of this pathway in tumors in which TP53 mutations are not observed (12, 13). TP53-dependent apoptosis induced by aberrant proliferative signals can also be interrupted by loss of p14ARF (14).
Mice genetically engineered to lack Tp53 are prone to early onset of multiple types of neoplasms, consistent with the role of Tp53 as a tumor suppressor (15-17). Although TP53 has been intensely investigated, questions remain regarding its function and downstream molecular pathways. Invertebrate TP53-deficient models in Caenorhabditis elegans and Drosophila do not exhibit increased tumor phenotypes and lack aspects of the TP53 responses (18-21). Thus, forward genetic analysis in a vertebrate model, such as the zebrafish, may yield important clues to the molecular pathways regulated by TP53 in mammals.
The zebrafish model system is amenable to whole-genome forward genetic approaches and has numerous additional advantages, including the relative ease of maintaining large stocks of animals and rapid embryonic development ex utero, which facilitates experimental manipulation and allows the direct observation of tissue formation and organogenesis in vivo. The organization of the human genome and the genetic pathways controlling signal transduction and development are highly conserved in zebrafish (22-26). These properties have established the zebrafish as an excellent model system that is relevant to studies of vertebrate developmental mechanisms as well as human diseases, including cancer (22, 27-33). Furthermore, studies examining the transient knockdown of tp53 in zebrafish have shown that the role of tp53 in apoptosis and other related cellular functions can be addressed in this organism (32).
We have isolated tp53-deficient zebrafish lines, which lack apoptosis and cell-cycle arrest responses to DNA damage. After 8.5 months of age, these animals spontaneously develop malignant peripheral nerve sheath tumors (MPNSTs), which are rarely observed in wild-type fish and indicate a tumor suppressor role for zebrafish tp53.
Materials and Methods
Materials and methods used for fish care, generation of zebrafish mutant lines, flow cytometry, paraffin embedding and sectioning, electron microscopy, and tumor transplantation can be found in Supporting Text, which is published as supporting information on the PNAS web site.
In Vitro Functional Reporter Assay. We generated a zebrafish tp53 cDNA construct replacing the GFP gene with tp53 in a pEGFP-N1 expression vector (BD Biosciences Clontech). Mutant tp53 constructs were generated by using the QuikChange Site-Directed Mutagenesis Kit following the manufacturer's instructions (Stratagene). We transfected SAOS2 cells with these constructs using the FuGENE 6 Transfection Reagent (Roche Applied Bioscience, Indianapolis, no. 1815091) according to the manufacturer's protocol. Additional details are described in Supporting Text.
Apoptosis Assay. Embryos were raised at 28°C and γ irradiated at 24 h postfertilization (hpf). At 30 hpf the embryos were fixed overnight in 4% paraformaldehyde followed by staining. Embryos were assayed by TUNEL using the In Situ Cell Death Detection Kit, TMR Red (Roche Applied Bioscience no. 2156792). Further details are described in Supporting Text.
Whole-Mount Phospho-H3 Antibody Staining. Embryos were γ irradiated at 24 hpf and 45 min later were dechorionated and fixed overnight in 4% paraformaldehyde. Embryos were stained with a polyclonal anti-phospho-H3 antibody (1:1,000) (Santa Cruz Biotechnology) and developed in diaminobenzidine solution for 5 min. Further details are described in Supporting Text.
Single-Embryo RT-PCR Gene Expression. Embryos were raised at 28°C and γ irradiated at 24 hpf. At 30 hpf, RNA was made from the embryos by using TRIzol (Invitrogen). One-step RT-PCR (Qiagen, Valencia, CA) was used. Additional details including primer sequences are described in Supporting Text.
Tumorigenesis. Time to tumor incidence was assessed in homozygous tp53M214K mutant (n = 105), heterozygous (n = 205), and wild-type (n = 107) zebrafish. Fish were genotyped at 8 months of age and followed for tumor incidence an additional 8.5 months. A cumulative incidence model was used to assess tumor incidence by genotype and differences between genotypes detected statistically by the Gray test.
Results
Isolating Zebrafish Lines with Point Mutations in the Zebrafish tp53 Gene. More than 90% of identified TP53 mutations in human tumors are found in the DNA-binding domain (DBD) of the protein (8, 9). As in humans, the highly conserved zebrafish tp53 DBD between codons 97 and 292 is encoded within exons 4-8 (34). Using a target-selected mutagenesis strategy, we examined these five exons by DNA sequencing in 2,679 individual N-ethyl-N-nitrosourea (ENU)-mutagenized F1 male fish, as described (35). Five zebrafish lines were identified that carried heterozygous ENU-induced mutations: one silent mutation was found in exon 7, whereas four missense mutations affecting the tp53 protein sequence were identified in exons 4-7 (Table 1, which is published as supporting information on the PNAS web site). After confirmation by resequencing, zebrafish lines carrying three of these mutations were recovered through in vitro fertilization of wild-type eggs by using cryopreserved sperm from each identified F1 male.
Two of the three recovered ENU-induced tp53 missense mutations affected the tp53 protein at amino acid positions that are orthologous to TP53 mutations found in human cancer cells (8, 9). In exon 6, the tp53N168K mutation affects the DBD and corresponds to human codon 200, which is mutated in nine reported cases of human tumors. The orthologous human codon for the tp53M214K mutation in exon 7, methionine-246, is mutated in 124 different human tumors, 8 of which exhibit the same amino acid change from a methionine to a lysine. In addition, this codon is positioned between other known mutation hot spots in the DBD of the human TP53 gene [codons 245, 248, and 249) (9)]. We also recovered a line of fish with a missense mutation in exon 4, but the tp53S26F mutation is located outside the DBD in the poorly conserved N-terminal region (34).
Functional Significance of Mutant Zebrafish tp53 Alleles. It is generally accepted that TP53 protein is first activated by various cellular stresses and then binds to its consensus DNA-binding sequence as a tetramer, leading to the transcriptional activation of target genes (1, 36, 37). If only one of the four protein subunits is mutated within the DBD, the transactivation function of the TP53 tetramer is believed to be at least partially lost (5). Thus, mutant TP53 proteins can inhibit wild-type protein function in a dominant-negative fashion. To assess the function of the mutated tp53 isoforms in zebrafish lines, we tested the cloned mutant proteins using tp53-mediated in vitro transactivation assays (Fig. 6, which is published as supporting information on the PNAS web site). In this assay, tp53 drives the expression of the luciferase gene using a human p21 response element containing a TP53 consensus-binding site that is conserved across species (19). The mutation in exon 4 did not affect the ability of the zebrafish tp53 protein to function in this assay. However, both the tp53N168K and tp53M214K mutated proteins were unable to activate transcription through the p21 response element in SAOS2 cells assayed at 37°C. In addition, the two mutations behaved in a dominant-negative manner, inhibiting the ability of wild-type tp53 to activate luciferase expression.
Development in tp53 Mutant Fish. To evaluate the role of tp53 in zebrafish development, we first identified heterozygous F2 offspring obtained from frozen sperm by in vitro fertilization and then outcrossed the heterozygotes to wild-type fish, creating an F3 generation. The F3 heterozygous fish were incrossed, and the progeny were used for all further experiments. These clutches were of normal size and exhibited Mendelian segregation for both the tp53N168K mutation (25.88% tp53+/+; 49.56% tp53+/N168K; 24.56% tp53N168K/N168K; n = 228) and the tp53M214K mutation (24.32% tp53+/+; 51.17% tp53+/M214K; 24.51% tp53M214K/M214K; n = 514). Homozygous and heterozygous mutant fish developed normally and were indistinguishable from their wild-type siblings in terms of viability and fertility. Thus, in contrast to an increased frequency of developmental abnormalities such as exencephaly and craniofacial malformations associated with Tp53 deficiency in mice, the embryonic development of the tp53 mutant zebrafish was not affected (38, 39).
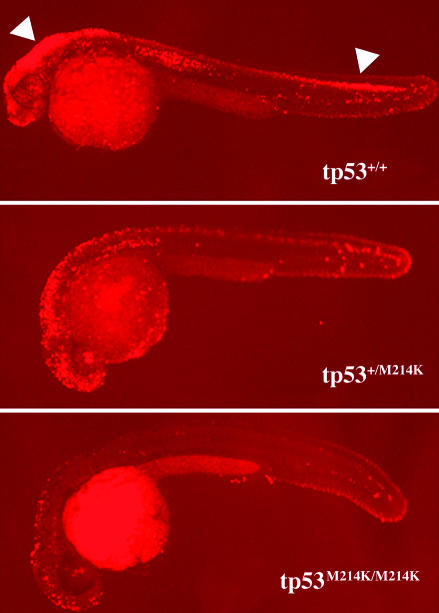
tp53 Mutations in Zebrafish Suppress Irradiation-Induced Apoptosis. As in mammals, the role of apoptosis in development is well documented in zebrafish, and tp53 has been shown to be a critical mediator of apoptosis in response to DNA damage (32, 40, 41). To investigate whether the tp53M214K mutation affected the apoptotic response in vivo, we irradiated embryos at 24 hpf and performed TUNEL assays at 30 hpf to detect apoptotic cells. Although irradiation resulted in massive apoptosis in wild-type embryos, the homozygous mutants exhibited baseline staining similar to nonirradiated control embryos (Fig. 1 and data not shown). Heterozygous mutant tp53M214K embryos exhibited intermediate levels of apoptosis, a state reminiscent of Tp53 γ irradiation induced apoptosis in mouse thymocytes carrying the Tp53-R172P DBD missense mutation (42). Interestingly, the tp53N168K mutation behaves in a temperature-sensitive manner, because the suppression of apoptosis could be seen only when embryos were maintained at 37°C and not at 28°C, the normal temperature for rearing zebrafish (Fig. 7, which is published as supporting information on the PNAS web site). Temperature-sensitive TP53 mutations have been described in mammals and Xenopus laevis (43-45). In addition, the orthologous TP53 mutation in humans has been shown to be temperature-sensitive in cell assays (46). Thus, the two ENU mutations we isolated in zebrafish, one temperature-sensitive and one temperature-independent, block the in vivo tp53-dependent apoptotic response. These findings are consistent with functional studies examining orthologous TP53 DBD mutations in human cancer cells and murine models (42, 47).
Fig. 1.
Induction of apoptosis by γ irradiation is suppressed in tp53 mutant embryos. Embryos were γ irradiated at 24 hpf (16 Gray) and fixed at 30 hpf for TUNEL assay. After the assay of multiple embryos resulting from the intercross of tp53M214K mutant heterozygous parents, the embryos were genotyped for their tp53 status. Embryos shown are representative of homozygous wild-type (tp53+/+), heterozygous (tp53+/M214K), and homozygous mutant (tp53M214K/M214K) genotypes. White arrows indicate widespread apoptosis in the brain and spinal cord. Embryos were raised at 28°C. All embryos are shown in lateral view with the head to the left.
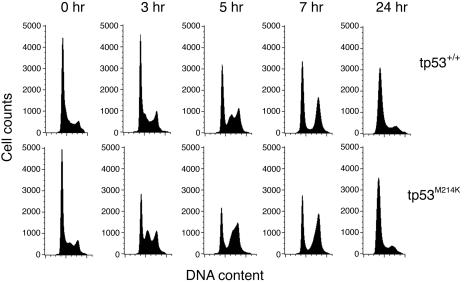
tp53 Mutation in Zebrafish Alters the Cell-Cycle Checkpoint Response. In mammalian cells, TP53 is required for the cellular G1-phase checkpoint in response to γ irradiation. To test the role of tp53 in cell-cycle checkpoint control, we examined the γ irradiation response of homozygous tp53M214K mutant embryos. Wild-type embryos (24 hpf) and their tp53M214K siblings were γ irradiated and analyzed for DNA content at 0, 3, 5, 7, and 24 h postirradiation (Fig. 2). The tp53M214K embryos showed an accumulation of cells in mid-S phase at 3-h postirradiation whereas wild-type embryos retained an increased percentage of cells in G1. Wild-type embryos had a similar accumulation of cells in mid-S phase at 5-h postirradiation, suggesting that the G1 checkpoint delays cells for ≈2 h. The increase in mid-S phase is presumably due to the radiation-induced intra-S-phase checkpoint, which is not affected by TP53 mutations (48). Mutant embryos showed a decreased percentage of G1-phase cells through 7 hours after irradiation, indicating that the G1 checkpoint is defective in tp53M214K embryos after irradiation.
Fig. 2.
tp53M214K embryos have defects in the γ irradiation-induced G1 checkpoint. DNA content analysis was performed on homozygous tp53M214K mutant (Lower) and wild-type sibling (Upper) embryos at 0, 3, 5, 7, and 24 h postirradiation (16 Gray). Results are representative of at least three experiments.
We also examined the γ irradiation-induced G2-phase checkpoint response in tp53M214K embryos. Wild-type embryos (24 hpf) displayed a transient decrease in the mitotic marker phosphorylated histone H3 (phospho-H3), when examined at 45 min after exposure to γ irradiation (J. F. Amatruda and J. L. Shepard, personal communication). In addition, cells in these embryos accumulated in G2/M by DNA content analysis 5 and 7 h postirradiation due to activation of the G2 checkpoint (Fig. 2). The tp53M214K embryos did not differ from wild-type embryos in phospho-H3 staining (data not shown) and showed similar percentages of cells in G2/M at 7 h postirradiation (Fig. 2), suggesting that the radiation-induced G2 checkpoint was intact in tp53M214K mutant embryos.
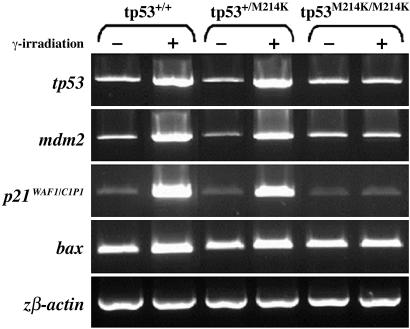
Downstream Effectors of tp53 Function. To further investigate downstream tp53-dependent transcriptional responses in the zebrafish tp53M214K mutant line, we examined the expression of tp53 target genes using single-embryo semiquantitative RT-PCR after γ irradiation. These tp53-response genes included p21WAF1/CIP-1, based on its central involvement in cell-cycle arrest; bax, for its role in the apoptosis pathway; and tp53 and mdm2 to explore the feedback loops downstream of tp53 (Fig. 3). All three tp53-target genes showed transcriptional up-regulation in irradiated wild-type embryos (Fig. 3, lanes 1 and 2); however, this response was not identified in homozygous tp53M214K siblings (Fig. 3, lanes 5 and 6). Consistent with the apoptosis data (Fig. 1), an intermediate up-regulation of these genes was observed in heterozygous mutant embryos (Fig. 3, lanes 3 and 4). tp53 mRNA is also up-regulated in wild-type and heterozygous embryos after γ irradiation but not in homozygous mutant embryos. Transcriptional regulation of TP53 is not thought to play a major role in mammalian apoptosis, where post-translational modification accounts for TP53 protein stabilization and the increase in TP53 protein levels after DNA damage (36). Consistent with these results, the up-regulation of the same genes was also observed in the tp53N168K mutant line when the assay was performed at 37°C (data not shown).
Fig. 3.
tp53 mutant embryos lack up-regulation of key downstream target genes after γ irradiation. Genes involved in the tp53 regulatory (tp53, mdm2), cell-cycle checkpoint (p21), and apoptotic (bax) pathways were analyzed in homozygous, heterozygous, and wild-type embryos with (+) or without (-) γ irradiation at 24 hpf (16 Gray). Gene expression was assayed at 30 hpf by single-embryo RT-PCR. Results are representative of at least five experiments. β actin was used to control gene expression.
Mutant tp53M214K Fish Spontaneously Develop MPNSTs. Tp53-deficient mice carrying a null allele are prone to develop tumors, leading to early death from cancer (16, 17). The vast majority of germ-line mutations found in Li-Fraumeni patients, or somatic mutations identified in human tumors, are missense mutations found in the TP53 DBD, particularly in exon 7 (9). To explore whether the tp53N168K and tp53M214K mutations increased their susceptibility to spontaneous tumors, we monitored homozygous and heterozygous mutant fish in parallel with their wild-type siblings. Although no sign of tumorigenesis could be detected in the tp53N168K mutant line when raised at 28°C, the tp53M214K mutant fish were prone to forming tumors.
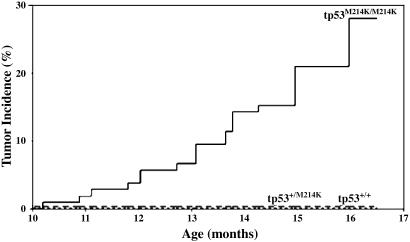
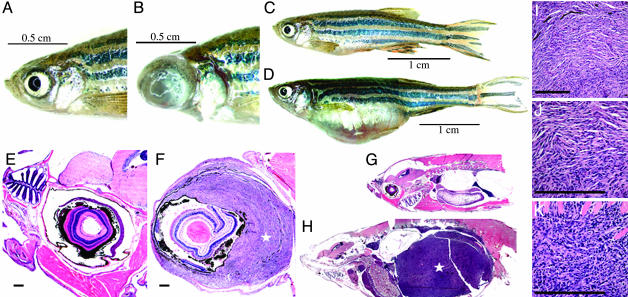
Although tumors were not observed in a first cohort of fish sectioned at 7 months of age, a cohort of 417 fish was examined between 8 and 16.5 months of age, and a total of 23 fish were identified with tumors. All of these tumor-bearing fish were identified as homozygous tp53 mutant zebrafish. Tumor incidence is estimated at 28% by 16.5 months (Fig. 4), and the Gray test for differences in time-to-tumor identification among genotypes is significant (P < 0.00001). Macroscopically, 13 of 23 fish (56.5%) showed a unilateral protrusion of the left or right eye due to tumorigenic overgrowth (Fig. 5 A, B and E, F), and 10 (43.5%) had a distended abdominal cavity due to the presence of a tumor mass (Fig. 5 C, D and G, H). Abdominal and periocular tumors demonstrated similar histological features and were comprised predominantly of spindle cells with tapered nuclei demonstrating uniform chromatin staining and moderately abundant palely eosinophilic cytoplasm (Fig. 5 I and J). A varying degree of epithelioid cell morphology was also noted in some tumors, with this cell type predominating in several cases (Fig. 5K). Overall, the morphologic features were most consistent with a zebrafish spindle and epithelioid cell neoplasm most similar to human malignant peripheral neural sheath tumor (49). No pigment was noted in definitive tumor cells, but pigmented cells were occasionally observed within a tumor, representing entrapped normal pigmented cells. Electron microscopy studies demonstrated tumor cells with elongated interdigitating cytoplasmic processes and reduplicated external lamina, features that support the morphologic characteristics of nerve sheath differentiation (Fig. 8, which is published as supporting information on the PNAS web site). No features of smooth or skeletal muscle differentiation were noted. In addition to the cohort of zebrafish described above, we have identified 20 other zebrafish MPNSTs (zMPNSTs) in homozygous tp53M214K mutant individuals that were raised in our aquariums, as well as one case of melanoma at 11 months of age (Fig. 9, which is published as supporting information on the PNAS web site). Histological examination revealed this melanoma to be a poorly differentiated highly invasive tumor with marked nuclear pleomorphism, prominent nucleoli, and moderate to large amounts of eosinophilic frequently pigmented cytoplasm similar to melanomas in humans. RNA in situ staining on zMPNST sections using a probe for the neuronal marker gfap demonstrated expression of this gene in tumors arising in tp53M214K mutant zebrafish (Fig. 10, which is published as supporting information on the PNAS web site).
Fig. 4.
tp53M214K mutant zebrafish develop zMPNST at high frequency. Tumor incidence curves show cohorts of homozygous mutant (tp53M214K/M214K), heterozygous (tp53M214K/+), and wild type (tp53+/+).
Fig. 5.
Tumorigenesis features of tp53M214K mutant zebrafish. (A-D) When compared with wild-type zebrafish (A and C), zMPNST development in tp53 mutant zebrafish is identifiable upon external observation because of ocular (B) or abdominal tumor localizations (D). (E-H) When compared with wild-type zebrafish (E and G), histopathology staining with hematoxylin/eosin reveals zMPNST in the eye (F) and abdominal cavity (H), as indicated by the stars (×4). (I-K) Histopathological features of tumors (I) composed predominantly of spindle cells (J) and to a varying degree of epitheloid cells (K) are consistent with the diagnosis of zMPNST. [Bar, 200 μm (E, F, I-K).]
We tested the ability to propagate zMPNST through cell transplant assays by injecting tumor cells intraperitoneally into irradiated 1-year-old wild-type zebrafish. Although no tumor was identified in mock-injected control fish, two of three surviving tumor cell recipients developed tumors with zMPNST characteristics (data not shown). Examination by sectioning at 23 days post-transplantation indicates that the tumors developed at the site of injection and spread throughout the abdominal cavity.
Flow cytometric measurements of the DNA content of tumor cells from eight zebrafish homozygous for the tp53M214K mutation showed that the tumor cells were clonally aneuploid, with gain or loss of chromosomes (Fig. 11, which is published as supporting information on the PNAS web site). The clonal origins of these tumors, combined with the long latency preceding tumor onset, suggest the need for additional mutations before malignant transformation in the tp53 mutant background.
Discussion
In this study, we used a reverse genetic strategy of target-selected inactivation to generate two mutant tp53 models in zebrafish. We isolated representative examples of the two classes of mutations described in human TP53 DBD (50). The tp53M214K mutation is an example of a DNA contact mutation, affecting a region of the protein that plays a role in DNA-binding to tp53-specific consensus sequences. In contrast, the tp53N168K mutation belongs to the class of structural mutations, which affect the ability of tp53 protein to bind DNA by modifying the protein conformation. The latter type of mutation is often temperature-sensitive, as observed in the tp53N168K zebrafish line.
In mammals, TP53 primarily exerts its role as a tumor suppressor through the maintenance of genomic integrity after DNA damage by its ability to arrest the cell cycle and induce apoptosis. Our stable mutant lines demonstrate the conservation of both of these critical tp53 functions in zebrafish, as earlier suggested by transient studies in zebrafish embryos (32). In addition, our results indicate that the mutations in these two zebrafish tp53 models can abolish these functions. Mutants exhibited a lack of p21 transcriptional activation and arrest at the G1 checkpoint after irradiation, indicating the absence of tp53-mediated cell-cycle control. Apoptosis assays demonstrated the absence of tp53-dependent apoptosis in irradiated mutant embryos. Wild-type and mutant zebrafish were raised between 26 and 28°C, a temperature range over which the tp53 protein is nonfunctional in the tp53M214K line, while retaining its function in the tp53N168K mutant line. The temperature-sensitive tp53N168K mutation exhibited abnormal apoptotic and cell-cycle phenotypes at 37°C and, as expected, did not develop spontaneous tumors in zebrafish raised at 26-28°C. The oncogenic potential of zebrafish mutant tp53 was demonstrated by mutant fish of the tp53M214K line, which developed tumors starting at 8.5 months with an incidence of 28% by 16.5 months.
Although the major functions of the tp53 pathway are conserved in zebrafish and mammals, it is important to note that differences do exist. The tumor spectrum found in our model differs from that found in human and mice with mutationally inactivated TP53 (9, 16, 17). With the exception of one case of melanoma, each of the 43 tumors identified in our homozygous tp53 mutant fish was classified as a zMPNST, which is a tumor that recapitulates neuroectodermal differentiation but is often thought of as a subtype of sarcoma. Individuals affected by the human Li-Fraumeni syndrome have high rates of sarcomas, principally osteosarcomas and rhabdomyosarcomas, and other types of sarcoma at lower frequencies. These patients also develop breast cancer, brain tumors, and leukemias at strikingly increased rates. Tp53 knockout mice show a high rate of tumorigenesis and die mainly of lymphomas and sarcomas. Nerve sheath sarcomas are uncommon in humans and mice with isolated TP53 deficiency; however, mice with both Nf1 and Tp53 mutations have also been shown to develop MPNSTs, supporting a cooperative and causal role for Tp53 mutations in MPNST development (51, 52). The reasons underlying the differences in tumor spectrum in zebrafish compared with humans and mice are unknown but presumably relate to tissue-specific differences in the response to oncogene-induced proliferative stimuli and DNA damage in fish compared with mammals. Because outbreeding of our tp53M214K line has so far progressed only to the F3 generation, we also cannot exclude the possibility that a cosegregating ENU mutation contributes to the predisposition to develop tumors.
A recent study has shown that zebrafish carrying mutations in a heterozygous state for 11 different ribosomal protein (RP) genes have a predisposition to develop tumors, with an 81% incidence of zMPNST (49). It is not known why zMPNST is the most prevalent cancer in both the RP and tp53 inherited loss-of-function models. It will be interesting to determine whether inactivating mutations of tp53, RP, and NF1 genes are synergistic in zMPNST tumorigenesis.
In humans, MPNST is the malignant counterpart of benign soft tissue tumors, such as neurofibromas and schwannomas. This rare tumor type represents ≈5% of malignant tumors of soft tissue, and ≈50% of these occur in patients with neurofibromatosis I (NF1; refs. 53 and 54). Although the molecular events leading to MPNST tumorigenesis have not been fully characterized, deletions and mutations in the TP53 gene have been identified, as well as nuclear accumulation of the TP53 protein in tumors with unsequenced TP53 genes, indicating a role for TP53 mutations in human MPNST (55, 56). Furthermore, the malignant transformation of neurofibromas in NF1, as well as in sporadic osteosarcomas, is associated with deletions of the p16-Ink4a locus, which uncouples aberrant cell proliferation from both RB- and TP53-mediated tumor suppressor pathways (57, 58). An MPNST cell line (HS-sch-2) has been shown to possess a point mutation in the TP53 DBD within codon 273 (59). In nf1-deficient mice, benign peripheral nerve sheath tumors develop and progress to MPNST when the mice also carry a germ-line mutation in the Tp53 gene (51, 52). In zebrafish, zMPNST is also a rare neoplasm with a low incidence in the wild-type population (49). The mutant tp53 zebrafish line we have developed provides a model for MPNST pathogenesis, and further analysis should help to elucidate molecular pathways involved in this tumor type.
Conclusion
The many advantages of the zebrafish vertebrate system are now available to further dissect TP53 functions in vivo, through analysis of the two zebrafish lines harboring mutant tp53 genes. The ability to analyze early embryonic phenotypes using apoptotic, cell-cycle, and γ irradiation assays will facilitate high-throughput small-molecule screens using efficient robotic handling of compounds and embryos (33, 60). In addition, our temperature-sensitive mutant line offers a vertebrate tp53-inducible system that can be regulated in vivo, by simply controlling the water temperature. These zebrafish tp53 models may provide new insights into the TP53 pathways that suppress tumorigenesis in vertebrates and can serve as the foundation for genetic and chemical screens to identify modifiers acting in downstream and parallel pathways.
Supplementary Material
Acknowledgments
We thank C. Jost (Dana-Farber Cancer Institute) and W. Kaelin (Dana-Farber Cancer Institute) for reagents; J. Aster, J. Glickman, R. Bronson, and H. Stern for tumor diagnosis; J. Vinokur, G. Kourkoulis, and W. Saganic for fish care; J. Williams and D. Skinner for histology services; C. Ford for electron microscopy services; N. Campisi for FACS analysis; and the Tübingen 2000 screen consortium for the mutagenesis library. This work was supported by National Institutes of Health Grant CA-68484 (to A.T.L.). Technical services for histology were supported by the Hematopathology Core Lab of the Dana-Farber/Harvard Cancer Center and National Institutes of Health Grant P30CA6516 (to J.L.K). S.B. was a fellow of the Lymphoma Research Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: hpf, hours postfertilization; DBD, DNA-binding domain; ENU, N-ethyl-N-nitrosourea; MPNST, malignant peripheral nerve sheath tumor; zMPNST, zebrafish MPNST.
References
- 1.Levine, A. J. (1997) Cell 88, 323-331. [DOI] [PubMed] [Google Scholar]
- 2.Vousden, K. H. (2000) Cell 103, 691-694. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein, B., Lane, D. & Levine, A. J. (2000) Nature 408, 307-310. [DOI] [PubMed] [Google Scholar]
- 4.Kastan, M. B., Onyekwere, O., Sidransky, D., Vogelstein, B. & Craig, R. W. (1991) Cancer Res. 51, 6304-6311. [PubMed] [Google Scholar]
- 5.Vousden, K. H. & Lu, X. (2002) Nat. Rev. Cancer 2, 594-604. [DOI] [PubMed] [Google Scholar]
- 6.Deng, C., Zhang, P., Harper, J. W., Elledge, S. J. & Leder, P. (1995) Cell 82, 675-684. [DOI] [PubMed] [Google Scholar]
- 7.Fridman, J. S. & Lowe, S. W. (2003) Oncogene 22, 9030-9040. [DOI] [PubMed] [Google Scholar]
- 8.Beroud, C. & Soussi, T. (2003) Hum. Mutat. 21, 176-181. [DOI] [PubMed] [Google Scholar]
- 9.Olivier, M., Eeles, R., Hollstein, M., Khan, M. A., Harris, C. C. & Hainaut, P. (2002) Hum. Mutat. 19, 607-614. [DOI] [PubMed] [Google Scholar]
- 10.Malkin, D., Li, F. P., Strong, L. C., Fraumeni, J. F., Jr., Nelson, C. E., Kim, D. H., Kassel, J., Gryka, M. A., Bischoff, F. Z., Tainsky, M. A., et al. (1990) Science 250, 1233-1238. [DOI] [PubMed] [Google Scholar]
- 11.Srivastava, S., Zou, Z. Q., Pirollo, K., Blattner, W. & Chang, E. H. (1990) Nature 348, 747-749. [DOI] [PubMed] [Google Scholar]
- 12.Oliner, J. D., Pietenpol, J. A., Thiagalingam, S., Gyuris, J., Kinzler, K. W. & Vogelstein, B. (1993) Nature 362, 857-860. [DOI] [PubMed] [Google Scholar]
- 13.Prives, C. (1998) Cell 95, 5-8. [DOI] [PubMed] [Google Scholar]
- 14.Lowe, S. W. & Sherr, C. J. (2003) Curr. Opin. Genet. Dev. 13, 77-83. [DOI] [PubMed] [Google Scholar]
- 15.Attardi, L. D. & Jacks, T. (1999) Cell Mol. Life Sci. 55, 48-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donehower, L. A., Harvey, M., Slagle, B. L., McArthur, M. J., Montgomery, C. A., Jr., Butel, J. S. & Bradley, A. (1992) Nature 356, 215-221. [DOI] [PubMed] [Google Scholar]
- 17.Jacks, T., Remington, L., Williams, B. O., Schmitt, E. M., Halachmi, S., Bronson, R. T. & Weinberg, R. A. (1994) Curr. Biol. 4, 1-7. [DOI] [PubMed] [Google Scholar]
- 18.Derry, W. B., Putzke, A. P. & Rothman, J. H. (2001) Science 294, 591-595. [DOI] [PubMed] [Google Scholar]
- 19.Brodsky, M. H., Nordstrom, W., Tsang, G., Kwan, E., Rubin, G. M. & Abrams, J. M. (2000) Cell 101, 103-113. [DOI] [PubMed] [Google Scholar]
- 20.Ollmann, M., Young, L. M., Di Como, C. J., Karim, F., Belvin, M., Robertson, S., Whittaker, K., Demsky, M., Fisher, W. W., Buchman, A., et al. (2000) Cell 101, 91-101. [DOI] [PubMed] [Google Scholar]
- 21.Jin, S., Martinek, S., Joo, W. S., Wortman, J. R., Mirkovic, N., Sali, A., Yandell, M. D., Pavletich, N. P., Young, M. W. & Levine, A. J. (2000) Proc. Natl. Acad. Sci. USA 97, 7301-7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langenau, D. M., Traver, D., Ferrando, A. A., Kutok, J. L., Aster, J. C., Kanki, J. P., Lin, S., Prochownik, E., Trede, N. S., Zon, L. I., et al. (2003) Science 299, 887-890. [DOI] [PubMed] [Google Scholar]
- 23.Postlethwait, J. H., Woods, I. G., Ngo-Hazelett, P., Yan, Y. L., Kelly, P. D., Chu, F., Huang, H., Hill-Force, A. & Talbot, W. S. (2000) Genome Res. 10, 1890-1902. [DOI] [PubMed] [Google Scholar]
- 24.Liu, T. X., Zhou, Y., Kanki, J. P., Deng, M., Rhodes, J., Yang, H. W., Sheng, X. M., Zon, L. I. & Look, A. T. (2002) Proc. Natl. Acad. Sci. USA 99, 6136-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett, C. M., Kanki, J. P., Rhodes, J., Liu, T. X., Paw, B. H., Kieran, M. W., Langenau, D. M., Delahaye-Brown, A., Zon, L. I., Fleming, M. D. & Look, A. T. (2001) Blood 98, 643-651. [DOI] [PubMed] [Google Scholar]
- 26.Hsu, K., Kanki, J. P. & Look, A. T. (2001) Curr. Opin. Hematol. 8, 245-251. [DOI] [PubMed] [Google Scholar]
- 27.Berman, J., Hsu, K. & Look, A. T. (2003) Br. J. Haematol. 123, 568-576. [DOI] [PubMed] [Google Scholar]
- 28.Zon, L. I. (1999) Genome Res. 9, 99-100. [PubMed] [Google Scholar]
- 29.Grunwald, D. J. & Eisen, J. S. (2002) Nat. Rev. Genet. 3, 717-724. [DOI] [PubMed] [Google Scholar]
- 30.Shin, J. T. & Fishman, M. C. (2002) Annu. Rev. Genom. Hum. Genet. 3, 311-340. [DOI] [PubMed] [Google Scholar]
- 31.Amatruda, J. F., Shepard, J. L., Stern, H. M. & Zon, L. I. (2002) Cancer Cell 1, 229-231. [DOI] [PubMed] [Google Scholar]
- 32.Langheinrich, U., Hennen, E., Stott, G. & Vacun, G. (2002) Curr. Biol. 12, 2023-2028. [DOI] [PubMed] [Google Scholar]
- 33.Langheinrich, U. (2003) BioEssays 25, 904-912. [DOI] [PubMed] [Google Scholar]
- 34.Cheng, R., Ford, B. L., O'Neal, P. E., Mathews, C. Z., Bradford, C. S., Thongtan, T., Barnes, D. W., Hendricks, J. D. & Bailey, G. S. (1997) Mol. Mar. Biol. Biotechnol. 6, 88-97. [PubMed] [Google Scholar]
- 35.Wienholds, E., Schulte-Merker, S., Walderich, B. & Plasterk, R. H. (2002) Science 297, 99-102. [DOI] [PubMed] [Google Scholar]
- 36.Lakin, N. D. & Jackson, S. P. (1999) Oncogene 18, 7644-7655. [DOI] [PubMed] [Google Scholar]
- 37.Cho, Y., Gorina, S., Jeffrey, P. D. & Pavletich, N. P. (1994) Science 265, 346-355. [DOI] [PubMed] [Google Scholar]
- 38.Sah, V. P., Attardi, L. D., Mulligan, G. J., Williams, B. O., Bronson, R. T. & Jacks, T. (1995) Nat. Genet. 10, 175-180. [DOI] [PubMed] [Google Scholar]
- 39.Armstrong, J. F., Kaufman, M. H., Harrison, D. J. & Clarke, A. R. (1995) Curr. Biol. 5, 931-936. [DOI] [PubMed] [Google Scholar]
- 40.Yamashita, M. (2003) Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 136, 731-742. [DOI] [PubMed] [Google Scholar]
- 41.Cole, L. K. & Ross, L. S. (2001) Dev. Biol. 240, 123-142. [DOI] [PubMed] [Google Scholar]
- 42.Liu, G., Parant, J. M., Lang, G., Chau, P., Chavez-Reyes, A., El-Naggar, A. K., Multani, A., Chang, S. & Lozano, G. (2004) Nat. Genet. 36, 63-68. [DOI] [PubMed] [Google Scholar]
- 43.Altura, R. A., Inukai, T., Ashmun, R. A., Zambetti, G. P., Roussel, M. F. & Look, A. T. (1998) Blood 92, 1397-1405. [PubMed] [Google Scholar]
- 44.Friedlander, P., Legros, Y., Soussi, T. & Prives, C. (1996) J. Biol. Chem. 271, 25468-25478. [DOI] [PubMed] [Google Scholar]
- 45.Ridgway, P. J., Soussi, T. & Braithwaite, A. W. (1994) J. Virol. 68, 7178-7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiraishi, K., Kato, S., Han, S. Y., Liu, W., Otsuka, K., Sakayori, M., Ishida, T., Takeda, M., Kanamaru, R., Ohuchi, N. & Ishioka, C. (2004) J. Biol. Chem. 279, 348-355. [DOI] [PubMed] [Google Scholar]
- 47.Slee, E. A., O'Connor, D. J. & Lu, X. (2004) Oncogene 23, 2809-2818. [DOI] [PubMed] [Google Scholar]
- 48.Bartek, J. & Lukas, J. (2001) Curr. Opin. Cell Biol. 13, 738-747. [DOI] [PubMed] [Google Scholar]
- 49.Amsterdam, A., Sadler, K. C., Lai, K., Farrington, S., Bronson, R. T., Lees, J. A. & Hopkins, N. (2004) PLoS Biol. 2, E139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bullock, A. N. & Fersht, A. R. (2001) Nat. Rev. Cancer 1, 68-76. [DOI] [PubMed] [Google Scholar]
- 51.Cichowski, K., Shih, T. S., Schmitt, E., Santiago, S., Reilly, K., McLaughlin, M. E., Bronson, R. T. & Jacks, T. (1999) Science 286, 2172-2176. [DOI] [PubMed] [Google Scholar]
- 52.Vogel, K. S., Klesse, L. J., Velasco-Miguel, S., Meyers, K., Rushing, E. J. & Parada, L. F. (1999) Science 286, 2176-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kourea, H. P., Bilsky, M. H., Leung, D. H., Lewis, J. J. & Woodruff, J. M. (1998) Cancer 82, 2191-2203. [DOI] [PubMed] [Google Scholar]
- 54.Lewis, J. J. & Brennan, M. F. (1996) Curr. Prob. Surg. 33, 817-872. [PubMed] [Google Scholar]
- 55.Kindblom, L. G., Ahlden, M., Meis-Kindblom, J. M. & Stenman, G. (1995) Virchows Arch. 427, 19-26. [DOI] [PubMed] [Google Scholar]
- 56.Menon, A. G., Anderson, K. M., Riccardi, V. M., Chung, R. Y., Whaley, J. M., Yandell, D. W., Farmer, G. E., Freiman, R. N., Lee, J. K., Li, F. P., et al. (1990) Proc. Natl. Acad. Sci. USA 87, 5435-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nielsen, G. P., Burns, K. L., Rosenberg, A. E. & Louis, D. N. (1998) Am. J. Pathol. 153, 159-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nielsen, G. P., Stemmer-Rachamimov, A. O., Ino, Y., Moller, M. B., Rosenberg, A. E. & Louis, D. N. (1999) Am. J. Pathol. 155, 1879-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sonobe, H., Takeuchi, T., Furihata, M., Taguchi, T., Kawai, A., Ohjimi, Y., Iwasaki, H., Kaneko, Y. & Ohtsuki, Y. (2000) Int. J. Oncol. 17, 347-352. [DOI] [PubMed] [Google Scholar]
- 60.Peterson, R. T., Shaw, S. Y., Peterson, T. A., Milan, D. J., Zhong, T. P., Schreiber, S. L., MacRae, C. A. & Fishman, M. C. (2004) Nat. Biotechnol. 22, 595-599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.