Abstract
Approximately 50 different chromosomal translocations of the human MLL gene are currently known and associated with high-risk acute leukemia. The large number of different MLL translocation partner genes makes a precise diagnosis a demanding task. After their cytogenetic identification, only the most common MLL translocations are investigated by RT-PCR analyses, whereas infrequent or unknown MLL translocations are excluded from further analyses. Therefore, we aimed at establishing a method that enables the detection of any MLL rearrangement by using genomic DNA isolated from patient biopsy material. This goal was achieved by establishing a universal long-distance inverse-PCR approach that allows the identification of any kind of MLL rearrangement if located within the breakpoint cluster region. This method was applied to biopsy material derived from 40 leukemia patients known to carry MLL abnormalities. Thirty-six patients carried known MLL fusions (34 with der(11) and 2 with reciprocal alleles), whereas 3 patients were found to carry novel MLL fusions to ACACA, SELB, and SMAP1, respectively. One patient carried a genomic fusion between MLL and TIRAP, resulting from an interstitial deletion. Because of this interstitial deletion, portions of the MLL and TIRAP genes were deleted, together with 123 genes located within the 13-Mbp interval between both chromosomal loci. Therefore, this previously undescribed diagnostic tool has been proven successful for analyzing any MLL rearrangement including previously unrecognized partner genes. Furthermore, the determined patient-specific fusion sequences are useful for minimal residual disease monitoring of MLL associated acute leukemias.
Keywords: acute leukemia, MLL translocations, translocation partner genes
Chromosomal translocations involving the human MLL gene are recurrently associated with high-risk acute leukemias (1–4). MLL translocations correlate with specific disease subtypes (acute myeloid and acute lymphocytic leukemias), a specific gene expression profile (5, 6), and outcome (favorable or poor), depending on the particular MLL fusion (7). Approximately 50 different MLL translocation partner genes have been identified, suggesting that the human MLL gene is a hot spot for illegitimate recombination events. During illegitimate recombination events, one MLL allele is reciprocally fused with one of the many translocation partner genes. The latter encode nuclear or cytosolic proteins that share only a little sequence homology; however, the fused portion of partner protein sequences is necessary to confer oncogenic potential.
The unambiguous identification of these MLL translocations is necessary to support rapid clinical decisions and specific therapy regimens. Current procedures to diagnose MLL rearrangements include cytogenetic analysis, FISH experiments (e.g., split-signal FISH) (8), and specific RT-PCR methods. However, results of RT-PCR analyses are influenced strongly by the quality of the investigated RNA samples. Furthermore, only the most frequent MLL fusions routinely are analyzed by single or multiplex RT-PCR approaches.
Diagnostic techniques using genomic DNA of leukemia patients have been established for the t(4;11) and t(9;11) translocations (9–12). The great benefit of these DNA-based methods is the determination of genomic sequences derived from the reciprocal chromosomal fusion sites. Because these sequences are patient-specific and exist in only one copy per leukemic cell, they can be used as reliable markers for minimal residual disease (MRD) studies.
Identifying unknown MLL translocation partner genes needs more sophisticated methods to uncover new fusion genes, e.g., inverse PCR in combination with isolated chromosomal patient DNA (13) or panhandle PCR using reverse-transcribed patient RNA (cDNA) (14). Both methods have been applied successfully in the past to identify chromosomal breakpoints (15–21), but they were never optimized for high-throughput analysis or for clinical use.
Here, we describe a universal method that uses long-distance inverse PCR (LDI-PCR) to identify MLL translocations independent of the involved partner gene or other MLL aberrations that occurred within the MLL breakpoint cluster region. This method allows high-throughput analyses because genomic MLL fusion sequences can be obtained with a minimum of only four PCR reactions. Moreover, this method requires only small quantities of genomic patient DNA (1 μg) and provides relevant genetic information that can be used directly for quantitative MRD analyses.
In this study, we present data on 40 leukemia patients who were prescreened in different European centers for MLL translocations. All cases were analyzed successfully, and the corresponding MLL fusions were identified. Thirty-six patients had fusions with known MLL partner genes, whereas three patient samples revealed previously unknown MLL translocation partners localized at chromosome bands 17q12 (ACACA), 3q21 (SELB), and 6q13 (SMAP1). One patient carried a previously undescribed fusion between MLL and TIRAP, resulting from a deletion of a 13-Mbp interval between 11q23 and 11q24. Because of the interstitial deletion, 123 genes and several pseudogenes were deleted from the derivative(11) [der(11)] chromosome.
Materials and Methods
Cell Lines and Patient Materials. The t(4;11) cell line SEM was used to establish the experimental procedures (22). Genomic DNA was isolated from bone marrow and/or peripheral blood samples of all patients and forwarded to our center. Patient samples were obtained from the Children's Cancer Research Institute, the Interfant-99 study group (Rotterdam), the German Acute Myeloid Leukemia Cooperative Group (Munich), and the German Multicenter Acute Lymphocytic Leukemia study group (Berlin). The karyotype of the four patients with the previously unrecognized MLL fusions were as follows: patient P03-160, carr ying the MLL·ACACA fusion, had the kar yotype 46,XY,t(11;17)(q23;q21)[14]/46,XY[6]; patient P03-169, carr ying the MLL·SELB fusion, had the kar yotype 46,XX,der(3)t(3;11)(q13;q23),der(3)t(3;?)x2, del(11)(q23); patient P03-216, carrying the MLL·SMAP1 fusion, had the karyotype 46,XX,t(6;11)(q13;q23)[9]/46,XX[1]; and patient P04-251, carrying the MLL·TIRAP fusion, had the karyotype 46,XX,del(5)(q13q31),t(10;14)(p13;q11),del(11)(q23)[19]/46,XX[3]. Informed consent was obtained from all patients' parents or legal guardians and control individuals.
LDI-PCR Experiments. From each patient, 1 μg of genomic DNA was digested with the restriction enzyme BamHI. Phenol extractions and ethanol precipitations were performed to remove residual enzymatic activity. Digested DNA samples were self-ligated at 16°C overnight in a total volume of 50 μl in the presence of 5 units of T4 DNA ligase. All ligation reactions were terminated at 65°C for 10 min. We used 5 μl of religated genomic DNA (100 ng) for all subsequent LDI-PCR analyses.
MLL gene-specific oligonucleotides were designed according to published DNA sequences (GenBank accession no. AJ235379). For BamHI-digested and religated genomic DNA, the five oligonucleotides A–E were used in four different combinations (A–B, A–C, A–D, and A–E; see Fig. 1). Each analysis included a positive control by using the oligonucleotides B and F that amplify a 7.9-kb DNA fragment of the MLL breakpoint cluster region, regardless of whether there are one or two germ-line MLL alleles present in a given patient sample. All LDI-PCR reactions were performed by using the TripleMaster PCR System (Eppendorf) according to the manufacturer's recommendations. PCR amplimers were separated on 0.8% agarose gels. Non-germ-line DNA amplimers were gelextracted and sequenced directly. If no der(11) alleles could be determined, a reciprocal approach was applied to identify the reciprocal allele by using the primer combinations G–L, H–L, I–L, and K–L (Fig. 1c). Annotations of fused MLL sequences were carried out by blasting the human genome database (Genomic blast, www.ncbi.nlm.nih.gov/genome/seq/Blast). DNA sequences of oligonucleotides A–L are available at www.biozentrum.unifrankfurt.de/PharmBiol/Mitarbeiter/Marschalek/download.html.
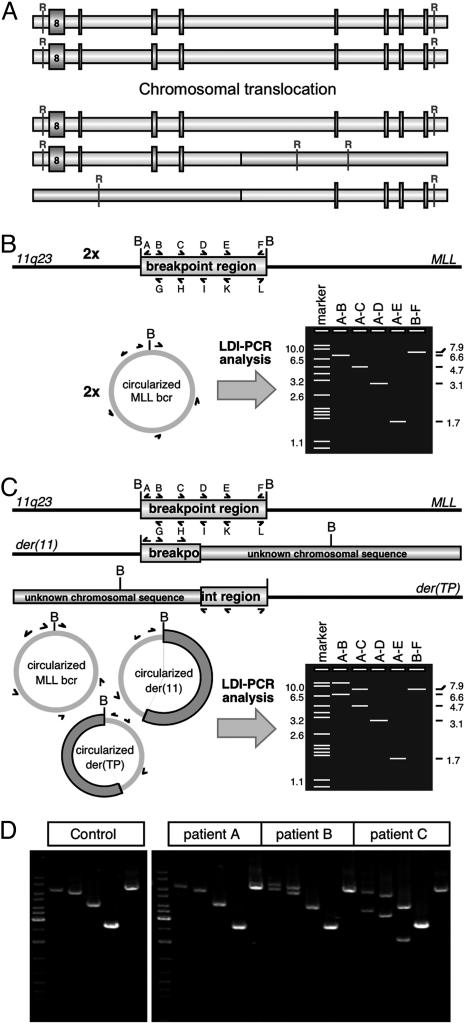
Fig. 1.
Principles of the LDI-PCR method and exemplary analyses. (A) Chromosomal translocations are creating restriction polymorphic DNA fragments that are targeted by the LDI-PCR approach. R, restriction site. (B) Nonrearranged MLL alleles: BamHI digestion and religation of the MLL bcr will lead to two DNA circles that can be amplified by the primer combinations A–B, A–C, A–D, and A–E. The primer combination B–F serves as internal control. B, BamHI restriction recognition site; bcr, breakpoint cluster region. (C) Presence of a rearranged MLL allele. BamHI digestion and religation of the two MLL alleles will lead to three different DNA circles [der(11) and der(TP); TP, translocation partner] that can be amplified by the designated primer combinations A–L. Non-germ-line PCR amplimers can be analyzed by sequence analysis using oligonucleotide A or L. (D) Genomic DNA of patients was tested with four different oligonucleotide combinations (A–B, A–C, A–D, and A–E). (Left) Size marker and control DNA (placenta). (Right) Analyses of different patients (A–C; not listed in Table 2). Non-germ-line amplimers can be isolated and analyzed by sequence analysis.
RT-PCR Analysis of MLL Fusion Alleles. Patients P03-160, P03-169, P03-216, and P04-251 were analyzed by specific RT-PCR experiments to verify the mRNA transcripts derived from the newly identified fusion alleles. All RT-PCR experiments were conducted by using touchdown PCR (40 cycles) as described in ref. 23. The following oligonucleotides were used in RT-PCR experiments: MLL4·3 (5′-GTTGTTGGTGAAGATGTTGC-3′), MLL6·3 (5′-CCTGTCACTAGAAACAAGGC-3′), MLL8·3 (5′-CCCAAAACCACTCCTAGTGAG-3′), MLL8F (5′-ATCCCGCCTCAGCCACCTAC-3′), MLL13·5 (5′-CAGGGTGATAGCTGTTTCGG-3′), MLL15·5 (5′-CATCATAACATTTGTCACAGAG-3′), MLL17·5 (5′-CTGGTGGATCAGGTCCTTC-3′), ACACA12·3 (5′-CTATCCGTAGGTGGTCTTATG-3′), ACACA17·5 (5′-CATACATCATACGGATATCC-3′), ACACA-R1 (5′-CCAAACAATCTCGTCATCTGGAGG-3′), SELB1·3 (5′-GTCGTCTTTGCCCGAGTTC-3′), SELB7·5 (5′-GTCAGGATCTTCTTGGACTC-3′), SMAP5·3 (5′-CTCCTCTGATGCTCCTCTTC-3′), SMAP7·5 (5′-GCTCCAGTTGCTGATCTATTC-3′), TIRAP4·31 (5′-CTAAGAAGCCTCTAGGCAG-3′), TIRAP5·3 (5′-CCCTGATGGTGGCTTTCGTC-3′), TIRAP5·51 (5′-GCAGACGTCATAGTCTTTGC-3′), TIRAP8·5 (5′-CAGAAGAAAGCAGATGTAGGG-3′), and DCPS2·5 (5′-CTGCAACTGGAGCTCAGG-3′).
Tissue-Specific Gene Expression Studies. Gene expression profiling was carried out by using human multiple tissue cDNA panels I and II (BD Biosciences) and oligonucleotides specific for the human genes MLL, ACACA, SELB, SMAP1, and TIRAP. Multiple tissue cDNA panels contain normalized cDNA probes derived from the following human tissues: heart, brain, placenta, lung, liver, skeletal muscle, kidney, pancreas, spleen, thymus, prostate, testis, ovary, small intestine, colon, and peripheral blood mononuclear cells. All RT-PCR experiments were performed as described above. The following oligonucleotides were used: MLL8·32 (5′-AGAACGTGGTGGACTCTAGTC-3′), MLL13·5, ACACA9·3 (5′-CTATCCGTAGGTGGTCTTATG-3′), ACACA12·5 (5′-GAAGAGTTGGGATACCTGCAG-3′), SELB1·3, SELB 3·5 (5′-CTCAATGAGCTCTGGAATGC-3′), SMAP2·3 (5′-CCTAGACCAATGGACAGCAG-3′), SMAP7·5, TIRAP4·31, and TIRAP 5·51. Control reactions (30 cycles) were performed using GAPDH-specific oligonucleotides.
Results
Identification of MLL Chromosomal Fusion Sites in Different DNA Samples of Leukemia Patients. Optimized reaction conditions were used to analyze genomic DNA samples derived from 40 leukemia patients who had been prescreened by FISH (e.g., MLL split-signal FISH) or cytogenetic analysis to carry a disrupted MLL allele. Each DNA sample was treated as described above and screened for germ-line and non-germ-line PCR amplimers. In all 40 cases, non-germ-line PCR amplimers were identified as well as the germ-line amplimers. Examples of LDI-PCR experiments are shown in Fig. 1. Non-germ-line PCR amplimers were subjected to sequence analysis, and the results are summarized in Table 2, which is published as supporting information on the PNAS web site, and Fig. 2. In two patients (P03-168 and P03-172), we were unable to detect the corresponding der(11) fusion site. In these cases, the genomic fusion sites of the reciprocal der(1) and der(6) chromosomes were identified by the reciprocal approach (see Materials and Methods). These two chromosomal translocations turned out to be fusions of EPS15(1p32) and MLLT4(6q27) with the MLL gene, respectively.
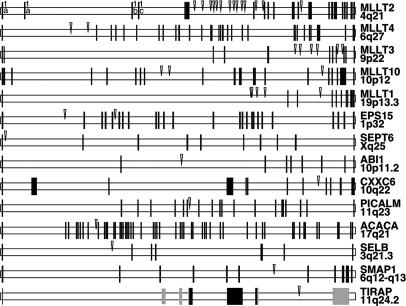
Fig. 2.
Genomic gene structures of MLL fusion genes and distribution of chromosomal breakpoints. Exon–intron structures of all MLL fusions identified in our study are shown. Gene names and chromosomal locations are given on the right. Approximate sizes of the genes are as follows: MLLT2, 210 kb; MLLT4, 140 kb; MLLT3, 280 kb; MLLT10, 210 kb; MLLT1, 330 kb; EPS15, 165 kb; SEPT6, 80 kb; ABI1, 115 kb; PICALM, 110 kb; CXXC6, 135 kb; ACACA, 350 kb; SELB, 260 kb; SMAP1, 200 kb; and TIRAP, 15 kb. Black and gray boxes represent coding and noncoding exons, respectively. Arrowheads mark the MLL fusion point.
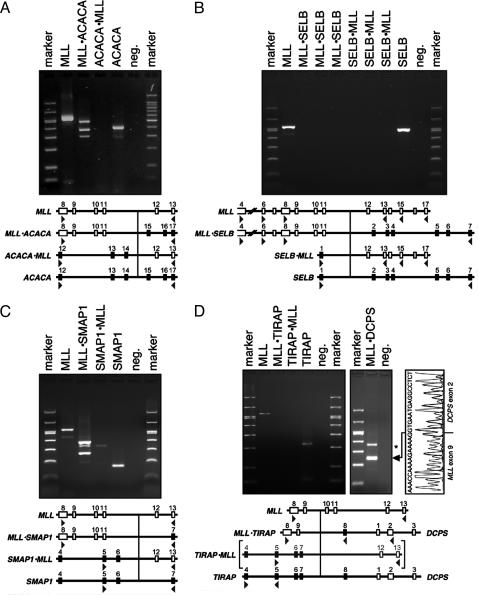
Previously Unrecognized MLL Fusion Genes. Three previously undescribed fusion partner genes were identified. The ACACA (acetyl-CoA carboxylase α) gene encompasses a genomic region of ≈250 kb and is located on band 17q12. The ACACA gene consists of 60 exons that encode a large protein of 2,347 aa (265 kDa). The ACACA protein is involved in fatty acid biosynthesis. The fusion between the MLL and ACACA genes occurred in introns 11 and 14, respectively. This particular chimeric transcript leads to a functional fusion gene that encodes a putative MLL·ACACA (3,489 aa) fusion protein. The presence of the MLL·ACACA fusion transcript was analyzed and verified by RT-PCR experiments, whereas the reciprocal ACACA·MLL could not be detected (Fig. 3A), although this experiment was repeated several times with different cDNA samples. Three different MLL·ACACA fusion mRNA species were identified (Fig. 3A), and sequence analysis showed that they correspond to fusions of MLL exons 8–11, 8–10, or 8–9 fused to exon 15 of the ACACA gene, respectively.
Fig. 3.
RT-PCR analyses of MLL fusion transcripts. (A) RT-PCR analysis of MLL·ACACA and ACACA·MLL fusion transcripts. Lanes 1 and 7, Size marker. Lanes 2–6, MLL (MLL8·3 × MLL13·5), MLL·ACACA (MLL8F × ACACA-R1), ACACA·MLL (ACACA12·3 × MLL13·5), ACACA (ACACA12·3 × ACACA17·5), and negative control. (B) RT-PCR analysis of MLL·SELB and SELB·MLL fusion transcripts. Lanes 1 and 11, Size marker. Lanes 2–10, MLL (MLL8·3 × MLL13·5), MLL·SELB (MLL4·3 × SELB7·5), MLL·SELB (MLL6·3 × SELB7·5), MLL·SELB (MLL8·3 × SELB7·5), SELB·MLL (SELB1·3 × MLL13·5), SELB·MLL (SELB1·3 × MLL15·5), SELB·MLL (SELB1·3 × MLL17·5), SELB (SELB1·3 × SELB7·5), and negative control. (C) RT-PCR analysis of MLL·SMAP1 and SMAP1·MLL fusion transcripts. Lanes 1 and 7, Size marker. Lanes 2–6, MLL (MLL8·3 × MLL13·5), MLL·SMAP1 (MLL8·3 × SMAP7·5), SMAP1·MLL (SMAP5·3 × MLL13·5), SMAP1 (SMAP5·3 × SMAP7·5), and negative control. (D) RT-PCR analysis of MLL·TIRAP and TIRAP·MLL fusion transcripts. Lanes 1, 8, and 9, Size marker. Lanes 2–7, MLL (MLL8·3 × MLL13·5), MLL·TIRAP (MLL8·3 × TIRAP8·5), TIRAP·MLL (TIRAP5·3 × MLL13·5), TIRAP (TIRAP4·31 × TIRAP5·51), and negative control. Lanes 10 and 11, MLL·DCPS (TIRAP4·31 × DCPS2·5) and negative control. Brackets indicate that the TIRAP·MLL allele is not expected because of the interstitial deletion of 13 Mbp. *, Head-to-head fused PCR artifact.
The gene coding for the elongation factor SELB is located on band 3q21 and covers an area of ≈260 kb. The SELB gene consists of seven exons and encodes a small protein of only 596 aa (65.3 kDa). The SELB protein is the mammalian homologue of an Escherichia coli elongation factor that is used to incorporate the amino acid selenocysteine in selenoproteins. The fusion between the MLL and SELB genes occurred in introns 9 and 1, respectively. This particular fusion would lead to reciprocal fusion mRNA species that are both out-of-frame. RT-PCR experiments testing for fusions between MLL exons 4, 6, and 8 and SELB exon 7 revealed no PCR amplimers. The same was true for SELB exon 1 in combination with oligonucleotides specific for MLL exons 13, 15, 17, or 16. In addition, reverse oligonucleotides specific for SELB exons 2–6 in combination with MLL forward primers were tested as well, but with the same negative result (data not shown). Any attempt to identify potential fusion mRNA species of this translocation failed, although both WT genes (SELB and MLL) were transcribed readily from the germ-line alleles (Fig. 3B). Assuming that there is no complex translocation where only parts of both genes are fused together, these data might indicate that the reciprocal fusion genes are not expressed.
The SMAP1 (stromal membrane associated protein 1) gene is located on band 6q13 and covers an area of ≈200 kb. The SMAP1 gene consists of 11 exons that encode a protein of 467 aa (50.4 kDa). The fusion between the MLL and SMAP1 genes occurred in introns 11 and 6, respectively. The SMAP1 protein is a membrane protein that facilitates erythropoietic development. This particular fusion leads to functional chimeric genes that encode the reciprocal fusion proteins MLL·SMAP1 (1,718 aa) and SMAP1·MLL (2,718 aa), respectively. The presence of both fusion transcripts was analyzed and verified by RT-PCR experiments (Fig. 3C).
The MLL–TIRAP Fusion Resulting from an Interstitial Deletion Generates Haploinsufficiency for 123 Genes. In patient P04-251, genomic MLL sequences were found to be fused to intron 7 of the TIRAP gene located at 11q24.2. To exclude the presence of a cryptic MLL translocation, we attempted to identify the reciprocal TIRAP·MLL allele. However, all attempts to detect a reciprocal genomic TIRAP·MLL fusion by LDI-PCR or RT-PCR failed so far. These data and the karyotype of this particular patient (see Materials and Methods) suggest an interstitial deletion on the long arm of chromosome 11 rather than a chromosomal translocation. The genomic fusion between MLL and TIRAP is accompanied by the deletion of a 13-Mbp interval between both gene loci. However, no fusion mRNA species between MLL exon 9 and TIRAP exon 8 (3′ nontranslated region) could be detected (Fig. 3D, lane3). The same was true for RT-PCR analyses using either TIRAP4·3 or TIRAP5·3 together with TIRAP8·5. This result indicated that TIRAP exon 8 might not be expressed in the leukemic cells. The WT TIRAP transcript was detected only with oligonucleotides that amplified the coding region. Therefore, we tested the possibility for a transcriptional fusion between MLL and the 3′-located DCPS gene. As shown in Fig. 3D, we were able to identify an MLL·DCPS fusion mRNA. This amplimer turned out to be a fusion between MLL exon 9 and DCPS exon 2.
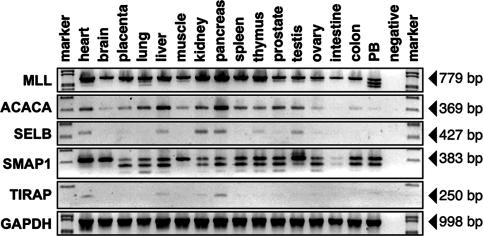
Expression of the Four Previously Undescribed MLL Partner Genes in Various Human Tissues. To gain information about the transcription profile of all four previously undescribed MLL partner genes, normalized cDNA preparations obtained from various human tissues were used for RT-PCR experiments. As shown in Fig. 4, transcripts of MLL were identified in nearly all examined tissues. The multiple MLL transcripts observed in peripheral blood mononuclear cells are due to skipping of MLL exon 10 or 10/11. The highest level of MLL gene transcription was identified in the heart, kidney, pancreas, and thymus. In general, under these conditions, ACACA gene expression seemed to be much weaker than that of MLL. Most transcripts were identified in the heart, liver, and pancreas. The ACACA gene was weakly transcribed only in peripheral blood mononuclear cells, and virtually no transcripts could be identified in the small intestine. Transcription of the SELB gene could be identified in the heart, liver, kidney, pancreas, thymus, and testis. Very low levels of transcription of the SELB gene were identified in the placenta, prostate, and ovary, and virtually no transcripts could be detected in the brain, lung, skeletal muscle, spleen, small intestine, colon, and peripheral blood mononuclear cells.
Fig. 4.
Tissue-specific expression of MLL, ACACA, SELB, SMAP1, and TIRAP. Steady-state transcription level of all four genes in different human tissues. Row 1 shows MLL-specific transcripts. Rows 2–5 show ACACA-, SELB-, SMAP1-, and TIRAP-specific transcripts. Row 6 shows GAPDH control reactions.
The SMAP1 gene was transcribed readily in all examined tissues, except for the small intestine. By using oligonucleotides specific for exons 2 and 7 of the SMAP1 gene, a single amplimer of 383 bp (representing exons 2–7) was detected in the heart and brain. In all other tissues, three different splice forms were observed, indicating that SMAP1 precursor RNAs are alternatively spliced. These splice forms are generated through the skipping of exon 6 or exons 5 and 6, both of which consist of 81 nt and therefore code for exactly 27 aa. Thus, both exons can be skipped without disrupting the SMAP1 ORF and encode smaller SMAP1 proteins.
Weak TIRAP transcripts were identified by the oligonucleotide combination TIRAP4·31 × TIRAP5·51. Amplimers were detected in the heart, liver, kidney, and pancreas.
Discussion
Here, we describe the identification of genetic aberrations of the human MLL gene, in particular chromosomal translocations and interstitial deletions by using LDI-PCR. This universal method allows the identification of unknown and known translocation partner genes and the establishment of patient-specific MLL fusion sequences.
Genomic DNA prepared from bone marrow and/or peripheral blood samples of 40 acute leukemia patients was successfully analyzed. The identified translocation partner genes are MLLT2 (14 cases), MLLT4 (2 cases), MLLT3 (6 cases), MLLT10 (3 cases), MLLT1 (5 cases), EPS15 (2 cases), SEPT6 (1 case), ABI1 (1 case), PICALM (1 case), and CXXC6 (1 case) (Fig. 2). In four other cases (P03-160, P03-169, P03-216, and P04-251), previously undescribed partner genes were identified. These genes were ACACA (17q25), SELB (3q25), SMAP1 (6q13), and TIRAP (11q24), respectively. In 2 of these 40 patients (P03-168 and P03-172), no der(11) fusion site could be identified. In these samples, the genomic fusion sites of the reciprocal alleles were identified by using reciprocal primer combinations (Fig. 1C) to identify the MLLT4·MLL and EPS15·MLL fusion alleles. This result could be due to the limitations of our technology (size of amplimers) or the fact that the chromosomal breakpoints of these particular der(11) alleles were located outside of the MLL breakpoint cluster region and, thus, cannot be analyzed by our approach.
The ACACA gene is regulated by thyroid hormone and encodes the ACACA subunit involved in fatty acid biosynthesis (24). It originally was identified in lipogenic tissue (liver and adipose tissue) (25) and is the rate-limiting enzyme in the biogenesis of long-chain fatty acids. As shown in Fig. 4, the ACACA gene is transcribed in most tissues, with the exception of the small intestine. ACACA gene expression in peripheral blood mononuclear cells seems to be very weak compared with other tissues examined.
In our RT-PCR experiments, only the MLL·ACACA transcript was identified, whereas attempts to identify an ACACA·MLL fusion mRNA species were unsuccessful. The absence of ACACA·MLL transcripts might be explained by a complex rearrangement that has occurred between the involved chromosomes. MLL and ACACA are located on the long arms of chromosome 11 and 17, respectively, but are transcribed from opposite DNA strands (MLL, telomeric; ACACA, centromeric). Therefore, gene material of both chromosomes must be inserted in inverse orientation into the partner chromosome to generate the appropriate fusion genes. Although both genomic fusion alleles were identified by our approach, we speculate that the reciprocal ACACA·MLL allele has lost some exons and/or the promoter region of the ACACA gene when fused to the 3′ MLL gene. Therefore, no chimeric transcript could be expressed from the reciprocal allele. Similar observations for inverse insertions were made for other MLL translocations that involved partner genes transcribed in centromeric direction, e.g., MLLT10 involved in t(10;11)(p12;q23) translocations and SEPT6 involved in t(X;11)(q24;q23) translocations (26, 27).
Interestingly, the ACACA protein also has been identified as the major binding partner protein of the human BRCA1 protein (28), a tumor-suppressor protein frequently mutated in breast cancer patients. Very recently, certain haplotypes of ACACA have been associated with an increased risk for breast cancer (29). Knockout experiments of the homologous ACACA gene in Saccharomyces cerevisiae demonstrated that the yeast ACACA protein also plays an important role in the cell cycle. Temperature-sensitive mutants of yeast show a G2/M cell-cycle arrest and are unable to complete mitosis at nonpermissive temperatures (30). Thus, the ACACA protein, besides fatty acid biosynthesis, seems to have additional biological features that might contribute to leukemogenesis.
The second identified gene, SELB, encodes an elongation factor necessary for the incorporation of selenocysteine at certain UGA stop codons (31). However, the fusion between MLL intron 9 and SELB intron 1 produce per se disrupted ORFs and, thus, incompatible exon–exon fusions for both the MLL·SELB and the SELB·MLL fusion genes. Any attempt to demonstrate the presence of specific fusion gene transcripts failed, although different primer combinations and different cDNA preparations were tested. However, both WT genes, MLL and SELB, were readily transcribed in leukemic patient cells. These data suggest that this particular translocation created nonfunctional fusion genes, because both fusion genes seem to be transcriptionally silenced. This result might be explained by incompatible chromatin imprints that led, after the fusion of the MLL and SELB genes, to a heterochromatinization effect. Another explanation would be a complex rearrangement where only a portion of the SELB gene is fused to the MLL gene. The identified genomic fusion contained a 2,222-bp DNA fragment of SELB intron 1 (out of 93,012 bp). The karyotype of that particular patient was described as the fusion of band 11q23 with 3q13. The SELB gene, however, is located at 3q21, and, thus, a fusion to other gene sequences derived from chromosome 3 cannot be excluded.
The third identified gene, stromal membrane associated protein 1 (SMAP1), is the second membrane protein that has been discovered as an MLL translocation partner gene. SMAP1 is involved in interactions between stromal cells and hematopoietic progenitors and seems to be important for the development of erythropoietic cells (32, 33). SMAP1 was the only gene identified by our screening that is expressed per se efficiently in cells of hematopoietic origin and with importance for developmental processes in the hematopoietic compartment.
The fourth genetic aberration was a fusion of MLL and TIRAP (34), a gene located ≈13 Mbp telomeric to the MLL gene. The TIRAP gene encodes an adapter molecule for the Toll signaling pathway that is necessary for Toll-like receptor 4 (TLR4) signaling and subsequent activation of the NF-κB pathway. In our RT-PCR experiments, no MLL·TIRAP chimeric RNA species was detected. Even WT TIRAP transcripts were not amplified when a TIRAP exon 8-specific oligonucleotide was used. Moreover, TIRAP exon 8 is part of the 3′ nontranslated region and, thus, does not encode any protein sequence. Therefore, we tested the possibility of whether MLL-derived trancripts were fused to a gene located ≈6 kb downstream of the TIRAP gene, the “mRNA decapping enzyme” gene DCPS. In fact, a transcript was identified that fused MLL exon 9 with DCPS exon 2. This unusual result might be explained by a transcriptional readthrough and subsequent splicing or by a trans-splicing mechanism. Because of the interstitial deletion, a loss of heterozygosity was created for ≈123 genes located between both loci. Interestingly, two other 11q deletions, resulting in the MLL·CBL and MLL·ARHGEF12 gene fusions, deleted 28 (35) and 44 (36) genes, respectively. The fusion between MLL and TIRAP is deleting a common chromosomal area shared by the MLL·CBL and MLL·ARHGEF12 gene fusions, indicating that leukemic diseases might be supported by the haploinsufficiency of several genes located in the 11q23–25 region (listed in Table 1). From these data, we conclude that all three fusions (i) create only one derivative allele, (ii) lead always to a functional MLL fusion gene, and (iii) lead to a loss of heterozygosity.
Table 1. Haploinsufficiency caused by MLL-qter deletions.
| GenBank code | Gene name |
|---|---|
| NM_005933 | MLL, myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog) |
| XM_084672 | Similar to cDNA sequence BC021608 |
| NM_032780 | TMEM25, transmembrane protein 25 |
| NM_020153 | Hypothetical protein FLJ21827 |
| NM_001655 | ARCN1, archain 1 |
| NM_015157 | PHLDB1, pleckstrin homology-like domain, family B, member 1 |
| NM_007180 | TREH, trehalase (brush-border membrane glycoprotein) |
| NM_004397 | DDX6, DEAD (Asp–Glu–Ala–Asp) box polypeptide 6 |
| LOC338656 | Pseudogene similar to protein phosphatase 2A inhibitor-2 I-2PP2A |
| NM_001716 | BLR1, Burkitt lymphoma receptor 1, GTP binding protein receptor |
| NM_182557 | BCL9L, B cell chronic lymphocytic leukemia/lymphoma 9-like =TLP |
| NM_006760 | UPK2, uroplakin 2 |
| XM_290498 | Hypothetical protein LOC283150 |
| NM_198489 | Similar to DLNB14 |
| NM_001028 | RPS25, ribosomal protein S25 |
| NM_016146 | TRAPPC4, trafficking protein particle complex 4 |
| NM_001467 | SLC37A4, solute carrier family 37 (glycerol-6-phosphate transporter) |
| XM_378313 | LOC399955 |
| NM_006389 | HYOU1, hypoxia up-regulated 1 |
| NM_021729 | VPS11, vacuolar protein sorting 11 (yeast) |
| NM_000190 | HMBS, hydroxymethylbilane synthase |
| NM_002105 | H2AFX, H2A histone family, member X |
| NM_001382 | DPAGT1, dolichyl-phosphate N-acetylglucosaminephosphotransferase 1 |
| NM_014807 | TMEM24, transmembrane protein 24 (KIAA0285) |
| NM_015517 | MIZF, MBD2 (methyl-CpG-binding protein)-interacting zinc finger protein |
| NM_022169 | ABCG4, ATP-binding cassette, subfamily G (WHITE), member 4 |
| NM_024618 | NOD9 protein |
| NM_024791 | PDZK2, PDZ domain containing 2 |
| XM_378314 | Hypothetical protein LOC283152 |
| NM_005188 | CBL, Cas-Br-M (murine) ecotropic retroviral transforming sequence |
The LDI-PCR method described here offers important benefits for detailed genetic unknown MLL fusion genes. The use of cytogenetic and FISH techniques in combination with our method seems to be sufficient to identify any MLL translocation or other MLL aberration (MLL deletions and gene-internal MLL duplications). In case of a complex rearrangement, the identified genomic fusion site can be used only as a starting point to identify the corresponding MLL fusion on the RNA level. Future analyses of leukemic biopsy material should allow the unraveling of the complete “MLL recombinome.” Secondly, the established genomic breakpoint fusions sequences can be used directly as targets for genomic MRD analyses. The use of chromosomal fusion sites for MRD analyses has several advantages over other techniques, in that chromosomal fusion sites are unique sequences for each patient and are present in exactly one copy per leukemic cell. This finding could greatly facilitate MRD studies and increase our understanding about treatment failure and relapse.
Supplementary Material
Acknowledgments
We thank all providers of patient DNA, especially Drs. J. Greil (University of Tübingen, Tübingen, Germany) and Th. Burmeister (University of Berlin, Berlin). This work was made possible by and was conducted within the framework of the International Berlin–Frankfurt–Münster Study Group. This work was supported in part by research program “Genome Research for Health” of the Austrian Ministry of Education, Science, and Culture Grant GEN-AU Child, GZ 200.071/3-VI/2a/2002 and by Wilhelm-Sander-Foundation Research Grant 2001.061.1 (to R.M., T.D., and T.K.).
Author contributions: T.D., T.K., and R.M. designed research; C.M., B.S., M.R., S.A., S. Strehl, O.A.H., and R.M. performed research; C.M., B.S., M.R., S. Strehl, O.A.H., T.D., T.K., and R.M. analyzed data; S. Schnittger, C.S., M.W.J.C.J., J.J.v.D., and R.P. contributed new reagents/analytic tools; and R.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: der(n), derivative(n); LDI-PCR, long-distance inverse PCR; MRD, minimal residual disease.
References
- 1.Bernard, O. A. & Berger, R. (1995) Genes Chromosomes Cancer 13, 75-85. [DOI] [PubMed] [Google Scholar]
- 2.Cimino, G., Rapanotti, M. C., Sprovieri, T. & Elia, L. (2001) Haematologica 83, 350-357. [PubMed] [Google Scholar]
- 3.Ayton, P. M. & Cleary, M. L. (2001) Oncogene 20, 5695-5707. [DOI] [PubMed] [Google Scholar]
- 4.Schoch, C., Schnittger, S., Klaus, M., Kern, W., Hiddemann, W. & Haferlach, T. (2003) Blood 102, 2395-2402. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong, S. A., Stounton, J. E., Silverman, L. B., Pieters, R., den Boer, M. L., Minden, M. D., Sallan, S. E., Lander, E. S., Golub, T. R. & Korsmeyer, S. J. (2002) Nat. Genet. 30, 41-47. [DOI] [PubMed] [Google Scholar]
- 6.Yeoh, E. J., Ross, M. E., Shurtleff, S. A., Williams, W. K., Patel, D., Mahfouz, R., Behm, F. G., Raimondi, S. C., Relling, M. V., Patel, A., et al. (2002) Cancer Cell 1, 133-143. [DOI] [PubMed] [Google Scholar]
- 7.Mrózek, K., Heinonen, K., Lawrence, D., Carroll, A. J., Koduru, P. R., Rao, K. W., Strout, M. P., Hutchison, R. E., Moore, J. O., Mayer, R. J., et al. (1997) Blood 90, 4532-4538. [PubMed] [Google Scholar]
- 8.van der Burg, M., Poulsen, T. S., Hunger, S. P., Beverloo, H. B., Smit, E. M., Vang-Nielsen, K., Langerak, A. W. & van Dongen, J. J. (2004) Leukemia 18, 895-908. [DOI] [PubMed] [Google Scholar]
- 9.Gillert, E., Leis, T., Repp, R., Reichel, M., Hösch, A., Breitenlohner, I, Angermüller, S., Borkhardt, A., Harbott, J., Lampert, F., et al. (1999) Oncogene 18, 4663-4671. [DOI] [PubMed] [Google Scholar]
- 10.Reichel, M., Hensel, J. P., Gillert, E., Breitenlohner, I., Repp, R., Greil, J., Beck, J. D., Fey, G. H. & Marschalek, R. (1999) Cancer Res. 59, 3357-3362. [PubMed] [Google Scholar]
- 11.Reichel, M., Gillert, E., Angermüller, S., Hensel, J. P., Heidel, F., Lode, M., Leis, T., Biondi, A., Haas, O. A., Strehl, S., et al. (2001) Oncogene 20, 2900-2907. [DOI] [PubMed] [Google Scholar]
- 12.Langer, T., Metzler, M., Reinhardt, D., Viehmann, S., Borkhardt, A., Reichel, M., Stanulla, M., Schrappe, M., Creutzig, U., Ritter, J., et al. (2003) Genes Chromosomes Cancer 36, 393-401. [DOI] [PubMed] [Google Scholar]
- 13.Ochman, H., Gerber, A. S. & Hartl, D. L. (1988) Genetics 120, 621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, D. H. & Winistorfer, S. C. (1992) Nucleic Acids Res. 20, 595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marschalek, R., Greil, J., Löchner, K., Nilson, I., Siegler, G., Zweckbronner, I. Beck, J. D. & Fey, G. H. (1995) Br. J. Haematol. 90, 308-320. [DOI] [PubMed] [Google Scholar]
- 16.Felix, C. A., Kim, C. S., Megonigal, M. D., Slater, D. J., Jones, D. H., Spinner, N. B., Stump, T., Hosler, M. R., Nowell, P. C., Lange, B. J. & Rappaport, E. F. (1997) Blood 90, 4679-4686. [PubMed] [Google Scholar]
- 17.Megonigal, M. D., Rappaport, E. F., Jones, D. H., Kim, C. S., Nowell, P. C., Lange, B. J. & Felix, C. A. (1997) Proc. Natl. Acad. Sci. USA 94, 11583-11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willis, T. G., Jadayel, D. M., Coignet, L. J., Abdul-Rauf, M., Treleaven, J. G., Catovsky, D. & Dyer, M. J. (1997) Blood 90, 2456-2464. [PubMed] [Google Scholar]
- 19.Felix, C. A. & Jones, D. H. (1998) Leukemia 12, 976-981. [DOI] [PubMed] [Google Scholar]
- 20.Raffini, L. J., Slater, D. J., Rappaport, E. F., Lo Nigro, L., Cheung, N. K., Biegel, J. A., Nowell, P. C., Lange, B. J. & Felix C. A. (2002) Proc. Natl. Acad. Sci. USA 99, 4568-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorsbach, R. B., Moore, J., Mathew, S., Raimondi, S. C., Mukatira, S. T. & Downing, J. R. (2003) Leukemia 17, 637-641. [DOI] [PubMed] [Google Scholar]
- 22.Greil, J., Gramatzki, M., Burger, R., Marschalek, R., Peltner, M., Trautmann, U., Hansen-Hagge, T. E., Bartram, C. R., Fey, G. H., Stehr, K. & Beck, J. D. (1994) Br. J. Haematol. 86, 275-283. [DOI] [PubMed] [Google Scholar]
- 23.Löchner, K., Siegler, G., Führer, M., Greil, J., Beck, J. D., Fey, G.H. & Marschalek, R. (1996) Cancer Res. 56, 2171-2177. [PubMed] [Google Scholar]
- 24.Zhang, Y., Yin, L. & Hillgartner, F. B. (2001) J. Biol. Chem. 276, 974-983. [DOI] [PubMed] [Google Scholar]
- 25.Kim, K. H. (1997) Annu. Rev. Nutr. 17, 77-99. [DOI] [PubMed] [Google Scholar]
- 26.Van Limbergen, H., Poppe, B., Janssens, A., De Bock, R., De Paepe, A., Noens, L. & Speleman, F. (2002) Leukemia 16, 344-351. [DOI] [PubMed] [Google Scholar]
- 27.Kim, H. J., Ki, C. S., Park, Q., Koo, H. H., Yoo, K. H., Kim, E. J. & Kim, S. H. (2003) Genes Chromosomes Cancer 38, 8-12. [DOI] [PubMed] [Google Scholar]
- 28.Magnard, C., Bachelier, R., Vincent, A., Jaquinod, M., Kieffer, S., Lenoir, G. M. & Venezia, N. D. (2002) Oncogene 21, 6729-6739. [DOI] [PubMed] [Google Scholar]
- 29.Sinilnikova, O. M., Ginolhac, S. M., Magnard, C., Leone, M., Anczukow, O., Hughes, D., Moreau, K., Thompson, D., Coutanson, C., Hall, J., et al. (2004) Carcinogenesis 25, 2417-2424. [DOI] [PubMed] [Google Scholar]
- 30.Al-Feel, W., DeMar, J. C. & Wakil, S. J. (2003) Proc. Natl. Acad. Sci. USA 100, 3095-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forchhammer, K., Leinfelder, W. & Bock, A. (1989) Nature 342, 453-456. [DOI] [PubMed] [Google Scholar]
- 32.Yanai, N., Sato, Y. & Obinata, M. (1997) Leukemia 11, 484-485. [PubMed] [Google Scholar]
- 33.Marcos, I., Borrego. S-, Rodriguez de Cordoba, S., Galan, J. J. & Antinolo, G. (2002) Gene 292, 167-171. [DOI] [PubMed] [Google Scholar]
- 34.Horng, T., Barton, G. M. & Medzhitov, R. (2001) Nat. Immunol. 2, 835-841. [DOI] [PubMed] [Google Scholar]
- 35.Fu, J. F., Hsu, J. J., Tang, T. C. & Shih, L. Y. (2003) Genes Chromosomes Cancer 37, 214-219. [DOI] [PubMed] [Google Scholar]
- 36.Kourlas, P. J., Strout, M. P., Becknell, B., Veronese, M. L., Croce, C. M., Theil, K. S., Krahe, R., Ruutu, T., Knuutila, S., Bloomfield, C. D. & Caligiuri, M. A. (2000) Proc. Natl. Acad. Sci. USA 97, 2145-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.