Abstract
Human pluripotent stem cells (hPSCs) are attractive sources for regenerative medicine and disease modeling in vitro. Directed hPSC differentiation approaches have derived from knowledge of cell development in vivo rather than from stochastic cell differentiation. Moreover, there has been great success in the generation of 3-dimensional organ-buds termed “organoids” from hPSCs; these consist of a variety of cell types in vitro to mimic organs in vivo. The organoid bears great potential in the study of human diseases in vitro especially when combined with CRISPR/Cas9-based genome editing approaches. We summarize the current literature describing organoid studies with a special focus on kidney organoids, and discuss goals and future opportunities for organoid-based studies.
Keywords: Organoids, Kidney, hPSCs
Human Pluripotent Stem Cells for Kidney Regenerative Medicine and Disease Modeling
Human pluripotent stem cells (hPSCs), by virtue of their unlimited self-renewal and ability to generate cells of all three embryonic germ layers, are ideally suited for the generation of functional human kidney cells and tissues [1, 2]. Human induced pluripotent stem cells (hiPSCs) can be readily generated from patients with kidney disease, enabling the development of immunocompatible tissues as well as patient-specific models of kidney disease [2]. Recent advances in directed differentiation protocols toward kidney lineage cells have resulted in the generation of nephron progenitor cells (NPCs) (see Glossary) and kidney (nephron) organoids from human embryonic stem cells (hESCs) and hiPSCs [3–7]. Chronic kidney disease (CKD) affects 9–14% of the U.S. adult population and the six regions of the world [8, 9]. Loss of functional nephrons and the development of tubulointerstitial fibrosis contribute to the progression of CKD, which impairs the regulation of fluid-electrolyte and acid-base balance, as well as the excretion of metabolic waste products with accumulation of uremic toxins. While adult kidneys possess an intrinsic capacity to self-repair following injury [10], the process of nephrogenesis, the formation of new nephrons, is limited to the period of embryonic development in humans [11]. NPCs and kidney organoids which can be generated now from hiPSCs derived from patients with kidney injury, might thus represent – as immunocompatible tissues – novel sources for ultimately generating new nephrons in CKD patients.
Other important applications which could lead to new insight and therapeutics stem from the use of hPSC-derived NPCs and kidney organoids to model kidney diseases in vitro. Animal models have contributed a great deal to our understanding of kidney physiology and pathophysiology. The models, however, have limitations in providing direction for therapeutic translation in humans. Recently, the mouse ENCODE Consortium reported that while there are many similarities, there are important differences between the mouse and human genomes [12–17]. Many DNA variations and differences in gene expression patterns have been uncovered, potentially limiting the usefulness of some disease models using mice. There are currently more than 160 inherited genetic kidney diseases [18]. By establishing hiPSCs from patients with a given genetic disease, it might be possible to study personalized disease mechanisms and to perform drug screening in vitro. Moreover, recent advances in genome editing have provided new approaches to modeling genetic kidney diseases using hPSCs in vitro [19]. CRISPR/Cas9 genome editing can be used to induce specific mutations at desired sites within the genome [20, 21]; therefore, it might be possible to study diseases without collecting somatic cells from patients. In this contribution, we review studies on differentiation approaches of hPSCs into kidney lineage cells, as well as on modeling kidney diseases. In addition, we discuss current trends and the future potential of using hPSC-derived kidney organoids to generate differentiated cells and structures which can be incorporated into bioengineered kidneys, hopefully replacing current dialysis and kidney transplantation therapies in the future.
The Development of Nephron Progenitor Cell Differentiation Protocols
Initially, differentiation protocols toward the kidney lineage were explored using mouse ESCs (mESCs) and/or mouse iPSCs (miPSCs) by testing growth factors with single-step or a few-step protocols [22–31]. From those mouse studies, a variety of growth factors were identified as potent inducers of kidney lineage cells: activin, bone morphogenetic protein 4 (BMP4), BMP7, retinoic acid, hepatocyte growth factor (HGF), and insulin-like growth factors (IGF). Most of these mouse studies, however, used fetal bovine serum (FBS) for the support of cell differentiation. Undefined components in FBS affected cell differentiation induced by defined growth factors. Some studies required transplantation of differentiated cells into mice in order to obtain kidney cell phenotypes [22, 26]. Most mouse studies used embryoid body (EB) formation in order to facilitate stochastic cell differentiation. Recently, published organoid differentiation methods have extended these approaches, applying EB formation methods to the generation of 3-dimensional (3D) structures [6, 32].
Following several studies with mESC and/or miPSC differentiation toward kidney lineage cells, research interest shifted towards using human pluripotent stem cells (hPSCs) and well-defined media components to achieve differentiation into kidney cells [4–6, 33–38]. Some directed differentiation approaches have attempted to mimic in vivo organ development step by step [39], in order to induce kidney lineage cells more efficiently, as well as to be able to induce more mature functional kidney tissues. Advances in our understanding of fundamental kidney development have guided directed differentiation protocols from hPSCs [6, 40–43]. In addition, the usage of small molecules for directed differentiation of hPSCs has also made these procedures more efficient, since small molecules typically yield highly penetrant effects across whole cell populations. For instance, usage of the glycogen synthase kinase 3 beta (GSK3β) inhibitor, CHIR99021 and 6-bromoindirubin-3'-oxime (BIO) have improved the differentiation efficiency of hPSCs into mesoderm and endoderm lineage cells by inducing primitive streak cells, the origin of mesendoderm [44–47].
It is known that kidneys arise from the intermediate mesoderm; however, the origin of functional kidneys, the metanephros, has not been clearly defined in the intermediate mesoderm, due to complexity of kidney development in humans. Three different kidney tissues, namely, pronephros, mesonephros, and metanephros form in humans during embryonic development. Only the metanephros survives and becomes a functional kidney while the pronephros and mesonephros degrade during embryonic development [48]. One of the most impactful studies in the development of kidney lineage differentiation protocols involved using lineage tracing techniques in mice to identify the precise origin of the metanephros, labeling specific cells to monitor subsequent differentiation [6]. The striking finding was that the origin of the metanephros was limited to the posterior area of the intermediate mesoderm where Osr1 and Wt1 were expressed, but Pax2 and Lim1 (LHX1 in humans) were not expressed. Pax2 and Lim1 have been used to specify the intermediate mesoderm in mouse embryos [49, 50], and have been used as markers to map the origin of kidney cells in studies attempting to induce kidney tubular cells from hPSCs [4, 5, 51]. Work from various laboratories, including ours, led to the generation of LTL+ (lotus tetragonolobus lectin) proximal tubular-like cells from hPSCs via induction of PAX2+LHX1+ cells [4, 5]; yet, the induction efficiency of SIX2+ nephron progenitor cells (NPCs) derived from PAX2+LHX1+ cells was low (~20%) [4, 5]. These findings were consistent with the previously mentioned study which redefined the origin of the metanephros to an Osr1+Wt1+Pax2−Lim1− posterior intermediate mesoderm in mice [6]. Thus, it was predicted that the induction of OSR1+WT1+PAX2−LHX1− posterior intermediate mesoderm cells from hPSCs would facilitate the differentiation into NPCs, and subsequently, into metanephros, i.e. functional kidneys.
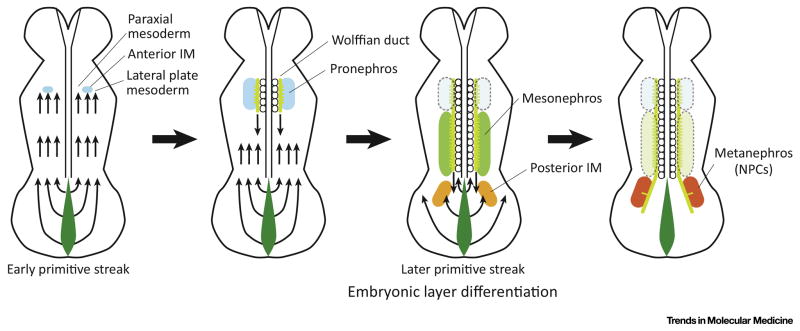
To induce the posterior intermediate mesoderm from hPSCs, it was important to precisely mimic the early patterning of mesoderm from the primitive streak [6]. The primitive streak is a structure that forms in the blastula during early stages of mammalian embryos, and appears as an elongating groove at the caudal or posterior end of the embryo (Fig. 1). The cells migrate from the primitive streak anteriorly and form mesoderm tissues from the paraxial to lateral plate mesoderm. Importantly, the cell locations in the primitive streak define subsequent differentiation into paraxial or lateral plate mesoderm (Fig. 1), as previously shown in chickens [43, 52]. Indeed, the cells located at the anterior part of the primitive streak differentiate into paraxial mesoderm while the posterior cells in the primitive streak become the lateral plate mesoderm. The origin of the intermediate mesoderm lies between the origins of the paraxial and lateral plate mesoderm. Thus, the progenitor cells of the intermediate mesoderm are located at the center of the primitive streak. The gradient of Wnt3a and Bmp4 guides the anterior-posterior axis of the primitive streak. Indeed, higher levels of BMP4 induce the posterior primitive streak in humans and mice [53, 54]. Hence, these results suggested that adjusting the BMP4 signal levels was important in order to induce cells that could mimic those at the center of the primitive streak, the origin of the intermediate mesoderm [3, 55, 56].
Figure 1. Mesoderm Patterning from the Primitive Streak in Mammals.
The location of cells in the primitive streak defines the subsequent differentiation into paraxial mesoderm, intermediate mesoderm, and lateral plate mesoderm. The early-stage primitive streak cells migrate anteriorly, and form mesoderm tissues at the anterior part of the embryo while the late-stage primitive streak cells forms the posterior mesoderm. The metanephros (containing NPCs) is derived from the late-stage mid-primitive streak cells. IM: intermediate mesoderm. NPCs: nephron progenitor cells.
The cells migrate from the primitive streak anteriorly, and form mesoderm tissues from the anterior part of embryos towards the posterior in mice [57]. The cells that migrate from the primitive streak at earlier stages of embryonic development subsequently differentiate into the more anterior mesoderm and those that migrate from the late stage of the primitive streak subsequently form the posterior mesoderm. This study also showed similar results using lineage tracing techniques to track the subsequent differentiation of Brachyury (T)+ primitive streak cells at different stages of embryonic development in mice [6]. T+ primitive streak cells at day E7.5 of mouse embryos were found to form the anterior intermediate mesoderm and subsequently differentiate into Wolffian ducts while T+ primitive streak cells at day E8.5 formed the posterior intermediate mesoderm and differentiated into the metanephric mesenchyme (NPCs). Collectively, results suggested that in order to induce the posterior intermediate mesoderm, it would be most efficient to induce the cells located at the center of the late-stage primitive streak, given that the intermediate mesoderm is derived from the center of the primitive streak, and the late-stage primitive streak gives rise to the posterior part of mesoderm. Since there are no specific markers to identify late-stage mid primitive streak cells during directed differentiation of hPSCs in vitro, the best timing and treatments of WNT and BMP4 modulators to be used were sought by examining the subsequent differentiation into WT1+HOXD11+ posterior intermediate mesoderm cells from hPSCs [3, 55]. Hoxd11 is also abundantly expressed in the lateral plate mesoderm in mice [58]; therefore, WT1 and OSR1 were used to specify the posterior intermediate mesoderm in combination with HOXD11 [3]. Initial screening experiments revealed that a longer treatment than previously used with the WNT activator CHIR99021 (CHIR) ((4 days versus 1–2 days) [4, 5], followed by subsequent differentiation induction with activin A, was more likely to induce HOXD11 expression [4, 5]. This suggested that the longer treatment with CHIR induced the late-stage primitive streak cells giving rise to posterior mesoderm cells. This finding was consistent with the developmental studies using mice, as discussed above [6]. A prolonged treatment of hPSCs with CHIR induced the later-stage of primitive streak cells which subsequently formed the posterior mesoderm, thus mimicking the anterior-posterior patterning of mesoderm in embryonic development in vivo [3]. In addition, subsequent treatment with activin A following CHIR treatment facilitated the expression of HOXD11. Very little is known about the regulation of HOXD11 expression. One study used a Hoxd11/lacZ reporter transgenic mouse to demonstrate that growth differentiation factor 11 (Gdf11, also known as BMP11) stimulated Hoxd11 gene expression via Smad2/3 binding to the Hoxd11 region VIII enhancer in cultured mouse tailbud fragments [59]. Taken together, these findings suggested that activin A might be capable of activating HOXD11 expression via activation of the SMAD2/3 signal in the protocol for directed differentiation of hPSCs, justifying the usage of activin A to induce WT1+HOXD11+ posterior intermediate mesoderm cells in Bonventre’s differentiation protocol [3].
Differentiation from the posterior intermediate mesoderm into SIX2+ NPCs (the metanephric mesenchyme) is better understood [4–6]. For instance, one study revealed the important role of Fgf9 and Fgf20 in the maintenance of NPC “stemness” in mice and humans by investigating the Six2+ NPC population with loss of Fgf9 or Fgf20 [60]. Moreover, a reciprocal interaction of the metanephric mesenchyme and ureteric buds was found to be a key component of kidney development in mice [61], indicating that two different lineage cells, and growth factor production from them were required for normal kidney formation. Both the metanephric mesenchyme and ureteric buds can produce growth factors that promote the differentiation of each other. Indeed, one study showed that a reduction in Fgf9 and Fgf20 levels led to a loss of NPCs in mice, suggesting that FGF proteins play critical signaling roles in NPC differentiation and maintenance [60]. The report also revealed that Fgf9 was produced by ureteric buds while Fgf20 was expressed in NPCs in mice. These results are significant in that they suggest that treatment of hPSCs with FGF9 might be required to induce NPCs from posterior intermediate mesoderm cells, unless ureteric bud cells are simultaneously induced by the directed differentiation protocol [3]. A low dose of FGF9 (10 ng/ml) was used to induce SIX2+SALL1+WT1+PAX2+ NPCs from WT1+HOXD11+ posterior intermediate mesoderm cells that had been derived from hPSCs [3]. However, the addition of FGF20 was not required, as predicted by another study [60]; this indicated that hPSC-derived NPCs presented similar characteristics to those of NPCs in vivo, capable of expressing FGF20 by themselves.
Comparison of Differentiation Protocols to Generate NPCs
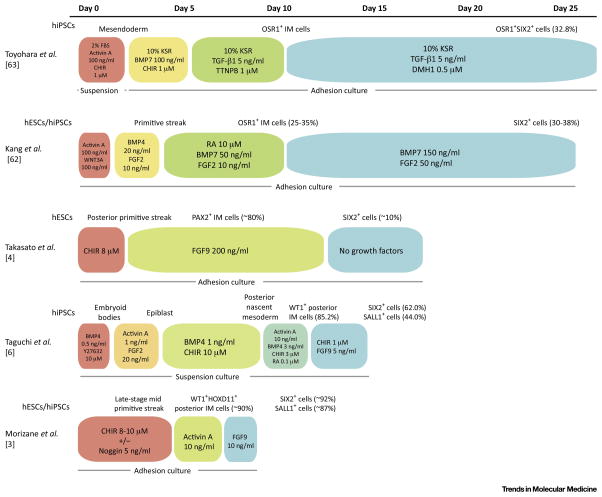
NPCs derived from hPSCs would be attractive sources for cell therapies and bioengineered kidney structures. In Fig 2 we have summarized 5 published protocols which have resulted in the generation of cells expressing NPC markers such as SIX2. Most of those studies employed adhesion cultures, which are suited for mass production of NPCs [3, 4, 62, 63] while one group used suspension cultures for the entire period of differentiation [6]. One key difference among these 5 protocols is whether posterior intermediate mesoderm cells could be induced as progenitors of NPCs, or not. The protocols from the Little, Han and Osafune laboratories attempted to induce PAX2+ or OSR+ intermediate mesoderm (IM) cells, and resulted in a relatively modest efficiency of NPC induction (SIX2+ cells: 10~38%) following longer differentiation periods (16~27 days) relative to Bonventre’s differentiation protocol (8~9 days) [3, 4, 55, 62, 63]. Protocols from the Nishinakamura and Bonventre laboratories attempted to induce posterior IM cells specifically [3, 6]. Nishinakamura’s approach resulted in a higher differentiation efficiency of IM cells (SIX2+ cells: 62%) with shorter differentiation periods (9~14 days) in humans [3]. In Bonventre’s study, IM cell induction efficiency was evaluated at each step of differentiation, relying on immunohistochemical assays of multiple protein markers so as to follow in vivo kidney development through the posterior intermediate mesoderm induction [6]. This resulted in a highly efficient (SIX2+ cells: ~75–92%) and rapid (~9 days) differentiation protocol deriving hESCs as well as hiPSCs into SIX2+SALL1+WT1+PAX2+ NPCs [3].
Figure 2. Comparison of Human Nephron Progenitor Cell Differentiation Protocols.
The depicted schematic summarizes recent protocols that have been used to differentiate nephron progenitor cells from hPSCs. Inducing factors, concentrations, and treatment duration in each step of differentiation protocols are shown with colored boxes. The culture method, and the suspension or adhesion culture, is indicated at the bottom of each protocol. The cell types induced by each step of differentiation are shown on the top of the protocols. IM: intermediate mesoderm. FBS: fetal bovine serum. CHIR: CHIR99021. KSR: knockout serum replacement. BMP: bone morphogenetic protein. TGF: transforming growth factor. TTNPB: 4-[(E)-2-(5,6,7,8-Tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl]benzoic acid, a retinoic acid receptor agonist. DMH1: 4-[6-(4-Isopropoxyphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, 4-[6-[4-(1-Methylethoxy)phenyl]pyrazolo[1,5-a]pyrimidin-3-yl]-quinoline, a selective inhibitor of BMP ALK2 receptor. Wnt3a: wingless-type MMTV integration site family, member 3A. FGF: fibroblast growth factor. RA: retinoic acid. Y27632: a selective p160ROCK inhibitor. OSR1: odd-skipped related transcription factor 1. SIX2: SIX homeobox 2. PAX2: paired box 2. WT1: Wilms tumor 1. SALL1: spalt like transcription factor 1. HOXD11: homeobox D11.
Comparison of Kidney Organoid Differentiation Protocols
Currently, there are several studies that have reported the differentiation of hPSCs into cells of the kidney lineage [3–7, 34, 35, 64]. Kidneys consist of many different cell types in the nephron, vasculature and interstitial compartments. Nephron epithelial cells occupy 80–90% of the kidney cortex. The nephron is the functional unit of the kidney, and consists of a structure responsible for filtration (a glomerulus), as well as the multisegmented tubule which is responsible for reabsorption and secretion of a large number of solutes and reabsorption of water. The glomerulus filters blood, driven by hydrostatic pressure, with passive selection by size and electrical charge of substances. This results in the production of a glomerular filtrate which then enters the proximal tubule as primitive urine. The proximal convoluted tubules, the loops of Henle -- containing a straight portion of the proximal and distal tubules, the distal convoluted tubules, as well as the connecting tubules and collecting ducts reabsorb most of the glomerular filtrate. The residues of the glomerular infiltrate are then excreted through the collecting ducts, renal pelvis and ureters into the bladder and expelled from human bodies as urine. Loss of nephrons is characteristic of chronic kidney disease (CKD), which eventually leads to end-stage kidney disease (ESKD) where patients require kidney replacement therapies such as hemodialysis and kidney transplantation to maintain healthy kidney function. To regenerate kidneys, it is therefore highly important to efficiently generate nephrons from hPSCs without contamination of other cell lineages.
Nephrons form complicated architectural 3D structures to function. Therefore, to recapitulate these structures ex vivo, the development of 3D culture systems is required. Organoids are 3D organ-like tissues which mimic structurally and functionally in vivo organs in culture plates in vitro. Although there is no clear definition as to how many cell types should be included, the following features are generally considered to constitute the main characteristics of organoids: i. 3D structures, ii. complex multicellular constructs, iii. self-organization, iv. in vitro culture, and v. recapitulation of developmental programs. For a recent summary of published organoid studies and categorization of organoids into two groups (organoids from primary tissues, and organoids from PSCs), see [65].
There are a number of published studies where organoids are generated from mouse and/or human primary tissues including lingual [66], salivary grand [67], esophagus [68, 69], stomach [70–77], intestine [68, 78–97], colon [68, 84, 98–101], liver [102, 103], pancreas [104–106], prostate [107–109], and lung [110, 111]. Notably, many primary tissue organoids have been generated from Lgr5+ stem cells from primary tissue using R-spondin, a LGR4/5 ligand [65]. These organoids have not been reconstructed by dissociation and reaggregation of the primary tissue, but instead, have been generated from the stem cell population residing within the tissue. In human adult kidneys, some studies previously suggested the presence of kidney stem cells in the kidney, as specified by co-expression of CD133 and CD24 [112, 113]. However, a number of reports have documented that terminally differentiated epithelia can re-express apparent stem-cell markers during injury-induced dedifferentiation and repair in mice, with the authors concluding that there is no evidence for a stem-cell population in adult kidneys [114–118]. It is therefore unlikely that there are kidney stem cells in adult mammalian kidneys. To date, there are no reports demonstrating kidney organoid generation from adult kidneys, possibly due to the absence of stem/progenitor cell populations in mammals.
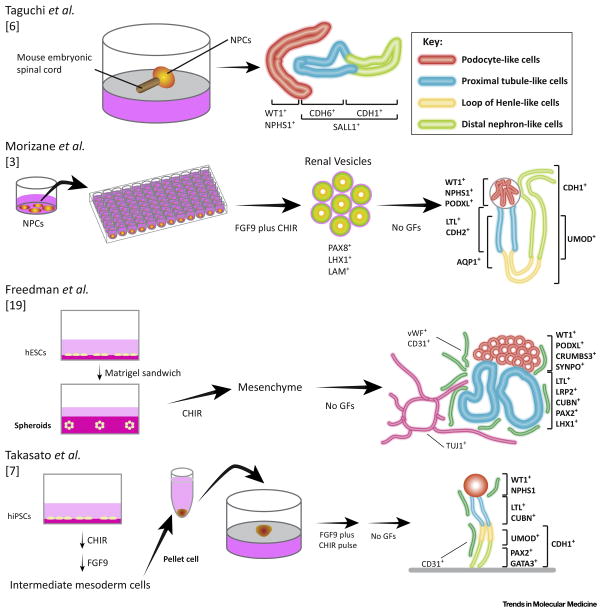
There are currently several published studies in which hPSCs are differentiated into organoids of various tissues including stomach [119], intestine [120–122], liver [46, 123, 124], lung [125], brain [126], anterior pituitary [127], and kidneys [3, 6, 7, 19, 51]. Here, we summarize four studies that generated kidney organoids from hPSCs (Key Figure, Figure 3, and Figure 4). Two of those 4 studies, including ours, first induced NPCs from hPSCs, and then generated kidney organoids from NPCs [3, 6]. Nishinakamura’s laboratory used mouse embryonic spinal cords to stimulate epithelialization of NPCs on a polycarbonate filter, demonstrating early-stage nephron structures resembling S-shaped bodies which expressed markers for podocytes (WT1 and NPHS1) and tubules (CDH1 and CDH6); however, NPC markers, such as SALL1 and PAX2, were still expressed in those structures, indicating that immature nephrons were being induced from the mouse embryonic spinal cords [6]. This work was the first to demonstrate the differentiation of hPSCs into kidney organoids which contained nephron-like structures; however, further refinement of the protocol was required to facilitate different biological applications of hPSCs.
Key Figure, Figure 3. Methods for Generating Kidney Organoids.
The diagram depicts recent protocols used to generate kidney organoids from human pluripotent stem cells. Taguchi [6], Morizane [3] and colleagues generated kidney organoids from nephron progenitor cells (NPCs) while Freedman [19], Takasato [7] and colleagues differentiated mesenchyme cells or intermediate mesoderm cells into kidney organoids. Red lines: podocyte-like cells. Light blue lines: proximal tubule-like cells. Yellow lines: loops of Henle-like cells. Light green lines: distal nephron-like cells. NPCs: nephron progenitor cells. WT1: Wilms tumor 1. NPHS1: nephrosis 1, nephrin. CDH6: cadherin 6. CDH1: cadherin 1. SALL1: spalt like transcription factor 1. PAX8: paired box 8. LHX1: LIM homeobox 1. LAM: laminin. PODXL: podocalyxin like. LTL: lotus tetragonolobus lectin. CDH1: cadherin 2. AQP1: aquaporin 1. UMOD: uromodulin. vWF: von Willebrand factor. CD31: cluster of differentiation 31. TUJ1: neuron-specific class 3 beta-tubulin. SYNPO: synaptopodin. LRP2: LDL receptor related protein 2. CUBN: cubilin. PAX2: paired box 2. GATA3: GATA binding protein 3. FGF: fibroblast growth factor. CHIR: CHIR99021.
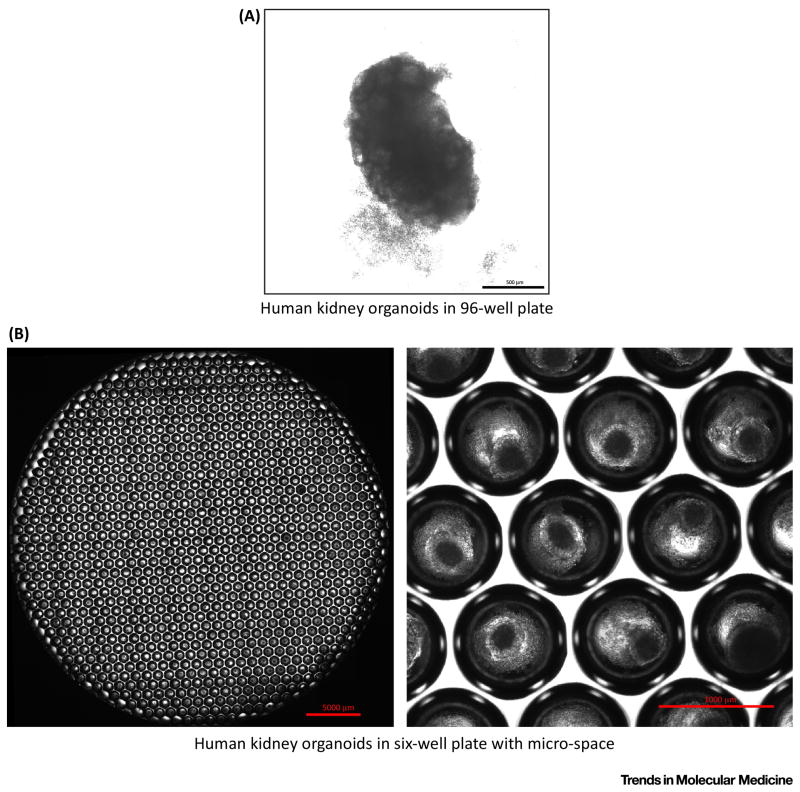
Figure 4. Human Kidney Organoids in Culture.
Representative micrograph images of human kidney organoids are shown in 3D culture.
(a) Human kidney organoids were generated in 96-well ultra-low attachment plates with U-shaped bottom on day 24 of initiation of differentiation. Scale bar: 500 μm. Images were taken with an inverted microscope (Nikon, Eclipse Ti) at 40x magnification. (b) Human kidney organoids were generated in 6-well plates with micro-space (Elplasia, #RB900700NA6) on day 22 of initiation of differentiation. Scale bars: 5 mm (left), and 1 mm (right). Photographs were taken with an inverted microscope (Nikon, Eclipse Ti) at 40x magnification. The left picture was generated by merging 40x pictures to show a whole well of a 6-well plate.
Important applications of hPSCs include the modeling of human diseases to gain insight into pathophysiological processes. Furthermore, hiPSCs derived from patients with a given disease, or CRISPR-Cas9 induced mutations of hPSCs have been used to model genetic abnormalities when testing potential therapeutic agents in drug discovery [128]. For this purpose, organoids have to be generated in small-scale cultures suited for high-throughput screening. Protocols that require co-cultures with mouse embryonic spinal cord to generate kidney organoids, may hinder drug screening, as the number of samples can be limited when collecting mouse embryonic spinal cord. Moreover, the use of mouse embryonic spinal cords with their undefined components (such as growth factors), represent a limitation in disease modeling and mechanistictic analysis; indeed, these undefined components might affect disease phenotypes in kidney organoids derived from patients with genetic kidney disease [6].
To address these issues, our laboratory developed a differentiation protocol for kidney organoids with fully defined components in small-scale cultures suitable for 96-well screening dishes [3, 55]. The protocol was designed to follow the in vivo development of metanephric functional kidneys, generating NPCs with 75~92% efficiency (Fig. 2) [3, 129]. Two methods for generating kidney organoids from NPCs have been developed: The first approach focuses on simply continuing hPSC differentiation from the NPC stage in regular culture plates as a 2D culture system which is suited for live cell monitoring. This is a standard adhesion culture, yet cells spontaneously form multiple layers and 3D structures in culture plates [3, 55]. This 2D system is well-suited for live cell monitoring and immunocytochemical analyses, since the structures are not too thick. The second approach involves a 3D culture system using ultra-low attachment 96-well round bottom plates [3, 55]. By re-plating NPCs onto ultra-low attachment plates, a large number of kidney organoids have been generated in small-scale 3D cultures. These kidney organoids were shown to contain segmented nephrons with regions bearing characteristics of podocytes (WT1+PODXL+NPHS1+, WT1: Wilms tumor 1, PODXL: poxocalyxin like, NPHS1: nephrin), proximal tubules (LTL+CDH2+AQP1+, LTL: lotus tetragonolobus lectin, CDH2: cadherin 2, AQP1: aquaporin 1), loops of Henle (CDH1+UMOD+, UMOD: uromodulin), and late distal tubules (CDH1+UMOD−) in an organized, continuous arrangement [3]. Since the induction efficiency of NPCs was found to be high, the derivation of kidney organoids in each small well was shown to be quite reproducible using this differentiation approach. Fully defined components (such as serum-free media) were used for the differentiation step and subsequently, in vivo development followed with high efficiency [3].
In terms of the utility of the organoid, this system facilitates a systematic developmental analysis of the kidneys. In one example, organoids were exposed to DAPT, the Notch inhibitor, after induction of renal vesicles[3]. The results implicated Notch signaling as critically important in the development of the proximal tubule during nephron formation [3]. DAPT significantly reduced the induction of LTL+ proximal tubules. And, this result was consistent with previous mouse studies [130], indicating that this organoid system could represent a useful platform for studies on human kidney development.
Unlike Nishinakamura’s study, the persistent expression of nephron progenitor markers such as SALL1 was not observed in the induced nephrons using our protocol, suggesting that the organoid protocol induced more mature nephrons [3]. Mature nephrons express a variety of transporters, and nephrotoxicants are taken up via those transporters [131]. For instance, cisplatin is a clinically-used anti-cancer drug that is known to cause kidney injury [132]. As such, proximal tubular cells take up cisplatin via organic cation transporters (OCTs) [133, 134]. When treated with cisplatin at 5 μM for 24 hours, kidney organoids were shown to respond to cisplatin by increasing the expression of kidney injury molecule-1 (KIM-1) -- a well-established injury marker for proximal tubules -- in LTL+ proximal tubules [3, 135]. γH2AX protein expression -- a DNA damage marker -- [136] was also evaluated to determine whether cisplatin-induced injury at 5 or 50 μM, could be specific to proximal tubules. At a cisplatin dose of 5 μM, these organoids showed specific upregulation of γH2AX only in LTL+ proximal tubules, indicating that LTL+ proximal tubules in organoids could actively take in cisplatin, presumably via OCTs. A cisplatin dose of 50 μM indeed resulted in global DNA damage in all cell types within the kidney organoids [3].
In two other studies, NPCs were not reported in the preliminary step of kidney organoid generation; but nevertheless, resulted in the generation of kidney organoids which included nephron-like cells [7, 19]. In another study, kidney organoids were generated using a unique approach [19]: cavitated spheroids were first generated from hPSCs by ‘sandwiching’ cells between two layers of Matrigel. Those spheroids possessed pluripotency, and the cells expressed pluripotent markers including OCT4, SOX2, TRA1–60, and NANOG (Yamanaka factors), even after several cell passages. Differentiation from those cavitated spheroids was initiated upon CHIR administration for 1.5 days, after which the cells were stochastically differentiated for up to 16 days [19]. Although foot processes were not observed by electron microscopy, PODXL+WT1+SYNPO+ podocyte-like cells were induced. There were also LTL+ tubular structures resembling proximal tubules, yet, immature markers including LHX1 and PAX2, were still expressed in these LTL+ tubular structures, suggesting that the nephron structures might have been less mature. Nephrotoxic assays were also undertaken, and KIM-1 was upregulated in LTL+ tubular structures after a 48-hour treatment with 50 μM cisplatin [19]. Importantly, the protocol induced other cell types along with nephron-like cells; vWF+CD31+ endothelial-like cells and TUJ1+ neuron-like cells were induced simultaneously, suggesting that multiple lineages could be generated, including lateral plate and ectoderm derivatives. Overall, the stochastic approach of differentiation into kidney lineages appeared to result in less specific differentiation into nephron-like cells, with possibly, less mature phenotypes.
In another study, Little and colleagues reported the generation of kidney organoids containing multiple lineages [7]. Interestingly, this study did not report NPCs as a preliminary stage of kidney organoid generation either. hiPSCs were differentiated with CHIR for 4 days, and the cells subsequently treated with 200 ng/ml FGF9 for 3 days. After dissociation into single cells, these were spun down to form a pellet which was then cultured on a transwell dish with 200 ng/ml FGF9 for 5 days, following CHIR pulse treatment. The cell pellet was then cultured without additional growth factors for an additional 13 days, and segmented nephron-like structures were generated. These included WT1+NPHS1+ podocytes, LTL+CUBN+ immature proximal tubules, LTL+CDH1+ mature proximal tubules, CDH1+UMOD+ loops of Henle, and CDH1+GATA3+PAX2+ collecting ducts. This study also observed CD31+ endothelial-like cells in kidney organoids. Nephrotoxic assays were performed using cisplatin at 5 μM and 20 μM for 24 hours, and LTL+CDH1− proximal tubules failed to respond, while LTL+CDH1+ mature proximal tubules underwent apoptosis, as evidenced by cleaved caspase 3 expression, suggesting that LTL+CHD1+ tubules, but not LTL+CDH1− tubules, expressed organic cation transporters for cisplatin uptake [7]. In mice, Cdh1 is expressed in proximal tubules when mature [137]; however, human, rat, and pig proximal tubules have been reported to not express CDH1 [138–140]. It is possible that LTL+CDH1+ tubules were present in the transitional segment from proximal tubules to distal tubules. The gene expression profiles of these kidney organoids were very similar to those of first trimester human kidneys. It is therefore possible that nephron epithelial cells represented an earlier maturation stage [7]. However, further studies are warranted to validate this supposition and to carry out similar studies using the other protocols we have described.
In conclusion, four studies have collectively demonstrated the successful generation of human kidney organoids using different approaches. Recognizing, however, that pronephros and mesonephros contain immature nephrons during embryogenesis in humans [48, 141] and kidney development is very complicated, it is quite difficult to distinguish metanephros-derived functional kidney structures, from pro/mesonephros based only on marker analyses. Therefore, further studies are desired to fully characterize the functionalities of such kidney organoids and to explore potential applications of kidney organoids for modeling kidney diseases.
Disease Modeling using Kidney Organoids
One of the most important applications of hPSCs is to model human diseases in vitro in order to study mechanisms of disease and to find new therapeutic approaches. One strength of this application is that a large number of chemicals can be screened in order to find new candidate therapeutic drugs, an application often not possible in animal studies, where the sample numbers are limited and there has been increasing concern about translatability of findings to humans. There are currently more than 160 known inherited kidney diseases [18]. These diseases affect mostly children and approximately 10% of adults develop end-stage kidney disease. Regrettably, there are currently few models that have led to new therapeutics for inherited kidney diseases. We suspect that there are currently many ongoing studies to model inherited kidney diseases using kidney organoid systems, and hope that disease mechanisms and potent therapeutic approaches will be elucidated in order to bring benefits to patients with kidney diseases.
Published kidney-related studies with iPSCs and organoids have focused on polycystic kidney disease (PKD), especially, autosomal dominant PKD (ADPKD). ADPKD is the most common inherited kidney disease and accounts for 7–10% of all patients on renal replacement therapy worldwide [142]. ADPKD, however, is a late-onset disease where cysts can be detected by ultrasound in approximately 68% in individuals of ages 30 years and older [143]. For this reason, it might be difficult to find cystic phenotypes in kidney organoids derived from cells of patients with ADPKD, yet early phenotypes of ADPKD might be potentially detected in kidney organoids or undifferentiated hiPSCs, particularly since the target genes, PKD1/PKD2, encode for ciliary proteins expressed in many cell types [144]. Our laboratory studied hiPSCs generated from three ADPKD patients harboring PKD1 mutations [145]. Specifically, protein localization of PKD1 and PKD2 in undifferentiated hiPSCs and hepatoblasts derived from hiPSCs were analyzed. The data revealed that the expression of PKD2 protein in cilia was reduced in cells from patients with PKD1 mutations relative to controls, suggesting that normal trafficking of PKD2 to the cilium was mediated by normal PKD1 protein [145]. This phenomenon might help explain the cause for cystogenesis in ADPKD patients; yet, further studies are required to elucidate the mechanisms of cystogenesis due to reduced PKD2 expression in the cilium and furthermore, on how this relates to why it takes a number of decades to form cysts in ADPKD patients. Another recent study on ADPKD using patient-derived hiPSCs attempted to explore risk factors for intracranial aneurysms [146]. The authors established hiPSCs from three ADPKD patients without intracranial aneurysms and four patients with aneurysms, and compared gene expression profiles between those two groups after hiPSC differentiation into endothelial cells. They found that the expression level of a metalloenzyme gene, matric metaloproteinase (MMP)-1 was specifically elevated in hiPSC-derived endothelial cells from ADPKD patients with aneurysms. In addition, they confirmed a positive correlation between serum MMP1 levels and the development of intracranial aneurysms in 354 ADPKD patients, indicating that high serum MMP1 levels might constitute a novel risk factor for ADPKD [146].
In both of these studies, clinical manifestations of ADPKD, such as kidney cysts and aneurysms were not reproduced in culture systems using hiPSCs derived from ADPKD patients [145, 146]. One reason might be that there were inadequate differentiation protocols to generate kidney organoids at that time. In addition, considering the long time course of disease progression of ADPKD, alternative approaches to modeling ADPKD might need to be developed beyond the study of patient-derived hiPSCs alone, so that clinical phenotypes can be reproduced in culture systems. We had hypothesized that a complete knock-out of the PKD1 or PKD2 genes in hiPSCs might facilitate cystic phenotypes in culture systems. To this end, PKD1 or PKD2 knock-out cell lines were generated in H9 hESCs using CRISPR/Cas9 genome editing approaches [19]. Although the differentiation protocol into kidney organoids might need further refinement, cyst formation was observed in 6% of the kidney organoids derived from CRISPR-mutants, while cyst formation was rarely detected in kidney organoids derived from wild-type H9 cells [19]. One important advantage of this CRISPR approach is that the genetic background is exactly the same in the non-mutated control parental line. Because of recent findings stating that a genetic background can dominate over variation due to cellular origin in hPSCs, or the reprogramming procedure to convert somatic cells into iPSCs using Sendai virus (SeV) [147], it may be desirable to use hPSCs with exactly the same genetic background as controls when analyzing phenotypes resulting from CRISPR/Cas9-induced genetic modifications. As such, two approaches are complementary and most informative: the comparison of hiPSCs from patients with and without disease, as well as the comparison of wild-type cells with CRISPR/Cas9 mutated cells derived from these wild-type cells. A recent study from Yoshida’s laboratory demonstrated the importance of epigenetic variation between hiPSC lines [148]. They used 35 hiPSC lines and four hESC lines to explore the effects of DNA methylation and chromatin status on the capacity to differentiate into hematopoietic cells. Their analyses revealed that commitment of hPSCs to hematopoietic precursors correlated with IGF2 gene expression levels, which in turn, was regulated by signaling-dependent chromatin accessibility at mesendodermal genes. Maturation capacity for conversion of hPSC-derived hematopoietic precursors to mature blood cells was associated with the amount and pattern of DNA methylation acquired during reprogramming [148]. Because variation in the differentiation capacity of hPSCs into specific lineages is a significant concern when considering their use in disease modeling, these current findings might also support the application of CRISPR-based approaches for disease modeling in order to minimize the extent of epigenetic variation in diseased as well as normal cell lines.
In summary, we will undoubtedly see many attempts to model kidney diseases using kidney organoids derived from hPSCs in the future. It is an attractive approach to studying mechanisms of human inherited kidney diseases which might then be applied to more common diseases as well as to the development of novel candidate therapies using human tissues which, we believe, may facilitate translation of this methodology to humans. However, in order to optimize this approach, differentiation protocols, genetic background, as well as epigenetic variation will need to be considered in depth when disease phenotypes are analyzed in kidney organoids.
Generation of Functional Bioengineered Kidneys
One goal of hPSC studies would be to regenerate kidney function. Kidneys form very complicated structures with blood filtration and urine reabsorption units needed for their function and homeostasis. There are many challenges to using organoids to generate functional bioengineered kidney tissues. One of the challenges relates to vascularization; vascularization of kidney organoids needs to be induced in an organized way to direct blood flow from arteries to then drain into venous structures. In one study, the authors demonstrated vascularization of glomeruli when hPSC-derived podocytes were transplanted into mouse kidney subcapsular spaces [149]. Outgrowth of host mouse endothelial cells into the region of transplanted human podocytes was then observed, though capillary loops--an important structure of glomeruli facilitating blood filtration-- were rarely formed [149]. Another study generated kidney buds by mixing human umbilical vein endothelial cells (HUVEC), mesenchymal stem cells (MSCs), and mouse embryonic kidney cells [150]. Kidney buds were then transplanted into a preformed cranial window of non-obese diabetic/severe combined immunodeficient (NOD/SCID) mice where immunorejection against kidney buds containing human cells was minimal [150]. Here again, vascularized glomerular structures were observed with outgrowth of host mouse endothelial cells [150]. These in vivo transplantation experiments are encouraging; however, further studies are required to incorporate vascular systems into kidney organoids in vitro, an important step which might facilitate the development of functional bioengineered kidneys in the future.
A second challenge involves the egress of formative urine. Current studies suggest resolving this challenge by inducing the formation of collecting duct cells in organoids [7, 37]; yet, the formed structures have not been sufficiently organized to excrete urine outside the kidney organoids. One alternative approach might be to co-culture hPSC-derived NPCs and ureteric buds in order to recapitulate renal structure development in vivo, where the reciprocal interactions of NPCs and ureteric buds could result in organized branching formation of collecting duct systems. Currently, one report has documented the induction of ureteric bud progenitor-like cells from hPSCs [37]; however, a subsequent directed differentiation into ureteric buds and collecting duct cells has not been established.
Considering Nishinakamura’s recent study [6], differentiation protocols into ureteric bud lineage cells need to be established independently from NPC differentiation, since the origins of ureteric buds and NPCs are distinct at the early stage of embryogenesis. It might then be reasonable to suppose that the establishment of differentiation protocols to produce ureteric bud cells from hPSCs could facilitate the generation of organized kidney structures with urinary exit.
A third challenge in scaling up the production of kidney tissues would be to improve the efficiency of the formation of nephrons. For example, one kidney contains approximately one million nephrons (although this number is highly variable); therefore, it is hardly possible that the small number of NPCs transplantated into kidneys will form sufficient nephrons to compensate for lost kidney function. One study showed improvement of kidney function after induction of acute kidney injury (AKI) with ischemia reperfusion, when OSR1+SIX2+ NPCs were transplanted into mouse kidney [63]. The authors concluded that the improvement in AKI was due to the trophic effects of NPCs providing renoprotective factors such as angiopoietin (ANG)-1 and vascular endothelial growth factor (VEGF)-A, given there was no new nephron formation from transplanted hiPSC-derived NPCs in host mouse kidneys. This may be a reasonable conclusion, and the approach might be applicable to patients with AKI. In ESKD, however, it will be necessary to generate a sufficient number of functional nephrons to replace at least 10–20% of kidney function since there are very few nephrons present to respond to the paracrine effects of NPCs.
Another possible approach might be to use decellularized kidneys and then repopulate these cell-free structures to generate functional kidneys [151]. As an example, a method was developed to decellularize whole kidneys from rats, swine and humans, by perfusing detergent into the renal artery [152]. Decellularized rat kidneys were seeded with rat neonatal kidney cells (NKC) via the ureters, and HUVECs, via the renal artery. Grafts produced rudimentary urine in vitro when perfused through their intrinsic vascular bed. The authors also transplanted the graft in an orthotopic position [152] in rat, which resulted in production of urine through the ureteral conduit in vivo. However, translation of this approach will clearly require the optimization of cell seeding methods to human-sized scaffolds, as well as the establishment of protocols to ensure that the cells are delivered to the entire nephron. In addition, upscaling of biomimetic organ cultures, as well as preparation of required cell types from hPSCs will also be needed.
Finally, another possible strategy for the generation of functional bioengineered kidneys includes the use of 3D bioprinting. Lewis’ laboratory has developed a new bioprinting method to fabricate 3D tissue constructs replete with vasculature, multiple cells types, including proximal tubular cells, and an extracellular matrix [153, 154]. This novel system is being used to generate heterogeneous structures in 3D by precisely co-printing multiple materials, known as bioinks. The system is capable of perfusing liquid into lumens of vasculature or tubule-like channels, which might be employed to mimic blood flow and intratubular flow in vascular and tubular channels, respectively [153–155].
Concluding Remarks
Significant advances have been made within the past decade to generate kidney organoids from hPSCs. The utility of kidney organoids is rapidly evolving to study kidney injury and repair, inherited kidney diseases, and nephrotoxicity of drugs in vitro. Advances in genome editing with CRISPR/Cas9 are further promoting the usage of kidney organoids to study inherited genetic kidney diseases. Despite progress in differentiation approaches and genome editing of hPSCs, several challenging questions need to be answered (see Outstanding Questions and Box 1). Modeling inherited kidney diseases is one of the most attractive applications of kidney organoids, yet some modifications to promote phenotypes might be required to model late-onset diseases such as ADPKD. In addition, the nature of the differentiation protocols into kidney organoids might affect disease phenotypes, considering that 4 different published approaches have generated kidney organoids with different characteristics. Quality control of kidney organoids will also be an important consideration for disease modeling in addition to the potential sources for developing kidney regenerative therapies. However, there is currently no consensus with regard to the methods that should be used for quality evaluation of kidney organoids, especially in terms of their functional characterization.
OUTSTANDING QUESTIONS BOX.
Recent progress in the differentiation of hPSCs towards kidney lineages has resulted in the generation of kidney-like tissues in vitro by different approaches, yet several challenging questions need to be answered:
What functionalities do kidney organoid structures possess and how well do they reproduce the functional characteristics of the human kidney in vivo?
How do we best take advantage of kidney organoids and genome editing to model kidney diseases and find new therapeutics for kidney diseases?
How can vascularization of kidney organoids be achieved in vitro?
How can functional integrated and vascularized bioengineered kidneys be generated with paths bringing blood supply as well as drainage, to eliminate waste products?
Box 1. CLINICIAN’S CORNER BOX.
Recent advances in stem cell biology have resulted in the generation of 3-D tissues of kidneys (kidney organoids) from human pluripotent stem cells (hPSCs) in vitro.
Kidney organoids have been demonstrated to model human kidney development, injury, as well as various kidney diseases.
Knock-out models of polycystin using CRISPR/Cas9 genome editing have shown the feasibility of using these approaches to model autosomal dominant polycystic kidney disease (ADPKD).
3D bioprinting will contribute to the development of functional bioengineered kidneys with hPSC
From the complex structural and functional nature of kidneys, which includes blood filtration and urine reabsorption, kidney organoid designs pose a difficult challenge. For urinary production, proper vascularization of kidney organoids will be required, as will “hooking up” the nephron to the drainage system. The presence of endothelial lineage cells in kidney organoids is encouraging, but it will be important to develop perfusable systems in vitro in order to mimic the in vivo kidney environment. Nevertheless, the translational journey of kidney organoids has only just begun.
Supplementary Material
TRENDS BOX.
Methods have been established to generate kidney organoids from human pluripotent stem cells (hPSCs). These organoids consist of cells with the characteristics of podocytes, proximal tubules, loops of Henle, and distal convoluted tubules in a contiguous arrangement resembling nephrons in vivo.
Polycystin knock-out mutants of hPSCs generated with CRISPR/Cas9 genome editing exhibit cystic phenotypes in kidney organoids. This specific mutagenesis approach will complement current efforts to use patient-derived human induced pluripotent stem cells (hiPSCs) in order to model human genetic diseases.
Whole rat, pig, and human kidneys have been decellularized with detergent perfusion, yielding acellular scaffolds with vascular, cortical and medullary tubules, as well as the collecting system architecture, intact.
Bioprinting methods have resulted in the fabrication of 3D vascular constructs with human umbilical vein endothelial cells. This approach may be applied to generate functional bioengineered kidneys.
Acknowledgments
This study was supported by National Institutes of Health grants R37 DK039773 and R01 DK072381 (to J.V.B.), a Grant-in-Aid for a Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship for Research Abroad (to R.M.), a ReproCell Stem Cell Research grant (to R.M.), a Brigham and Women’s Hospital Research Excellence Award (to R.M.), a Brigham and Women’s Hospital Faculty Career Development Award (to R.M.) and a Harvard Stem Cell Institute Seed Grant (to R.M.).
Glossary
- Nephron progenitor cells (NPCs)
exist in metanephric mesenchyme in embryonic kidneys and have the capacity to differentiate into nephrons consisting of the renal corpuscle (podocytes and Bowman’s capsules), proximal tubules, loops of Henle, and distal tubules. A transcription factor, Six2 defines and regulates the multipotent self-renewing NPC population throughout mammalian kidney development.
- Tubulointerstitial fibrosis
the pathogenesis of chronic kidney disease, characterized by continuous accumulation of extracellular matrix leading to a diffuse fibrosis.
- CRISPR/Cas9 genome editing
a gene editing platform derived from Streptococcus Pyogenes using an endonuclease (Cas9) and a synthetic guide RNA to introduce a double strand DNA break at a specific location within the genome.
- Activin
growth factor belonging to the transforming growth factor (TGF)-β superfamily, participatng in the regulation of biological processes including cell differentiation and proliferation. Activin is used for differentiation of pluripotent stem cells into mesoderm and endoderm lineages.
- BMP4, BMP7
growth factors belonging to the bone morphogenetic protein family (part of the TGF-β superfamily). The gradient of BMP4 levels during embryogenesis patterns mesoderm compartments including paraxial mesoderm, intermediate mesoderm, and lateral plate mesoderm. BMP4 is involved in development of a variety of tissues including the muscle, bone, heart, eye, and ureteric bud. BMP7 is involved in kidney development through induction of mesenchymal-epithelial transition of metanephric mesenchyme.
- Retinoic acid
metabolite of vitamin A mediating the functions of vitamin A required for growth and development. Retinoic acid is involved in the determination of the embryonic anterior-posterior axis.
- HGF
growth factor involved in cell growth, motility, and morphogenesis. HGF is secreted by mesenchymal cells and acts as a multi-functional cytokine on cells of mainly epithelial origin.
- IGF
growth factor with high sequence similarity to insulin. IGF is essential for development and function of organs such as the brain, liver, and kidney.
- Embryoid body
3D aggregates of pluripotent stem cells, generated by non-adherent (suspension) culture. The differentiation of pluripotent stem cells through embryoid body formation exhibit heterogeneous patterns of differentiated cell types, yet 3D structures can be generated in embryoid bodies.
- Primitive streak
transient structure whose formation marks the start of gastrulation, the process in which the inner cell mass is converted into the trilaminar embryonic disc, and comprised of the three germ layers (ectoderm, mesoderm, and endoderm).
- Metanephros
the most developed and functional kidney, which persists as the definitive adult kidney in mammals. The metanephros arises at the posterior part of intermediate mesoderm in embryos.
- Pronephros
the most primitive kidney, which cannot excrete urine outside embryos in mammals. The pronephros first arises at the anterior part of intermediate mesoderm, yet the pronephros disappears during the embryonic development in mammals.
- Mesonephros
the second kidney, which arises at the mid part of intermediate mesoderm after formation of pronephros in mammals. The mesonephros contains tubules and glomeruli which produce urine, yet the mesonephros degenerates during embryonic development in mammals.
- Kidney tubular cells
epithelial cells representing the majority of cells in kidneys. Kidney tubular cells reabsorb water, electrolytes, and nutrients from primitive urine which is produced by glomeruli. Kidney tubular cells include different segments of nephrons, namely proximal tubules, loops of Henle, and distal tubules.
- Lotus tetragonolobus lectin (LTL)
a family of closely related glycoproteins that appear to have similar specificities toward α-linked L-fucose containing oligosaccharises. LTL binds to brush borders which exist at the luminal surface of proximal tubules in kidneys; therefore, LTL is used as a marker of proximal tubular cells.
- SIX2
transcription factor which defines and regulates a multipotent self-renewing NPC population throughout mammalian kidney development. SIX2 is used to identify NPCs in embryonic development and pluripotent stem cell differentiation.
- Brachyury (T)
transcription factor found in all bilaterian animals that have been screened. T is expressed in primitive streak cells and required for mesoderm formation and cellular differentiation.
- Wolffian ducts
urogenital structures arising in intermediate mesoderm. They subsequently differentiate into collecting ducts of kidneys and ureters.
- HOXD11
transcription factor which belongs to the homeobox family of genes. HOXD11 is mainly expressed in posterior intermediate mesoderm and lateral plate mesoderm, involving in development of limbs and kidneys.
- FGF9, FGF20
members of the fibroblast growth factor family, involved in a variety of biological processes, including embryonic development, cell growth, morphogenesis, tissue repair, tumor growth and invasion. FGF9 is produced by ureteric buds, and required for the induction and maintenance of NPCs during embryonic kidney development. FGF20 is produced by NPCs, and maintain the stemness of NPCs in cooperation with FGF9.
- Stemness
the ability of cells to self-renew and differentiate into different cell types.
- Ureteric buds
a derivative of Wolffian ducts, which interact with NPCs and differentiate these into nephron epithelial cells. Ureteric buds differentiate into collecting ducts via reciprocal interaction with NPCs.
- Intermediate mesoderm (IM)
type of mesoderm located between the paraxial mesoderm and the lateral plate mesoderm. Anterior IM cells give rise to Wolffian ducts while posterior IM cells differentiate into NPCs.
- Proximal tubules
epithelial cells located between the glomerulus and the loop of Henle in the nephron. Proximal tubules reabsorb most of substances from primitive urine.
- Loop of Henle
portion of a nephron that leads from proximal tubules to the distal tubule. Loops of Henle reabsorb water and create concentrated urine.
- Distal tubules
a portion of a nephron that is located between the loop of Henle and the collecting duct. Distal tubules are partly responsible for the regulation of electrolytes and acid-base balance.
- Ureters
tubes made of smooth muscle fibers that propel urine from kidneys to the urinary bladder.
- End-stage kidney disease (ESKD)
the last stage of chronic kidney disease where patients need to receive kidney replacement therapies such as dialysis to survive.
- Lgr5+ stem cells
tissue stem cells expressing Lgr5, which possess stemness, and can be differentiated into different types of cells to form organoids.
- S-shaped body
early structure of the nephron which forms an S-shape. These bodies can be divided into distinct proximal, medial, and distal segments which subsequently differentiate into proximal tubules, loops of Henle, and distal tubules, respectively.
- Podocytes
epithelial cells located in Bowman's capsules in kidneys. Podocytes wrap around capillaries of glomeruli, and filter blood retaining large molecules such as proteins, while smaller molecules such as water, salts, and sugars are filtered as the first step in the formation of urine.
- NPHS1
one of the podocyte-specific genes, encoding a protein called nephrin. Nephrin is an important protein involved in blood filtration in glomeruli. Loss of nephrin leads to proteinuria.
- CDH1, CDH6
epithelial cadherin (E-cad) and K-cadherin, respectively. CDH1 is expressed in a variety of epithelial cells. Loops of Henle, distal tubules, and collecting ducts express CDH1 in human kidneys. CDH6 is expressed in proximal tubules in human kidneys.
- SALL1
transcription factor expressed in NPCs. Loss of SALL1 leads to kidney agenesis.
- Renal vesicles
the earliest structure of the nephron during kidney development, forming a round cyst-like structure with polarized epithelial cells.
- Cavitated spheroid
cyst-like structure with a lumen formed by epithelial cells. HPSCs form cavitated spheroids when cultured in Matrigel.
- Matrigel
trade name for a gelatinous protein mixture secreted by Engelbreth-Holm-Swarm (EHS) mouse sarcoma cells. Matrigel is used for 3D culture of cells.
- Yamanaka factors
four transcriptional factors, OCT4, SOX2, KLF4, and c-MYC used to reprogram somatic cells into embryonic stem cell-like cells termed induced pluripotent stem cells.
- Foot processes
foot-like structures formed by podocytes, which cover capillaries to filtrate blood. Disruption of foot processes (foot process effacement) leads to proteinuria.
- KIM-1 (kidney injury molecule-1)
marker of proximal tubular injury. Injured proximal tubules express KIM-1 at the apical surface.
- vWF (von Willebrand factor)
blood glycoprotein involved in hemostasis. vWF is also expressed in vascular endothelial cells.
- CD31
protein encoded by the PECAM1 gene. CD31 is found on the surface of platelets, monocytes, neutrophils, and endothelial cells.
- TUJ1
neuron-specific class III beta-tubulin. TUJ1 is used as a marker for neuronal cells.
- Ciliary proteins
form cilia, hairlike organelles. Mutations in ciliary proteins cause certain genetic kidney diseases.
- Kidney subcapsular spaces
spaces under kidney capsules located at the surface of kidneys. Kidney subcapsular spaces are used for cell transplantation, since cells exhibit better growth in these spaces.
- Orthotopic position
location where the organ normally exists in the body. A heterotopic position, in contrast, indicates the sites are different from the normal organ position.
- 3D bioprinting
technology to print cells and/or tissues in 3D structures.
Footnotes
COMPETING FINANCIAL INTERESTS
J.V.B. is a co-inventor on KIM-1 patents that have been licensed by Partners Healthcare to several companies. He has received royalty income from Partners Healthcare. J.V.B. and R.M. are co-inventors on patents (PCT/US16/52350) on organoid technologies that are assigned to Partners Healthcare. J.V.B. or his family has received income for consulting from companies interested in biomarkers: Sekisui, Millennium, Johnson & Johnson and Novartis. J.V.B. is a co-founder, consultant to, and owns equity in, Goldfinch Bio.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Morizane R, et al. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nature biotechnology. 2015 doi: 10.1038/nbt.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takasato M, et al. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nature cell biology. 2014;16:118–126. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- 5.Lam AQ, et al. Rapid and efficient differentiation of human pluripotent stem cells into intermediate mesoderm that forms tubules expressing kidney proximal tubular markers. Journal of the American Society of Nephrology: JASN. 2014;25:1211–1225. doi: 10.1681/ASN.2013080831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taguchi A, et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014;14:53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Takasato M, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564–568. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- 8.Coresh J, et al. Prevalence of chronic kidney disease in the United States. Jama. 2007;298:2038–2047. doi: 10.1001/jama.298.17.2038. [DOI] [PubMed] [Google Scholar]
- 9.Ene-Iordache B, et al. Chronic kidney disease and cardiovascular risk in six regions of the world (ISN-KDDC): a cross-sectional study. The Lancet Global health. 2016;4:e307–319. doi: 10.1016/S2214-109X(16)00071-1. [DOI] [PubMed] [Google Scholar]
- 10.Humphreys BD, et al. Intrinsic epithelial cells repair the kidney after injury. Cell Stem Cell. 2008;2:284–291. doi: 10.1016/j.stem.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Little MH, McMahon AP. Mammalian kidney development: principles, progress, and projections. Cold Spring Harbor perspectives in biology. 2012:4. doi: 10.1101/cshperspect.a008300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yue F, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stergachis AB, et al. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature. 2014;515:365–370. doi: 10.1038/nature13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Y, et al. Principles of regulatory information conservation between mouse and human. Nature. 2014;515:371–375. doi: 10.1038/nature13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pope BD, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vierstra J, et al. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science. 2014;346:1007–1012. doi: 10.1126/science.1246426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin S, et al. Comparison of the transcriptional landscapes between human and mouse tissues. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:17224–17229. doi: 10.1073/pnas.1413624111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devuyst O, et al. Rare inherited kidney diseases: challenges, opportunities, and perspectives. Lancet. 2014;383:1844–1859. doi: 10.1016/S0140-6736(14)60659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freedman BS, et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nature communications. 2015;6:8715. doi: 10.1038/ncomms9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen B, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nature methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 22.Kim D, Dressler GR1. Nephrogenic factors promote differentiation of mouse embryonic stem cells into renal epithelia. Journal of the American Society of Nephrology: JASN. 2005;16:3527–3534. doi: 10.1681/ASN.2005050544. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi T, et al. Wnt4-transformed mouse embryonic stem cells differentiate into renal tubular cells. Biochemical and biophysical research communications. 2005;336:585–595. doi: 10.1016/j.bbrc.2005.08.136. [DOI] [PubMed] [Google Scholar]
- 24.Bruce SJ, et al. In vitro differentiation of murine embryonic stem cells toward a renal lineage. Differentiation; research in biological diversity. 2007;75:337–349. doi: 10.1111/j.1432-0436.2006.00149.x. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto M, et al. Branching ducts similar to mesonephric ducts or ureteric buds in teratomas originating from mouse embryonic stem cells. American journal of physiology Renal physiology. 2006;290:F52–60. doi: 10.1152/ajprenal.00001.2004. [DOI] [PubMed] [Google Scholar]
- 26.Vigneau C, et al. Mouse embryonic stem cell-derived embryoid bodies generate progenitors that integrate long term into renal proximal tubules in vivo. Journal of the American Society of Nephrology: JASN. 2007;18:1709–1720. doi: 10.1681/ASN.2006101078. [DOI] [PubMed] [Google Scholar]
- 27.Nakane A, et al. Pax2 overexpression in embryoid bodies induces upregulation of integrin alpha8 and aquaporin-1. In vitro cellular & developmental biology Animal. 2009;45:62–68. doi: 10.1007/s11626-008-9151-8. [DOI] [PubMed] [Google Scholar]
- 28.Morizane R, et al. Differentiation of murine embryonic stem and induced pluripotent stem cells to renal lineage in vitro. Biochemical and biophysical research communications. 2009;390:1334–1339. doi: 10.1016/j.bbrc.2009.10.148. [DOI] [PubMed] [Google Scholar]
- 29.Mae S, et al. Combination of small molecules enhances differentiation of mouse embryonic stem cells into intermediate mesoderm through BMP7-positive cells. Biochemical and biophysical research communications. 2010;393:877–882. doi: 10.1016/j.bbrc.2010.02.111. [DOI] [PubMed] [Google Scholar]
- 30.Nishikawa M, et al. Stepwise renal lineage differentiation of mouse embryonic stem cells tracing in vivo development. Biochemical and biophysical research communications. 2012;417:897–902. doi: 10.1016/j.bbrc.2011.12.071. [DOI] [PubMed] [Google Scholar]
- 31.Morizane R, et al. Kidney specific protein-positive cells derived from embryonic stem cells reproduce tubular structures in vitro and differentiate into renal tubular cells. PloS one. 2014;8:e64843. doi: 10.1371/journal.pone.0064843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakano T, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Lin SA, et al. Subfractionation of differentiating human embryonic stem cell populations allows the isolation of a mesodermal population enriched for intermediate mesoderm and putative renal progenitors. Stem cells and development. 2010;19:1637–1648. doi: 10.1089/scd.2010.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song B, et al. The directed differentiation of human iPS cells into kidney podocytes. PloS one. 2012;7:e46453. doi: 10.1371/journal.pone.0046453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narayanan K, et al. Human embryonic stem cells differentiate into functional renal proximal tubular-like cells. Kidney international. 2013;83:593–603. doi: 10.1038/ki.2012.442. [DOI] [PubMed] [Google Scholar]
- 36.Mae S, et al. Monitoring and robust induction of nephrogenic intermediate mesoderm from human pluripotent stem cells. Nature communications. 2013;4:1367. doi: 10.1038/ncomms2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia Y, et al. The generation of kidney organoids by differentiation of human pluripotent cells to ureteric bud progenitor-like cells. Nature protocols. 2014;9:2693–2704. doi: 10.1038/nprot.2014.182. [DOI] [PubMed] [Google Scholar]
- 38.Araoka T, et al. Efficient and rapid induction of human iPSCs/ESCs into nephrogenic intermediate mesoderm using small molecule-based differentiation methods. PloS one. 2014;9:e84881. doi: 10.1371/journal.pone.0084881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irion S, et al. Directed differentiation of pluripotent stem cells: from developmental biology to therapeutic applications. Cold Spring Harbor symposia on quantitative biology. 2008;73:101–110. doi: 10.1101/sqb.2008.73.065. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi A, et al. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mugford JW, et al. Hoxd11 specifies a program of metanephric kidney development within the intermediate mesoderm of the mouse embryo. Developmental biology. 2008;319:396–405. doi: 10.1016/j.ydbio.2008.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi A, et al. Identification of a multipotent self-renewing stromal progenitor population during mammalian kidney organogenesis. Stem cell reports. 2014;3:650–662. doi: 10.1016/j.stemcr.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iimura T, Pourquie O1. Collinear activation of Hoxb genes during gastrulation is linked to mesoderm cell ingression. Nature. 2006;442:568–571. doi: 10.1038/nature04838. [DOI] [PubMed] [Google Scholar]
- 44.Sumi T, et al. Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/beta-catenin, Activin/Nodal and BMP signaling. Development. 2008;135:2969–2979. doi: 10.1242/dev.021121. [DOI] [PubMed] [Google Scholar]
- 45.Pagliuca FW, et al. Generation of functional human pancreatic beta cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogawa M, et al. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nature biotechnology. 2015;33:853–861. doi: 10.1038/nbt.3294. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi S, et al. Generation of kidney tubular organoids from human pluripotent stem cells. Scientific reports. 2016;6:38353. doi: 10.1038/srep38353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCrory WW1. The normal embryologic development of the kidney: a basis for understanding structural abnormalities. Birth defects original article series. 1974;10:3–11. [PubMed] [Google Scholar]
- 49.Tsang TE, et al. Lim1 activity is required for intermediate mesoderm differentiation in the mouse embryo. Developmental biology. 2000;223:77–90. doi: 10.1006/dbio.2000.9733. [DOI] [PubMed] [Google Scholar]
- 50.Bouchard M, et al. Nephric lineage specification by Pax2 and Pax8. Genes & development. 2002;16:2958–2970. doi: 10.1101/gad.240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia Y, et al. Directed differentiation of human pluripotent cells to ureteric bud kidney progenitor-like cells. Nature cell biology. 2013;15:1507–1515. doi: 10.1038/ncb2872. [DOI] [PubMed] [Google Scholar]
- 52.Sweetman D, et al. The migration of paraxial and lateral plate mesoderm cells emerging from the late primitive streak is controlled by different Wnt signals. BMC Dev Biol. 2008;8:63. doi: 10.1186/1471-213X-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lengerke C, et al. BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell. 2008;2:72–82. doi: 10.1016/j.stem.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 54.Liu P, et al. Requirement for Wnt3 in vertebrate axis formation. Nature genetics. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- 55.Morizane R, Bonventre JV1. Generation of nephron progenitor cells and kidney organoids from human pluripotent stem cells. Nature protocols. 2017;12:195–207. doi: 10.1038/nprot.2016.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chal J, et al. Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nature biotechnology. 2015;33:962–969. doi: 10.1038/nbt.3297. [DOI] [PubMed] [Google Scholar]
- 57.Deschamps J, van Nes J1. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development. 2005;132:2931–2942. doi: 10.1242/dev.01897. [DOI] [PubMed] [Google Scholar]
- 58.Patterson LT, et al. Hoxa11 and Hoxd11 regulate branching morphogenesis of the ureteric bud in the developing kidney. Development. 2001;128:2153–2161. doi: 10.1242/dev.128.11.2153. [DOI] [PubMed] [Google Scholar]
- 59.Gaunt SJ, et al. Direct activation of a mouse Hoxd11 axial expression enhancer by Gdf11/Smad signalling. Developmental biology. 2013;383:52–60. doi: 10.1016/j.ydbio.2013.08.025. [DOI] [PubMed] [Google Scholar]
- 60.Barak H, et al. FGF9 and FGF20 maintain the stemness of nephron progenitors in mice and man. Developmental cell. 2012;22:1191–1207. doi: 10.1016/j.devcel.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Majumdar A, et al. Wnt11 and Ret/Gdnf pathways cooperate in regulating ureteric branching during metanephric kidney development. Development. 2003;130:3175–3185. doi: 10.1242/dev.00520. [DOI] [PubMed] [Google Scholar]
- 62.Kang M, Han YM1. Differentiation of human pluripotent stem cells into nephron progenitor cells in a serum and feeder free system. PloS one. 2014;9:e94888. doi: 10.1371/journal.pone.0094888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toyohara T, et al. Cell Therapy Using Human Induced Pluripotent Stem Cell-Derived Renal Progenitors Ameliorates Acute Kidney Injury in Mice. Stem cells translational medicine. 2015;4:980–992. doi: 10.5966/sctm.2014-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ciampi O, et al. Generation of functional podocytes from human induced pluripotent stem cells. Stem cell research. 2016;17:130–139. doi: 10.1016/j.scr.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fatehullah A, et al. Organoids as an in vitro model of human development and disease. Nature cell biology. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 66.Hisha H, et al. Establishment of a novel lingual organoid culture system: generation of organoids having mature keratinized epithelium from adult epithelial stem cells. Scientific reports. 2013;3:3224. doi: 10.1038/srep03224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nanduri LS, et al. Purification and ex vivo expansion of fully functional salivary gland stem cells. Stem cell reports. 2014;3:957–964. doi: 10.1016/j.stemcr.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 69.DeWard AD, et al. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell reports. 2014;9:701–711. doi: 10.1016/j.celrep.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barker N, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 71.Stange DE, et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 2013;155:357–368. doi: 10.1016/j.cell.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schlaermann P, et al. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut. 2016;65:202–213. doi: 10.1136/gutjnl-2014-307949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bartfeld S, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–136. e126. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wroblewski LE, et al. Helicobacter pylori targets cancer-associated apical-junctional constituents in gastroids and gastric epithelial cells. Gut. 2015;64:720–730. doi: 10.1136/gutjnl-2014-307650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bertaux-Skeirik N, et al. CD44 plays a functional role in Helicobacter pylori-induced epithelial cell proliferation. PLoS pathogens. 2015;11:e1004663. doi: 10.1371/journal.ppat.1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nadauld LD, et al. Metastatic tumor evolution and organoid modeling implicate TGFBR2 as a cancer driver in diffuse gastric cancer. Genome biology. 2014;15:428. doi: 10.1186/s13059-014-0428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schumacher MA, et al. The use of murine-derived fundic organoids in studies of gastric physiology. The Journal of physiology. 2015;593:1809–1827. doi: 10.1113/jphysiol.2014.283028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 79.Dekkers JF, et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nature medicine. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 80.Yin X, et al. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nature methods. 2014;11:106–112. doi: 10.1038/nmeth.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Drost J, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 82.Finkbeiner SR, et al. Transcriptome-wide Analysis Reveals Hallmarks of Human Intestine Development and Maturation In Vitro and In Vivo. Stem cell reports. 2015 doi: 10.1016/j.stemcr.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Finkbeiner SR, et al. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. mBio. 2012;3:e00159–00112. doi: 10.1128/mBio.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Matano M, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nature medicine. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 85.Fordham RP, et al. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell. 2013;13:734–744. doi: 10.1016/j.stem.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mustata RC, et al. Lgr4 is required for Paneth cell differentiation and maintenance of intestinal stem cells ex vivo. EMBO reports. 2011;12:558–564. doi: 10.1038/embor.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mustata RC, et al. Identification of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium. Cell reports. 2013;5:421–432. doi: 10.1016/j.celrep.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 88.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Simmini S, et al. Transformation of intestinal stem cells into gastric stem cells on loss of transcription factor Cdx2. Nature communications. 2014;5:5728. doi: 10.1038/ncomms6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van Es JH, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nature cell biology. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang YG, et al. Salmonella-infected crypt-derived intestinal organoid culture system for host-bacterial interactions. Physiological reports. 2014:2. doi: 10.14814/phy2.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Horita N, et al. Fluorescent labelling of intestinal epithelial cells reveals independent long-lived intestinal stem cells in a crypt. Biochemical and biophysical research communications. 2014;454:493–499. doi: 10.1016/j.bbrc.2014.10.091. [DOI] [PubMed] [Google Scholar]
- 93.Wang X, et al. Cloning and variation of ground state intestinal stem cells. Nature. 2015;522:173–178. doi: 10.1038/nature14484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li X, et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nature medicine. 2014;20:769–777. doi: 10.1038/nm.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gracz AD, et al. A high-throughput platform for stem cell niche co-cultures and downstream gene expression analysis. Nature cell biology. 2015;17:340–349. doi: 10.1038/ncb3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fukuda M, et al. Small intestinal stem cell identity is maintained with functional Paneth cells in heterotopically grafted epithelium onto the colon. Genes & development. 2014;28:1752–1757. doi: 10.1101/gad.245233.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ootani A, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nature medicine. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jung P, et al. Isolation and in vitro expansion of human colonic stem cells. Nature medicine. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 99.Okamoto R, Watanabe M1. Role of epithelial cells in the pathogenesis and treatment of inflammatory bowel disease. Journal of gastroenterology. 2016;51:11–21. doi: 10.1007/s00535-015-1098-4. [DOI] [PubMed] [Google Scholar]
- 100.Yui S, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell. Nature medicine. 2012;18:618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- 101.van de Wetering M, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huch M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huch M, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huch M, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. The EMBO journal. 2013;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Boj SF, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Greggio C, et al. Artificial three-dimensional niches deconstruct pancreas development in vitro. Development. 2013;140:4452–4462. doi: 10.1242/dev.096628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Karthaus WR, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159:163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gao D, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chua CW, et al. Single luminal epithelial progenitors can generate prostate organoids in culture. Nature cell biology. 2014;16:951–961. 951–954. doi: 10.1038/ncb3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mondrinos MJ, et al. Engineering de novo assembly of fetal pulmonary organoids. Tissue engineering Part A. 2014;20:2892–2907. doi: 10.1089/ten.tea.2014.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mondrinos MJ, et al. Engineering three-dimensional pulmonary tissue constructs. Tissue engineering. 2006;12:717–728. doi: 10.1089/ten.2006.12.717. [DOI] [PubMed] [Google Scholar]
- 112.Sagrinati C, et al. Isolation and characterization of multipotent progenitor cells from the Bowman's capsule of adult human kidneys. Journal of the American Society of Nephrology: JASN. 2006;17:2443–2456. doi: 10.1681/ASN.2006010089. [DOI] [PubMed] [Google Scholar]
- 113.Ronconi E, et al. Regeneration of glomerular podocytes by human renal progenitors. Journal of the American Society of Nephrology: JASN. 2009;20:322–332. doi: 10.1681/ASN.2008070709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kusaba T, et al. Differentiated kidney epithelial cells repair injured proximal tubule. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1527–1532. doi: 10.1073/pnas.1310653110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kramann R, et al. Who regenerates the kidney tubule? Nephrology, dialysis, transplantation: official publication of the European Dialysis and Transplant Association - European Renal Association. 2015;30:903–910. doi: 10.1093/ndt/gfu281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rinkevich Y, et al. In vivo clonal analysis reveals lineage-restricted progenitor characteristics in mammalian kidney development, maintenance, and regeneration. Cell reports. 2014;7:1270–1283. doi: 10.1016/j.celrep.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Berger K, Moeller MJ1. Mechanisms of epithelial repair and regeneration after acute kidney injury. Seminars in nephrology. 2014;34:394–403. doi: 10.1016/j.semnephrol.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 118.Kumar S, et al. Sox9 Activation Highlights a Cellular Pathway of Renal Repair in the Acutely Injured Mammalian Kidney. Cell reports. 2015;12:1325–1338. doi: 10.1016/j.celrep.2015.07.034. [DOI] [PubMed] [Google Scholar]
- 119.McCracken KW, et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Spence JR, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cao L, et al. Intestinal lineage commitment of embryonic stem cells. Differentiation; research in biological diversity. 2011;81:1–10. doi: 10.1016/j.diff.2010.09.182. [DOI] [PubMed] [Google Scholar]
- 122.Watson CL, et al. An in vivo model of human small intestine using pluripotent stem cells. Nature medicine. 2014;20:1310–1314. doi: 10.1038/nm.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Takebe T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 124.Sampaziotis F, et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nature biotechnology. 2015;33:845–852. doi: 10.1038/nbt.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dye BR, et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife. 2015:4. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ozone C, et al. Functional anterior pituitary generated in self-organizing culture of human embryonic stem cells. Nature communications. 2016;7:10351. doi: 10.1038/ncomms10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sterneckert JL, et al. Investigating human disease using stem cell models. Nature reviews Genetics. 2014;15:625–639. doi: 10.1038/nrg3764. [DOI] [PubMed] [Google Scholar]
- 129.Ryuji Morizane JB. Generation of nephron progenitor cells and kidney organoids from human pluripotent stem cells. Nature protocols. 2016 doi: 10.1038/nprot.2016.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cheng HT, et al. Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development. 2007;134:801–811. doi: 10.1242/dev.02773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nigam SK, et al. Handling of Drugs, Metabolites, and Uremic Toxins by Kidney Proximal Tubule Drug Transporters. Clinical journal of the American Society of Nephrology: CJASN. 2015;10:2039–2049. doi: 10.2215/CJN.02440314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Perazella MA, Moeckel GW1. Nephrotoxicity from chemotherapeutic agents: clinical manifestations, pathobiology, and prevention/therapy. Seminars in nephrology. 2010;30:570–581. doi: 10.1016/j.semnephrol.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 133.Ludwig T, et al. Nephrotoxicity of platinum complexes is related to basolateral organic cation transport. Kidney international. 2004;66:196–202. doi: 10.1111/j.1523-1755.2004.00720.x. [DOI] [PubMed] [Google Scholar]
- 134.Yonezawa A, et al. Association between tubular toxicity of cisplatin and expression of organic cation transporter rOCT2 (Slc22a2) in the rat. Biochemical pharmacology. 2005;70:1823–1831. doi: 10.1016/j.bcp.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 135.Vaidya VS, et al. Kidney injury molecule-1 outperforms traditional biomarkers of kidney injury in preclinical biomarker qualification studies. Nature biotechnology. 2010;28:478–485. doi: 10.1038/nbt.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen HT, et al. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science. 2000;290:1962–1965. doi: 10.1126/science.290.5498.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cho EA, et al. Differential expression and function of cadherin-6 during renal epithelium development. Development. 1998;125:803–812. doi: 10.1242/dev.125.5.803. [DOI] [PubMed] [Google Scholar]
- 138.Nouwen EJ, et al. Differential expression and function of cadherin-6 during renal epithelium development. (1993) Stage- and segment-specific expression of cell-adhesion molecules N-CAM, A-CAM, and L-CAM in the kidney. Kidney international. 1998;44:147–158. doi: 10.1038/ki.1993.225. [DOI] [PubMed] [Google Scholar]
- 139.Nurnberger J, et al. Differential expression and function of cadherin-6 during renal epithelium development. (2010) N-cadherin is depleted from proximal tubules in experimental and human acute kidney injury. Histochemistry and cell biology. 1998;133:641–649. doi: 10.1007/s00418-010-0702-1. [DOI] [PubMed] [Google Scholar]
- 140.Lee SY, et al. Expression of E-cadherin in pig kidney. Journal of veterinary science. 2013;14:381–386. doi: 10.4142/jvs.2013.14.4.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Georgas KM, et al. Expression of metanephric nephron-patterning genes in differentiating mesonephric tubules. Developmental dynamics: an official publication of the American Association of Anatomists. 2011;240:1600–1612. doi: 10.1002/dvdy.22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ong AC, et al. Autosomal dominant polycystic kidney disease: the changing face of clinical management. Lancet. 2015;385:1993–2002. doi: 10.1016/S0140-6736(15)60907-2. [DOI] [PubMed] [Google Scholar]
- 143.Nahm AM, et al. Renal cystic disease (ADPKD and ARPKD) Nephrology, dialysis, transplantation: official publication of the European Dialysis and Transplant Association - European Renal Association. 2002;17:311–314. doi: 10.1093/ndt/17.2.311. [DOI] [PubMed] [Google Scholar]
- 144.Van Adelsberg JS, Frank D1. The PKD1 gene produces a developmentally regulated protein in mesenchyme and vasculature. Nature medicine. 1995;1:359–364. doi: 10.1038/nm0495-359. [DOI] [PubMed] [Google Scholar]
- 145.Freedman BS, et al. Reduced ciliary polycystin-2 in induced pluripotent stem cells from polycystic kidney disease patients with PKD1 mutations. Journal of the American Society of Nephrology: JASN. 2013;24:1571–1586. doi: 10.1681/ASN.2012111089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ameku T, et al. Identification of MMP1 as a novel risk factor for intracranial aneurysms in ADPKD using iPSC models. Scientific reports. 2016;6:30013. doi: 10.1038/srep30013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Choi J, et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nature biotechnology. 2015 doi: 10.1038/nbt.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nishizawa M, et al. Epigenetic Variation between Human Induced Pluripotent Stem Cell Lines Is an Indicator of Differentiation Capacity. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 149.Sharmin S, et al. Human Induced Pluripotent Stem Cell-Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation. Journal of the American Society of Nephrology: JASN. 2015 doi: 10.1681/ASN.2015010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Takebe T, et al. Vascularized and Complex Organ Buds from Diverse Tissues via Mesenchymal Cell-Driven Condensation. Cell Stem Cell. 2015;16:556–565. doi: 10.1016/j.stem.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 151.Caralt M, et al. Optimization and critical evaluation of decellularization strategies to develop renal extracellular matrix scaffolds as biological templates for organ engineering and transplantation. American journal of transplantation: official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2015;15:64–75. doi: 10.1111/ajt.12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Song JJ, et al. Regeneration and experimental orthotopic transplantation of a bioengineered kidney. Nature medicine. 2013;19:646–651. doi: 10.1038/nm.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kolesky DB, et al. 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs. Advanced materials. 2014;26:3124–3130. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- 154.Homan KA, et al. Bioprinting of 3D Convoluted Renal Proximal Tubules on Perfusable Chips. Scientific reports. 2016;6:34845. doi: 10.1038/srep34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kolesky DB, et al. Three-dimensional bioprinting of thick vascularized tissues. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:3179–3184. doi: 10.1073/pnas.1521342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.