Abstract
The stability of an immunogen against enzymatic degradation is considered an important factor for the design of synthetic vaccines. For our studies, we have selected an epitope from the tandem-repeat unit of the high-molecular-weight MUC2 mucin glycoprotein, which can be underglycosylated in case of colon cancer. In this study, we prepared a MUC2 peptide containing the PTGTQ epitope of a MUC2 protein backbone-specific mAb 996 and its derivatives. In these peptides, the N- and C-terminal flanking regions were systematically substituted by up to three d-amino acids. Peptides prepared by solid-phase synthesis were tested for their mAb 996 binding in competitive ELISA experiments, and their stability was studied in serum and lysosomal preparation. Our data show that the epitope function of peptide 15TPTPTGTQTPT25 is retained even in the presence of two d-amino acid residues at its N-terminal flanking region and up to three at its C-terminal flanking region (tpTPTGTQtpt). Also, this partly d peptide shows high resistance against proteolytic degradation in diluted human serum and in lysosomal preparation. These findings suggest that, by appropriate combination of structural modifications (namely, d-amino acid substitution) in the flanks of an Ab epitope, it is feasible to construct a synthetic antigen with preserved recognition properties and high stability against enzymatic degradation. Peptides tPTPTGTQTpt and tpTPTGTQTpt derived from this study can be used for immunization experiments and as potential components of synthetic vaccines for tumor therapy.
Keywords: human serum, rat liver lysosome preparation
The design of appropriate immunogen and its delivery, among other factors (e.g., schedule and route of immunization), are essential for the development of an effective peptide-based subunit vaccine. A major problem limiting the use of peptides is their instability, which is mainly due to the rapid degradation in vivo by proteases. There are different approached for protecting biologically active peptides from enzymatic decomposition, such as alteration of the amide bond (1), cyclization, conjugation to carrier molecule (2), and incorporation of nonproteinogenic amino acids such as β-Ala (3) or d-amino acids (4, 5).
Sela and Zisman (as reviewed in ref. 6) clearly demonstrated that the presence of d-amino acid residues in linear and multichain polypeptide antigens could improve their stability to proteolysis and alter the biological half-life but also influence B or T cell immunrecognition. The d-amino acids were incorporated into epitope peptides mainly to investigate their effect on peptide-specific B cell and/or T cell immunogenicity and/or to analyze the fine specificity and cross-reactivity of Abs/T cells induced by the all-l, all-d peptides (7). Correlation between the characteristics of immune response and extended biological half-life, as well as their stability to proteolysis, were described (8-10). However, most of the available literature reports on peptides in which all l-amino acid residues were replaced by their d-enantiomers, leading to normal (in all-d-isomer peptides) or reversed (in retro-all-d-isomer peptides) amide linkage (11, 12). Only a few studies have been published on immune recognition of linear oligopeptides partially substituted by d-amino acid residue(s) in the epitope sequence (13, 14). Apart from our previous studies (15, 16), to the best of our knowledge, no data are available on the Ab binding to epitope peptides containing d-amino acid residues that are not in the core but in the neighboring N- and/or C-terminal flanking region. According to our results published in those communications, partial replacement of l-amino acids by the respective d-isomers in the close vicinity of the B-cell epitope was tolerated, and no significant decrease in the Ab recognition was observed.
High-molecular-weight MUC2 mucin glycoproteins that may be expressed in elevated level in human colonic carcinoma are also under-glycosylated or aberrantly glycosylated (17). This alteration makes it possible to differentiate immunologically between mucins from healthy and tumor tissues by using protein core-specific Abs. Peptides representing epitope(s) of the protein core, if attached to carrier, or their analogues (e.g., d-amino acid substituted versions) could also be considered as synthetic vaccines in active specific immunotherapy to stimulate the B cell-specific immune responses of patients with MUC2 related disease (e.g., colon carcinoma). We have identified (18, 19) an epitope (18PTGTQ22) (18, 19) within the 1PTTTPITTTTTVTPTPTPTGTQT23 tandem-repeat unit of MUC2 glycoprotein (20) by using protein core-specific mAb 996 (21). We have also demonstrated that the N-terminal part of peptide 16PTPTGTQ22 forms β-turn secondary structure required for the Ab binding (22). Experiments with peptides comprising d-amino acids on the N terminus of peptide 16PTPTGTQ22 suggested that Pro-16 and Thr-17 could be substituted without significant loss of Ab recognition (15).
The aim of this study is to create peptides that are stable toward proteolysis and display the same, if not enhanced, binding properties compared to the original all-l peptides. Therefore, we have studied the antigenic properties and enzymatic stability of several MUC2 peptides partially substituted by d-amino acids in the flanking regions only. All peptides contained the core epitope sequence (18PTGTQ22) and flanking regions both at the C and N termini (15TPTPTGTQTPT25) with native or one to three d-amino acids. The peptides were tested for their binding activities with epitope-specific mAb 996. Their resistance against enzymatic degradation was investigated in human serum and also by using rat liver lysosomal preparation. Considering the lack of consensus in the literature about optimal pH in lysosomes, we performed these studies at two pH values (pH 3.5 and 5). Our data show that peptide tpTPTGTQtpt with d-amino acids in the flanks exhibited fully retained Ab binding and high resistance against proteolytic degradation in diluted human serum and lysosomal preparation.
Materials and Methods
Peptide Synthesis and Characterization. Peptides were prepared by solid-phase methodology by using p-benzyloxy-benzylalcohol resin (Wang resin; capacity, 0.6 mmol/g) (23) for free C terminus peptides with manual synthesis. All amino acids were coupled as 9-fluorenylmethoxycarbonyl (Fmoc)-derivatives (Nova Biochem). The tert-butyl group was applied as a protecting group for the side chain of Thr. Coupling was carried out by using 1-hydroxybenzotriazole/N,N′-diisopropylcarbodiimide in situ active ester methodology in N,N-dimethylformamide (DMF). Fmoc groups were removed by 20% piperidine in DMF, or 2% piperidine and 2% diazabicyclo[5.4.0]undec-7-ene in DMF, respectively. The success of the coupling and deprotection steps was monitored by ninhydrin test (24) and/or isatine assay (25). After the completion of the synthesis, the peptides were cleaved from the resin with trifluoroacetic acid containing 5% water. The crude products were purified by RP-HPLC on a Supelcosil C18 column (12 μm, 250 × 10 mm, Supelco) by using gradient elution with the following eluents: A, 0.1% trifluoroacetic acid in water; and B, 0.1% trifluoroacetic acid in acetonitrile/water (80:20, vol/vol). After sample application an isocratic elution with 0 or 5% eluent B for 5 min, a linear gradient from 0-25% B or 5-30% B was generated over 25 min at room temperature and with a flow rate of 4 ml/min. UV detection was performed at λ = 214 nm. The purity of the peptides was investigated by analytical RP-HPLC on a Synergi (4.6 mm × 25 cm, MAX-RP 80 Å, 4 μm, Phenomenex, Torrance, CA) column.
The relative molecular mass of the peptides was determined by plasma desorption or electronspray mass spectrometry (BIO-ION 2, Applied Biosystems; VG-ZA-2SEQ, Fisons, Loughborough, U.K.; or Esquire 3000 Iontrap MS, EU, Bruker). The peptides were hydrolyzed with 6 mol/dm3 HCl for 24 h, and the amino acid composition was then determined on a Beckmann 6300 amino acid analyzer after ninhydrin derivatization.
Inhibition ELISA. The Ab binding of peptides was measured by the competitive ELISA method as described (26). [K12VTPTPTPTGTQTPT25-OH]-BSA conjugate as target antigen at 2 μg/ml in PBS (0.1 mol/dm3, pH 7.3) containing 0.02% NaN3 was added to 96-well Enzyplate Microtest plates (well capacity, 250 μl; Propilén Kft, Pécs, Hungary) at 100 μl per well. Plates were incubated overnight at room temperature. The wells were washed twice with PBS/0.1% Tween 20 washing buffer, and the nonspecific adsorption sites were then blocked with PBS containing 1% BSA. We added mAb 996 at 1 μg/ml concentration, 50 μl per well, or PBS alone in negative controls to the wells in which 50 μl of the test peptide solution had been added previously (concentration range, 3.2 μmol/dm3 to 2 mmol/dm3). After incubation for 1.5 h at room temperature, the wells were aspirated and washed four times with washing buffer. Peroxidase-labeled rabbit anti-mouse Ig (DAKO) was added at a 1/2,000 dilution (100 μl per well) and incubated for 1.5 h. The wells were washed again six times with washing buffer, and 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS), and H2O2 in citrate phosphate buffer (pH 4) was then added (100 μl per well). After 10 min, the intensity of the resulting green color was measured at λ = 405 nm in an ELISA reader (Labsystems, Helsinki, Finland). Binding of Ab to target antigen without inhibiting peptide (B0) was considered as 100% binding. For each peptide concentration, the inhibition percentage (Ic%) was calculated as Ic% = (1 - Bc/B0) × 100%, where Bc is the Ab binding at c peptide concentration. Based on c and Ic% values, a c vs. Ic% curve was constructed, and the amount of added peptide required to inhibit Ab binding by 50% (IC50) was determined by interpolation as a measure of peptide-Ab binding.
Enzymatic Digestion
Lysosome Preparation. Livers from two male rats were collected and homogenized in two volumes of ice-cold 0.3 mol/dm3 sucrose with 10 strokes at 15 × g. The homogenate was diluted with three volumes of 0.3 mol/dm3 sucrose. The nuclei and cell debris were centrifuged at 700 × g for 10 min. The supernatant was centrifuged at 10,000 × g for 10 min to sediment the crude lysosomal-mitochondrial fraction. The sediment was rehomogenized in 10 ml of 0.3 mol/dm3 sucrose, containing 1 mmol/dm3 CaCl2. Homogenate was incubated at 37°C for 5 min for the mitochondria swallowing. We added 10 ml of 50% Percoll solution, and the homogenate was centrifuged at 10,000 × g for 10 min. The hard brown pellet at the bottom was the lysosomal fraction. For the best pipetting, it was diluted 1:2 by 0.3 mol/dm3 sucrose. The enzymatic activity of the preparation was determined with BSA as substrate according to Dingle (27).
Digestion of Peptides in Lysosome Homogenates. We dissolved 1 mg of peptide in 400 μl of 0.1 mol/dm3 acetate buffer (pH 3.5 or 5.0; c = 2.5 mg/ml), and 8 μl of the lysosome fraction was added (1:50 dilution). Peptides were incubated at room temperature for 180 min. Samples of 100 μl were taken at 30 sec and 60, 120, and 180 min. The enzymatic reaction was stopped by the addition of 5% (vol/vol) perchloric acid. Samples were centrifuged at 3,500 × g for 5 min at -4°C, and the concentration of the intact peptide in the supernatant was determined by RP-HPLC (Symmetry C18 3.5 μm; 4.6 × 150 mm column), based on calibration curves. For the RP-HPLC analysis, gradient elution was used with the following eluents: A, 0.1% TFA in water; and B, 0.1% TFA in acetonitrile/water (80:20, vol/vol). A gradient of 0-20% B in 20 min was used. Each sample was injected twice. Each peptide was digested in two independent experiments. Standard deviation of HPLC data were negligible, and standard deviation for the independent studies was calculated.
Digestion of Peptides in Human Serum. Human serum was obtained from the National Institute of Hematology and Immunology (Budapest). We dissolved 1.75 mg of peptide in 630 μl 0.1 mol/dm3 phosphate buffer (pH 7.2), and 70 μl of human serum was added (10% serum, vol/vol), or it was dissolved in 350 μl buffer with an additional 350 μl of serum (50% serum, vol/vol), resulting in 2.5 mg/ml peptide concentration. Peptide solution in the same concentration, but without added serum was used as control. Peptides were incubated for 96 h at 37°C and samples (100 μl for 0% and 10% human serum and 140 μl for 50% human serum) were taken after 0, 24, 48, 72, and 96 h. The termination of the enzymatic reaction as well as the determination of intact peptide concentration was performed as described above.
Results
Peptides. For the experiments described in this article, we have designed synthetic peptides containing the 18PTGTQ22 epitope sequence derived from the 1PTTTPITTTTTVTPTPTPTGTQT23 tandem-repeat unit of MUC2 glycoprotein (19). Our earlier data showed that replacement of Pro-16 and Thr-17 by their d-isomer in peptide comprising the epitope (16PTPTGTQ22) had no significant effect on the epitope recognition (15). Therefore, 11 peptides with the sequence of 15TPTPTGTQTPT25 were produced. These compounds contain the PTGTQ epitope, but the native C- and N-terminal flanking regions were systematically substituted by one to three d-amino acids (Table 1). The amino acid analysis and the mass spectrometry of the peptides show the expected composition. We found also that the l- and d-amino acid ratio of the synthetic peptides does not change during the synthesis and purification (15). According to the HPLC chromatograms of the final products, the purity of the peptides is >95%. Characteristic data of the peptides, including HPLC retention time (Rt), relative molecular mass, and amino acid composition values, are given in Table 1.
Table 1. Characteristics of peptides corresponding to 15TPTPTGTQTPT25 of MUC2 glycoprotein.
| Peptide | Amino acid composition* (calculated) | Rt,† min | Calculated relative molecular mass [M+H]+‡ | Observed relative molecular mass [M+H]+‡ | IC50 μM |
|---|---|---|---|---|---|
| TPTPTGTQTPT | T 6.03 (6), P 2.59 (3), G 1.17 (1), E 1.20 (1) | 26.2 | 1,101.5 | 1,101.5 | 60 |
| t p t PTGTQTPT | T 5.65 (6), P 3.03 (3), G 1.07 (1), E 1.26 (1) | 26.0 | 1,101.5 | 1,101.5 | 392 |
| TPTPTGTQ t p t | T 5.76 (6), P 2.97 (3), G 1.09 (1), E 1.17 (1) | 25.8 | 1,101.5 | 1,101.8 | 61 |
| t PTPTGTQ t p t | T 5.79 (6), P 3.50 (3), G 1.04 (1), E 0.83 (1) | 27.3 | 1,101.5 | 1,102.0 | 40 |
| t p TPTGTQ t p t | T 5.76 (6), P 3.20 (3), G 1.00 (1), E 1.18 (1) | 26.1 | 1,101.5 | 1,102.3 | 50 |
| t p t PTGTQ t p t | T 5.79 (6), P 2.98 (3), G 1.07 (1), E 1.17 (1) | 25.9 | 1,101.5 | 1,101.8 | 792 |
| t p t PTGTQTP t | T 6.09 (6), P 2.70 (3), G 1.10 (1), E 1.07 (1) | 26.8 | 1,101.5 | 1,101.5 | 515 |
| t p t PTGTQT p t | T 6.06 (6), P 2.80 (3), G 1.10 (1), E 1.06 (1) | 26.9 | 1,101.5 | 1,101.5 | 1,670 |
| t PTPTGTQTPT | T 5.95 (6), P 3.02 (3), G 0.88 (1), E 1.10 (1) | 22.1 | 1,101.5 | 1,102.3 | 55 |
| TPTPTGTQTP t | T 5.70 (6), P 3.01 (3), G 1.08 (1), E 1.12 (1) | 24.2 | 1,101.5 | 1,102.3 | 63 |
| t PTPTGTQTP t | T 5.72 (6), P 3.20 (3), G 0.96 (1), E 1.10 (1) | 24.6 | 1,101.5 | 1,102.3 | 50 |
Gln was measured as Glu
HPLC retention time. Gradient was 0–5 min with 5% B and 5–35 min with 5–35% B. [A, 0.1% TFA/water; and B, 0.1% TFA/acetonitril/water (80:20, vol/vol)]
Electrospray ionization mass spectometry. Protonated molar mass
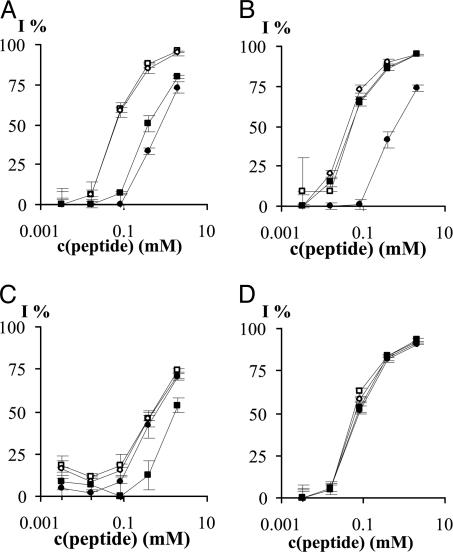
Binding of 996 mAb to MUC2 Peptides in Solution. In the competitive ELISA experiments, we first studied the ability of peptides with all l-amino acids (TPTPTGTQTPT), with three d-amino acids at the N (tptPTGTQTPT), or C terminus (TPTPTGTQtpt) and at both termini (tptPTGTQtpt) to inhibit the binding of MAb 996 to the [K12VTPTPTPTGTQTPT25-OH]-BSA target antigen. We observed that peptide TPTPTGTQTPT and TPTPTGTQtpt had similar Ab-binding profile (IC50 = 60 and 61 μmol/dm3), whereas peptides tptPTGTQTPT and tptPTGTQtpt show binding of approximately one order of magnitude weaker (IC50 = 392 and 792 μmol/dm3) (Fig. 1A). These findings indicate that substitution with d-amino acids on the C terminus has no or small effect on the Ab binding but that the same replacement at the N terminus has an important role in the peptide-Ab interaction.
Fig. 1.
Inhibition of 996 Ab binding to BSA-[K12VTPTPTPTGTQTPT25-OH] target antigen with synthetic MUC2 peptides in ELISA competition assays, inhibition percentage vs. peptide concentration. (A) TPTPTGTQTPT (□), TPTPTGTQtpt (○), tptPTGTQTPT (▪), and tptPTGTQtpt (•). (B) TPTPTGTQtpt (□), tPTPTGTQtpt (○), tpTPTGTQtpt (▪), and tptPTGTQtpt (•). (C) tptPTGTQTPT (□), tptPTGTQTPt (○), tptPTGTQTpt (▪) and tptPTGTQtpt (•). (D) TPTPTGTQTPT (□), TPTPTGTQTPt (○), tPTPTGTQTPT (▪), and tPTPTGTQTPt (•).
In the next series of experiments, we compared the binding properties of peptides with three d-amino acid residues on their C termini and containing zero to three d-amino acids on the N termini. Fig. 1B shows that peptide with one (tPTPTGTQtpt) or two d-amino acids at the N terminus (tpTPTGTQtpt) has very similar Ab binding to peptide TPTPTGTQtpt (IC50 = 40 and 50 μmol/dm3). However, the presence of the d-isomer of Thr-17 resulted in one order of magnitude decrease of Ab binding. This result indicates that only the substitution of Thr-17 to its enantiomer is responsible for the reduced Ab binding.
The effect of the number of d-amino acid substitutions in the C-terminal flanking region was also analyzed. We have compared the Ab binding of peptides with zero to three d-amino acids on their C termini possessing three d-amino acids on their N termini. Fig. 1C shows that the enantiomer composition of the C-terminal flanking region of the peptide has no significant influence on the mAb 996-peptide binding. The peptides show IC50 values of 400-600 μmol/dm3, except for tptPTGTQTpt, which shows weaker binding (IC50 = 1,700 μmol/dm3). This feature may have conformational reasons, and it needs further examination.
Based on these data, we have investigated the Ab binding of peptides with only one d-amino acid on either or both termini. Fig. 1D shows that all three peptides (tPTPTGTQTPt, tPTPTGTQTPT, and TPTPTGTQTPt) and the all-l isomer exhibited very similar curves. This result and the similarity of IC50 values (all 50-60 μmol/dm3) suggest that the replacement of one amino acid at both termini essentially does not diminish Ab-epitope interaction.
Enzymatic Stability of TPTPTGTQTPT Peptides. We have studied the enzymatic stability of all-l-amino acid and partly d-amino acid substituted peptides derived from the TPTPTGTQTPT sequence in human serum and in rat liver lysosomal preparation. The data are given in Tables 2 and 3, and the degradation profiles of selected peptides in serum (Fig. 2) or in lysosomal preparation (Fig. 3) are also given.
Table 2. Effect of d-amino acid substitution in the flanking region of TPTPTGTQTPT peptide in human serum.
| In 10% human serum
|
In 50% human serum
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Peptides | 0 h | 24 h | 48 h | 72 h | 96 h | 0 h | 24 h | 48 h | 72 h | 96 h |
| TPTPTGTQTPT | 100 | 42 ± 4 | 14 ± 1 | 3 ± 1 | 0 | 100 | 0 | 0 | 0 | 0 |
| TPTPTGTQTPt | 100 | 46 ± 4 | 17 ± 1 | 4 ± 1 | 0 | 100 | 0 | 0 | 0 | 0 |
| TPTPTGTQtpt | 100 | 36 ± 1 | 1 ± 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 |
| tPTPTGTQTPT | 100 | 83 ± 12 | 68 ± 3 | 61 ± 2 | 49 ± 0 | 100 | 85 ± 10 | 69 ± 3 | 61 ± 7 | 50 ± 1 |
| tptPTGTQTPT | 100 | 95 ± 4 | 95 ± 4 | 90 ± 7 | 84 ± 0 | 100 | 89 ± 4 | 80 ± 5 | 74 ± 1 | 55 ± 3 |
| tptPTGTQTPt | 100 | 100 ± 1 | 100 ± 0 | 97 ± 2 | 97 ± 2 | 100 | 97 ± 2 | 86 ± 2 | 79 ± 6 | 71 ± 8 |
| tptPTGTQTpt | 100 | 98 ± 3 | 96 ± 1 | 93 ± 0 | 92 ± 4 | 100 | 99 ± 1 | 100 ± 1 | 96 ± 6 | 97 ± 3 |
| tptPTGTQtpt | 100 | 100 ± 1 | 100 ± 0 | 99 ± 2 | 100 ± 1 | 100 | 96 ± 4 | 99 ± 2 | 97 ± 3 | 96 ± 4 |
| tpTPTGTQtpt | 100 | 100 ± 0 | 97 ± 1 | 97 ± 0 | 94 ± 3 | 100 | 99 ± 1 | 100 ± 0 | 99 ± 2 | 98 ± 4 |
| tPTPTGTQtpt | 100 | 92 ± 5 | 81 ± 9 | 66 ± 5 | 60 ± 8 | 100 | 93 ± 3 | 90 ± 3 | 84 ± 1 | 81 ± 4 |
| tPTPTGTQTPt | 100 | 75 ± 8 | 63 ± 1 | 56 ± 6 | 53 ± 7 | 100 | 83 ± 1 | 68 ± 1 | 55 ± 1 | 50 ± 0 |
*Data are percentage of peptide calculated as cintact/ctotal. The concentration of intact peptide was calculated from the AUC of the corresponding peak obtained after HPLC analysis, as described in Materials and Methods.
Table 3. Effect of d-amino acid substitution in the flanking region of TPTPTGTQTPT peptide in lysosomal preparation.
| In lysosomes, pH 3.5
|
In lysosomes, pH 5.0
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Peptide | 0.5 min | 60 min | 120 min | 180 min | 0.5 min | 60 min | 120 min | 180 min |
| TPTPTGTQTPT | 100 | 62 ± 8 | 31 ± 4 | 20 ± 10 | 100 | 56 ± 0 | 25 ± 3 | 18 ± 2 |
| tPTPTGTQTPT | 100 | 57 ± 7 | 24 ± 4 | 10 ± 1 | 100 | 58 ± 0 | 25 ± 1 | 18 ± 2 |
| tptPTGTQTPT | 100 | 62 ± 8 | 14 ± 8 | 3 ± 2 | 100 | 52 ± 11 | 14 ± 10 | 2 ± 2 |
| TPTPTGTQTPt | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| TPTPTGTQtpt | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| tptPTGTQTPt | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| tptPTGTQTpt | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| tptPTGTQtpt | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| tpTPTGTQtpt | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| tPTPTGTQtpt | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| tPTPTGTQTPt | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
*Data are percentage of peptide calculated as cintact/ctotal. The concentration of intact peptide was calculated from the AUC of the corresponding peak obtained after HPLC analysis, as described in Materials and Methods.
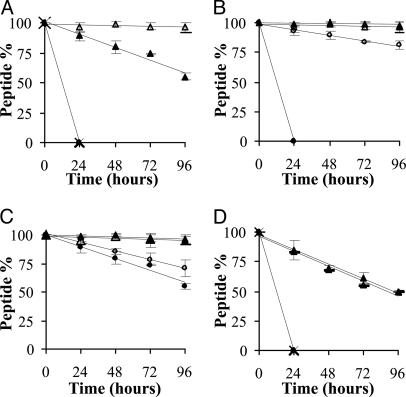
Fig. 2.
Decomposition of synthetic MUC2 peptides in 50% human serum. (A) TPTPTGTQTPT (•), TPTPTGTQtpt (×), tptPTGTQTPT (▪), and tptPTGTQtpt (□). (B) TPTPTGQTtpt (•), tPTPTGQTtpt (○), tpTPTGTQtpt (▪), and tptPTGTQtpt (□). (C) tptPTGTQTPT (•), tptPTGTQTPt (○), tptPTGTQTpt (▪), and tptPTGTQtpt (□). (D) TPTPTGTQTPT (•), TPTPTGQTTPt (×), tPTPTGTQTPT (▪), and tPTPTGTQTPt (□).
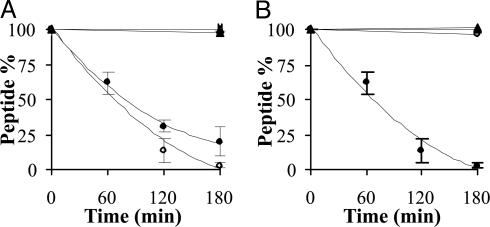
Fig. 3.
Decomposition of synthetic MUC2 peptides in lysosomal preparation (pH 3.5). (A) TPTPTGTQTPT (•), TPTPTGTQtpt (○), tptPTGTQTPT (▪), and tptPTGTQtpt (□). (B) tptPTGTQTPT (•), tptPTGTQTPt (○), tptPTGTQTpt (▪), and tptPTGTQtpt (□).
Stability in Human Serum. First, we compared the stability of peptides with all-l-amino acids (TPTPTGTQTPT) and three d-amino acids at the N terminus (tptPTGTQTPT), C terminus (TPTPTGTQtpt), and at both termini (tptPTGTQtpt) in 10% and 50% human serum. Native peptide and peptide with three d-amino acids at the C terminus (TPTPTGTQtpt) were decomposed within 24 h in 50% serum (Fig. 2 A) and within ≈2 days in 10% serum (Table 2). Peptide containing three d-amino acids only at the N terminus (tptPTGTQTPT) was more stable, only 16% and 45% of it degraded in 10% and 50% serum, respectively (Fig. 2 A and Table 2). Peptide with three d-amino acids on both N and C termini was stable even after 96 h in 50% human serum (Fig. 2 A).
Next, we investigated peptides with three d-amino acids on their C termini and zero to three d-amino acids on their N termini. Fig. 2B shows that the presence of at least one d-amino acid is important in this flank. Introduction of one d-amino acid (tPTPTGTQtpt) markedly enhanced the stability of the peptide compared with TPTPTGTQtpt in human serum. However, note that tPTPTGTQtpt decomposed more rapidly in 10% than in 50% human serum (Table 2). However, peptide tpTPTGTQtpt was found to be completely stable under both conditions.
The influence on serum stability of zero to three d-amino acids at the C terminus of peptides with three d-amino acids on their N termini was studied. Fig. 2C shows that the presence of a single d-amino acid at the end position partly prevented degradation (tptPTGTQTPt vs. tptPTGTQTPT), but 30% of the peptide still decomposed in 96 h in 50% human serum. Peptide tptPTGTQTpt was fully stable under these conditions.
Last, we have examined the effect of the presence of a single d-amino acid on the C and/or N termini of the peptide. Data show that the native peptide as well as peptide TPTPTGTQTPt decomposed in 24 h in 50% human serum. Inclusion of one d-amino acid at the N terminus or both termini (tPTPTGTQTPT and tPTPTGTQTPt) created partially enzyme-resistant analogues (Fig. 2D and Table 2).
Stability in Lysosomal Preparation. In the next series of experiments, we studied the stability of the peptides in lysosomal preparation, containing various proteolytic enzymes, at pH 3.5 and 5.0. We found no significant difference between the stability profiles of the peptides studied at these pH values (Table 3). Fig. 3A shows that, contrary to the serum stability, peptides containing three d-amino acids on their C terminus (TPTPTGTQtpt and tptPTGTQtpt) were stable after incubation with lysosome preparation for 3 h. Peptides with one or two d-amino acids at the N terminus (tPTPTGTQtpt and tpTPTGTQtpt) showed similar stability (Table 3). In contrast, the all-l and tptPTGTQTPT peptides degraded completely under the same conditions.
Peptides with three d-amino acids at the N terminus, and zero to three C-terminal d-amino acid(s) were free of enzymatic degradation (Fig. 3B). These data also show that even a single d-amino acid substitution on the C terminus (tptPTGTQTPt) is enough to provide protection against enzymatic hydrolysis of the peptide.
Last, we examined the influence of one d-amino acid at the C and/or N termini on the stability of peptides. Results given in Table 3 clearly demonstrate that the presence of even a single d-amino acid on the C terminus (TPTPTGTQTPt) is satisfactory to attain the complete stability. Note that the peptide tptPTGTQTPT containing three d-amino acids on its N terminus decomposed more rapidly than the native peptide at both pH values, which might have conformational reasons.
Discussion
We synthesized several peptides corresponding to the 15TPTPTGTQTPT25 antigenic part of the MUC2 tandem-repeat unit. The peptides contained the 18PTGTQ22 epitope of mAb 996 and native flanks resulting in 15TPTPTGTQTPT25. Within this sequence, the three C- and N-terminal flanking residues were systematically substituted by their d-amino acid counterparts. We tested these peptides in competitive ELISA experiments to determine whether they retain Ab binding, and we performed enzymatic digestion studies to ascertain their increased stability. Based on these experiments, it was our aim to produce a synthetic peptide to be used as potential immunogen exhibiting preserved Ab recognition and increased stability against proteolytic enzymes.
Our results indicated that the C-terminal flanking region of the PTGTQ epitope could be substituted with d-amino acids, resulting in no loss of Ab binding [TPTPTGTQTPT (IC50 = 60 μmol/dm3) vs. TPTPTGTQtpt (IC50 = 61 μmol/dm3)]. A similar tendency was observed even with peptides containing three d-amino acids in the N-terminal flank (tptPTGTQTPT, IC50 = 392 μmol/dm3 vs. tptPTGTQtpt, IC50 = 792 μmol/dm3). [Although peptide tptPTGTQTpt showed even weaker binding (IC50 = 1670 μmol/dm3) than either peptide tptPTGTQTPt (IC50 = 515 μmol/dm3) or tptPTGTQtpt, the strength of the binding is one-third of that of the other peptides. The reason for this feature may be conformational, and it should be investigated.] These data indicate that the presence of even three d-amino acids at the C terminus cause no significant change in Ab recognition. However, the N-terminal flanking region is more sensitive for alteration: tPTPTGTQTPT (IC50 = 55 μmol/dm3) vs. tptPTGTQTPT (IC50 = 392 μmol/dm3). This phenomenon could also be demonstrated with peptides containing three d-amino acids in the C-terminal flank [tPTPTGTQtpt (IC50 = 40 μmol/dm3) vs. tptPTGTQtpt (IC50 = 792 μmol/dm3)]. This result is in accordance with our previous findings (15) in which the substitution of the immediate neighboring amino acid of the PTGTQ epitope within the PTPTGTQ peptide caused approximately one order of magnitude reduction in the binding ability of the peptides. Together, we conclude that replacement of three amino acids at the C terminus or single amino acid at the N terminus has no influence on Ab binding. Furthermore, simultaneous substitutions of flanks at both termini up to two and three d-enantiomer were fully tolerated, and they essentially did not change the mAb 996 binding.
In parallel with the binding studies, we also compared the stability of the native MUC2 peptide TPTPTGTQTPT and its N- and/or C-terminally d-amino acid-substituted analogues in rat lysosomal preparation and in human serum. For in vitro enzymatic stability studies, mainly isolated enzymes like carboxypeptidase A, aminopeptidase M, proteinase A, carboxypeptidase Y (28), α-chymotrypsin, and carboxypeptidase Y (29), or complex biological fluids, like human serum and urine (30), human plasma (31), or rat liver lysosomes (32), are in use. For this study, we used diluted human serum and rat liver lysosome preparation. Considering the lack of consensus in the literature about the pH within the lysosome, we have selected two pH values to perform the assay. Harada et al. (33) reported on a value of pH 4; Bach et al. (34) observed that the pH in lysosomes varies between 4.3 and 4.5. Other groups found different level of acidity between pH 4.5 and 5.0 [pH 4.5 (35, 36); 4.67 (37), 4.75 (38), and 5.0 (39)]. It is also known that the optimal pH for the main lysosomal enzyme, cathepsin D is active at pH 3.5 (40). Based on these data, our experiments were performed at pH 3.5 and 5.
Our results show that in lysosomal preparation the presence of d-amino acids at the N terminus provides no protection against enzymatic hydrolysis, the peptides tPTPTGTQTPT and tptPTGTQTPT are degraded with the same rate as the native all-l peptide (Table 3). In contrast, to our surprise, peptides containing at least one d-amino acid at their C terminus are completely resistant against lysosomal enzymes. Note that this observation was independent of the pH values used (pH 3.5 and 5). These results might indicate that under these circumstances perhaps only carboxypeptidases were active, and it is likely that other proteolytic enzymes, such as dipeptidyl peptidase or endopeptidases, exhibited negligible activity. For example, it has been reported that dipeptidyl peptidase (cleaving N-terminal XP dipeptides) has optimum pH under alkaline conditions (41). It is also attractive to speculate that perhaps the Thr- and Pro-rich sequence may not be an adequate substrate for endopeptidases.
The stability of the all-l peptide and the d-amino acid-containing peptides was studied in 10% and 50% human serum. We found that the presence of d-amino acids only at the C terminus does not influence the degradation kinetics of the peptide (Table 2). The d-amino acid(s) present at the N terminus (tPTPTGTQTPT and tptPTGTQTPT) provided improved, but not complete, stability both in 10% and 50% human serum (Table 2). However, substitutions simultaneously at both ends of the peptide resulted in compounds with almost full protection against serum degradation. For example, peptide tPTPTGTQtpt exhibited enhanced stability compared with TPTPTGTQtpt, and tpTPTGTQtpt was fully stable during the assay. Note that the stability of the peptide increases with the number of d-amino acids at the C terminus: one d-amino acid at the C terminus (tptPTGTQTPt) generates a higher stability, although the peptide is still degradable; two d-amino acids at the C terminus (tptPTGTQTpt) produce a completely stable peptide in 50% human serum during the time of the assay. Our study shows that in the serum the N-terminal degradation of the peptides is significantly faster than the C-terminal one. It might indicate that mainly aminopeptidases are active in the diluted serum.
Together, our results indicate that the epitope function (namely, specific binding to mAb 996) of peptide 15TPTPTGTQTPT25 is retained even in the presence of two d-amino acid residues at its N-terminal flanking region, and up to three at its C-terminal flanking region (tpTPTGTQtpt). This substituted peptide also shows high resistance against proteolytic degradation in diluted human serum and in lysosomal preparation. These findings also indicate that, by appropriate combination of structural modification (namely d-amino acid substitution) in the flanks of an Ab epitope, it is feasible to construct a synthetic antigen with preserved recognition properties and high stability against enzymatic degradation. Further studies are needed to identify enzymes responsible for degradation in diluted human serum as well as in the lysosome preparation and to gain more detailed information on the mechanism of action. Also, we consider peptides tPTPTGTQTpt and tpTPTGTQTpt as lead compounds for immunization experiments and use them as potential component of synthetic vaccines for tumor therapy.
Acknowledgments
This work was supported by Hungarian Research Fund Grants OTKA T037749, T043576, and F034886 and Ministry of Education Grant FKFP 0153/2001.
Author contributions: F.H. designed research; R.T., K.U., D.I., and E.F. performed research; A.C.P. contributed new reagents; R.T. and K.U. analyzed data; and R.T., K.U., and F.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Meyer, J. P., Gillespie, T. J., Hom, S., Hruby, V. J. & Davis, T. P. (1995) Peptides 16, 1215-1219. [DOI] [PubMed] [Google Scholar]
- 2.Hudecz, F. (2001) Biologicals 29, 197-207. [DOI] [PubMed] [Google Scholar]
- 3.Steer, D., Lew, R., Perlmutter, P., Smith, A. I. & Aguilar, M. I. (2002) Biochemistry 41, 10819-10826. [DOI] [PubMed] [Google Scholar]
- 4.Briand, J. P., Benkirane, N., Guichard, G., Newman, J. F., Van Regenmortel, M. H., Brown, F. & Muller, S. (1997) Proc. Natl. Acad. Sci. USA 94, 12545-12550, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamamoto, K., Kida, Y., Zhang, Y., Shimizu, T. & Kuwano, K. (2002) Microbiol. Immunol. 46, 741-749. [DOI] [PubMed] [Google Scholar]
- 6.Sela, M. & Zisman, E. (1997) FASEB J. 11, 449-456. [DOI] [PubMed] [Google Scholar]
- 7.Van Regenmortel, M. H. & Muller, S. (1998) Curr. Opin. Biotechnol. 9, 377-382. [DOI] [PubMed] [Google Scholar]
- 8.Guichard, G., Benkirane, N., Zeder-Lutz, G., Van Regenmortel, M. H., Briand, J. P. & Muller, S. (1994) Proc. Natl. Acad. Sci. USA 91, 9765-9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dintzis, H. M., Symer, D. E., Dintzis, R. Z., Zawadzke, L. E. & Berg, J.M. (1993) Proteins 16, 306-308. [DOI] [PubMed] [Google Scholar]
- 10.Miller, S. M., Simon, R. J., Ng, S., Zuckermann, R. N., Kerr, J. M. & Moos, W. H. (1995) Drug Dev. Res. 35, 20-32. [Google Scholar]
- 11.Goodman, M. & Chorev, M. (1979) Accounts Chem. Res. 12, 1-7. [Google Scholar]
- 12.Ben-Yedidia, T., Beignon, A. S., Partidos, C. D., Muller, S. & Arnon, R. (2002) Mol. Immunol. 39, 323-331. [DOI] [PubMed] [Google Scholar]
- 13.Lasonder, E., Schellekens, G. A., Koedijk, D. G., Damhof, R. A., Welling-Wester, S., Feijlbrief, M., Scheffer, A. J. & Welling, G. W. (1996) Eur. J. Biochem. 240, 209-214. [DOI] [PubMed] [Google Scholar]
- 14.Kramer, A., Stigler, R. D., Knaute, T., Hoffmann, B. & Schneider-Mergener, J. (1998) Protein Eng. 11, 941-948. [DOI] [PubMed] [Google Scholar]
- 15.Uray, K., Kajtár, J., Vass, E., Price, M.R. Hollósi, M. & Hudecz, F. (2000) Arch. Biochem. Biophys. 378, 25-32. [DOI] [PubMed] [Google Scholar]
- 16.Uray, K., Schlosser, G. & Hudecz, F. (2002) in: Peptides 2002, eds. Benedetti, E. & Pedone, C. (Edizione Ziino, Naples, Italy) pp. 644-645.
- 17.Gold, D. & Mill, F. (1975) Nature 255, 85-87. [DOI] [PubMed] [Google Scholar]
- 18.Price, M. R., Sekowski, M., Ladányi, A., Uray, K., Ma, Y., Durrant, L.G. & Tendler, S. J. B. (1993) Int. J. Cancer 55, 753-759. [DOI] [PubMed] [Google Scholar]
- 19.Uray, K., Price, M. R. & Hudecz, F. (1998) J. Pept. Sci. 4, 319-326. [DOI] [PubMed] [Google Scholar]
- 20.Gum, J. R., Byrd, J. C., Hicks, J. W., Toribara, N. W., Lamport, D. T. A. & Kim, Y. S. (1989) J. Biol. Chem. 264, 6480-6487. [PubMed] [Google Scholar]
- 21.Durrant, L. G., Jacobs, E. & Price, M. R. (1994) Eur. J. Cancer 30A, 355-363. [DOI] [PubMed] [Google Scholar]
- 22.Uray, K., Kajtár, J., Vass, E., Price, M. R., Hollósi, M. & Hudecz, F. (1999) Arch. Biochem. Biophys. 361, 65-74. [DOI] [PubMed] [Google Scholar]
- 23.Wang, S. S. (1973) J. Am. Chem. Soc. 95, 1328-1333. [DOI] [PubMed] [Google Scholar]
- 24.Kaiser, E., Colescott, R. L., Bossinger, C. D. & Cook, P. I. (1970) Anal. Biochem. 34, 595-598. [DOI] [PubMed] [Google Scholar]
- 25.Kaiser, E., Bossinger, C. D., Colescott, R. L. & Olser, D. B. (1980) Anal. Biochem. 118, 149-151. [DOI] [PubMed] [Google Scholar]
- 26.Uray, K., Price, M. R., Majer, Z., Vass, E., Hollósi, M. & Hudecz, F. (2003) Arch. Biochem. Biophys. 410, 254-260. [DOI] [PubMed] [Google Scholar]
- 27.Dingle, J. T., ed. (1972) Lysosomes. A Laboratory Handbook (North-Holland, Amsterdam) p. 46.
- 28.Péter, A., Tóth, G., Tömböly, Cs., Laus, G. & Tourwe, D. (1999) J. Chromatog. A 846, 39-48. [DOI] [PubMed] [Google Scholar]
- 29.Cardillo, G., Gentilucci, L., Qasem, A. R., Sgarzi, F. & Spampinato, S. (2002) J. Med. Chem. 45, 2571-2578. [DOI] [PubMed] [Google Scholar]
- 30.Georga, K. A., Samanidou, V. F. & Papadoyannis, I. N. (2001) J. Chromatog. B 759, 209-218. [DOI] [PubMed] [Google Scholar]
- 31.Terenius, L., Sandin, J. & Sakurada, T. (2000) Peptides 21, 919-922. [DOI] [PubMed] [Google Scholar]
- 32.Trouet, A., Masquelier, M., Baurain, R. & Deprez-De Campeneere, D. (1982) Proc. Natl. Acad. Sci. USA 79, 626-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harada, M., Sakakibara, H., Yano, T., Suzuki, T. & Okuno, S. (2000) J. Control. Release 69, 399-412. [DOI] [PubMed] [Google Scholar]
- 34.Bach, G., Chen, C.-S. & Pagano, R. E. (1999) Clin. Chim. Acta 280, 173-179. [DOI] [PubMed] [Google Scholar]
- 35.Yu, Z., Persson, H. L., Eaton, J. W. & Brunk, U. T. (2003) Free Radic. Biol. Med. 34, 1243-1252. [DOI] [PubMed] [Google Scholar]
- 36.Zatta, P., Taylor, A., Zambenedetti, P., Milacic, R. & dell'Antone, P. (2000) Life Sci. 66, 2261-2266. [DOI] [PubMed] [Google Scholar]
- 37.Myers, B. M., Tietz, P. S., Tarara, J. E. & Larusso, N. F. (1995) Hepatology 22, 1519-1526. [PubMed] [Google Scholar]
- 38.Pshezhetsky, A.V. & Potier, M. (1993) Biochem. Biophys. Res. Commun. 195, 354-362. [DOI] [PubMed] [Google Scholar]
- 39.King, H. D., Staab, A. J., Pham-Kaplita, K., Yurgaitis, D., Firestone, R. A., Lasch, S.J. & Trail, P. A. (2003) Bioorg. Med. Chem. Lett. 13, 2119-2122. [DOI] [PubMed] [Google Scholar]
- 40.Hurley, M. J., Larsen, L. B., Kelly, A. L. & McSweeney, P. L. H. (2000) Int. Dairy J. 10, 673-681. [Google Scholar]
- 41.Mentlein, R., Galwitz, B. & Schmidt, W. E. (1993) Eur. J. Biochem. 214, 829-835. [DOI] [PubMed] [Google Scholar]