Summary
Understanding the mechanisms driving tissue and organ formation requires knowledge across scales. How do signaling pathways specify distinct tissue types? How does the patterning system control morphogenesis? How do these processes evolve? The Drosophila egg chamber, where EGF and BMP signaling intersect to specify unique cell types that construct epithelial tubes for specialized eggshell structures, has provided a tractable system to ask these questions. Work there has elucidated connections between scales of development, including across evolutionary scales, and fostered the development of quantitative modeling tools. These tools and general principles can be applied towards understanding other developmental processes across organisms.
eTOC/In-Brief
Understanding the mechanisms driving tissue/organ formation requires knowledge across scales. How do signaling pathways pattern tissue that control morphogenesis, and how do these processes evolve? Work answering these questions in Drosophila eggshell structure formation has uncovered broadly applicable general principles and developed modeling tools for understanding other developmental processes.
Introduction
The generation of complex tissue structures is an important part of development. In many instances, tissues form through epithelial morphogenesis; that is, simple, flat epithelial sheets serve as starting materials that deform into three-dimensional structures. Developmental processes that occur through epithelial morphogenesis range from early embryonic events, such as gastrulation and neural-tube formation, to later events, such as the formation of kidneys, lungs, or other mature organs.
At its simplest, epithelial morphogenesis involves a few distinct steps. First, morphogen gradients or other signals establish spatial information across the tissue. Next, cells interpret these spatial cues and differentiate into distinct cell types with specific gene-expression profiles. Finally, changes in tissue shape emerge from the collective effect of different cells expressing genetically determined behaviors and mechanical properties.
Epithelial morphogenesis is similar to origami (Zartman and Shvartsman, 2010), since in both processes a two-dimensional sheet (of cells, or paper) is patterned (with gene-expression patterns, or folds) to guide its transformation into a 3D structure. However, epithelial morphogenesis is clearly more complex. For example, it enlists processes that have no analog in origami, such as cell division and cell-neighbor exchange. Furthermore, multiple iterations of patterning and morphogenesis can occur during the development of a single tissue, with each round affecting the starting conditions for the next. This complexity is one reason we still can’t answer the fundamental question: how would one pattern an epithelial sheet in order to generate a specific desired shape?
The Drosophila eggshell (Figure 1A) provides an attractive model system for studying epithelial patterning and morphogenesis, due to its relative simplicity and experimental tractability (Hudson and Cooley, 2014). Each eggshell is produced by an individual egg chamber (Figure 1B,C), which consists of 15 germline-derived nurse cells, a single oocyte, and a surrounding follicular epithelium. The egg chamber is a self-contained unit that can easily be cultured in vitro (Cliffe et al., 2007; Dorman et al., 2004; Manning and Starz-Gaiano, 2015; Peters and Berg, 2016a; Prasad et al., 2007). The development of an egg chamber proceeds through 14 stages (Spradling, 1993); during the stages in which the eggshell is formed (stages 10–14), there is neither cell division nor significant cell death among the follicle cells. Furthermore, relatively little feedback occurs from morphogenesis back to patterning (Yakoby et al., 2008a).
Figure 1.
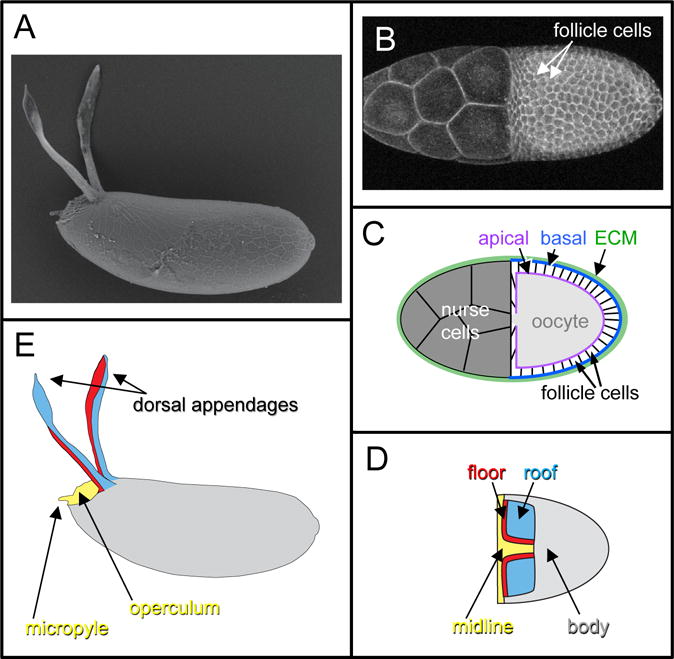
The formation of Drosophila eggshells from egg chambers is a model for epithelial patterning and morphogenesis. A) Scanning Electron Micrograph (SEM) of an eggshell from Drosophila melanogaster. B) A stage-10 D. melanogaster egg chamber, visualized with a fluorescent membrane marker. In the right half of the image, the columnar follicle cells surround and thus obscure the underlying oocyte. In the left half of the egg chamber are 15 nurse cells; these are covered by a thin layer of squamous “stretch” follicle cells that are not readily apparent under these visualization conditions. C) Schematic of a stage-10 egg chamber, in cross-section. The stretch cells, not shown, do not produce eggshell material. D) Schematic of a stage-10 egg chamber (dorsal view), showing the four distinct cell types of columnar follicle cells. E) Schematic of an eggshell (lateral view), color-coded to show which eggshell structures are formed by which follicle-cell type in (D). Figures C-E are adapted from (Osterfield et al., 2015).
The eggshell is formed during oogenesis by the columnar follicle cells, which initially surround the oocyte in a single-layered epithelial sheet (Figure 1C). The follicle cells secrete eggshell components apically, that is, into the space between the follicle cells and the oocyte itself, where they are cross-linked to form the mature eggshell (Waring, 2000). Therefore, the shape of the eggshell directly reflects the final shape of the apical surface of the follicular epithelium, which acts as a mold. The apical surface of the follicular epithelium at stage 10 (Figure 1B,C) is shaped like half of an ellipsoid. It transforms into a full ellipsoid during stage 11, when the nurse cells transfer their contents through cytoplasmic bridges into the oocyte in a process termed nurse-cell dumping (Mahajan-Miklos and Cooley, 1994). Building on this ellipsoid base, specific subpopulations of follicle cells form specialized eggshell structures. The most prominent eggshell structures, called the dorsal appendages or respiratory filaments, reflect the formation of two tubes in the follicular epithelium. The dorsal appendages are thought to facilitate gas exchange by the embryo in part by projecting above the substrate (for example, decomposing fruit) in which the developing egg is embedded; furthermore, when immersed in water, their porous material retains a layer of air that functions as a plastron to increase oxygen uptake (Hinton, 1960). This review focuses on the formation of these structures.
Although the stage-10 egg chamber exhibits an elliptical shape, it did not always have that form. Early egg chambers are round, but beginning around stage 5, the egg chamber changes shape from a sphere into an ellipsoid with an elongated anterior-posterior axis. This process provides another system in which the conversion of gene expression to tissue shape is being actively studied. Strikingly, this shape change is partly due to a global rotation of the egg chamber cell mass within the surrounding extracellular matrix; the follicle cells remodel the extracellular matrix as they migrate on it, and the increasingly polarized extracellular matrix fibers constrict the egg chamber as it grows (Haigo and Bilder, 2011). We direct the reader to several references for information on other molecular mechanisms implicated in this very interesting process (Cetera et al., 2014; Gates, 2012; He et al., 2010; Horne-Badovinac, 2014; Isabella and Horne-Badovinac, 2015; Lerner et al., 2013). Other aspects of eggshell morphogenesis, including the formation of eggshell structures such as the micropyle and operculum, are also described elsewhere (Horne-Badovinac and Bilder, 2005; Levine et al., 2010; Montell et al., 2012; Wu et al., 2008).
Introducing the Dorsal Appendage as a Model System
The biological processes underpinning dorsal-appendage formation are found in a wide variety of developing epithelial tissues. Therefore, insights gained from this system should be generally applicable. For example, patterning of the initially naive follicular epithelium into distinct cell types is initiated by morphogen gradients. As discussed below, cellular interpretation of these gradients is context-dependent and requires integration of multiple pathways. Likewise, morphogenesis in this system exhibits many of the typical cellular behaviors found in other systems, including cell neighbor rearrangements, apical constriction and other cell-shape changes, and cell migration. The only common epithelial behaviors missing from dorsal-appendage formation are cell division and cell death. Furthermore, dorsal-appendage morphogenesis offers the opportunity to study a simple but still truly three-dimensional system, unlike other popular models such as Drosophila ventral- furrow formation or germ-band extension, which exemplify 2D systems.
The dorsal appendage provides a well-developed model system for investigating tissue formation, as there exists a basic framework for understanding key processes, from initiating signals through shaping of the final structure. This framework includes 1) patterning of the follicular epithelium by signaling gradients; 2) translating these signals into gene-expression patterns that specify the four major subtypes of follicle cells; and 3) converting these gene-expression patterns into changes in tissue morphology. We review these topics below and discuss how studies of divergent Drosophilid species have deepened our understanding of this system. Furthermore, we highlight how computational modeling has contributed to our understanding of all of these processes. To conclude, we compare eggshell formation to other developmental processes.
Patterning
One striking observation about metazoan development is that a small number of signaling pathways can generate an enormous diversity of cell types, all arranged in highly ordered spatial patterns (Gerhart, 1999; Pires-daSilva and Sommer, 2003). Part of the reason so few signaling pathways are required for development is that activation of a single pathway can be interpreted in many different ways. Varying interpretations can be due to differences in signal amplitude; for example, uniform low levels of DPP promote wing-disc growth in Drosophila, but the stripe of high expression in the center of the wing imaginal disc is essential for patterning rather than growth (Akiyama and Gibson, 2015). Varying interpretations can also be due to differences in developmental timing or the identity of the cell receiving the signal; for example, Wingless (Wnt) signaling is required for cell-fate decisions throughout Drosophila development in tissues ranging from the embryonic epidermis to the wing imaginal disc (Swarup and Verheyen, 2012). Additionally, combinatorial signaling, or the integration of multiple pathways, is another well-established means to increase the information available to cells from limited numbers of signaling pathways. Combinatorial signaling is used repeatedly in metazoan development, from cell-fate specification to axon guidance (Briscoe and Small, 2015; Cornell and Kimelman, 1994; Flores et al., 2000; Morales and Kania, 2016).
All of these mechanisms facilitate the patterning of the Drosophila follicular epithelium. Here, we will review how signaling pathways are interpreted and integrated to establish specific cell fates, but first we will describe the results of these patterning events. Central to our understanding of eggshell formation is the fate map detailing which follicle-cell populations form which eggshell structures (Dorman et al., 2004). During mid-oogenesis, the follicle cells surrounding the oocyte can be classified into four types based on their eventual fates; these cell types and the structures they form are indicated by color-coding in Figure 1D–E. Each dorsal appendage is formed from a primordium that contains two types of cells, the “floor” and “roof” cells, which form the floor and roof, respectively, of the appendage tube. The operculum (which facilitates larval hatching) and part of the micropyle (which allows sperm entry for fertilization) are formed by the “midline” cells, which are also called the “operculum-forming” cells. These cells are initially found in a continuous T-shaped domain that includes the region between the two appendage primordia and the anterior-most follicle cells. The remaining follicle cells, which we will call the “body” cells, produce the rest of the eggshell. Note that while this division of the follicle cells into four types is useful for describing dorsal-appendage formation, it is a simplification. For example, the body cells consist of at least three distinct cell types: the main body cells, which form the majority of the eggshell, along with the posterior terminal and polar cells at the posterior extreme (Horne-Badovinac and Bilder, 2005). Furthermore, within a specific cell type, subpopulations exist with distinct gene-expression profiles or cell behaviors (Boyle et al., 2010; Yakoby et al., 2008a).
Each of the two appendage primordia consists of approximately 65 cells and is composed of two adjacent subdomains, the roof and the floor (Dorman et al., 2004; Ward and Berg, 2005). Cells within each roof domain express high levels of Broad (Br), a Zn-finger transcription factor (Dorman et al., 2004), and in fact, high levels of Br are sufficient for inducing some follicle cells to adopt a roof-like fate (Tzolovsky et al., 1999). As a result, the question of how roof cells are specified can be restated as the question of how Br is regulated. We therefore begin by examining the molecular mechanisms regulating Br.
Patterning of the Br-positive roof domain results from the integration of three signaling events (Figure 2). The first event occurs early in oogenesis between stage 1 and stage 6), when the oocyte nucleus is positioned at the posterior extreme of the oocyte. Low levels of Gurken (Grk), a TGFα-like ligand, are synthesized near the oocyte nucleus and secreted by adjacent regions of the oocyte cortex; therefore, the Grk/Epidermal Growth Factor Receptor (EGFR) signaling in these stages is found in a posterior-to-anterior gradient (Figure 2A). One function of this early phase of Grk signaling is to prepattern the follicular epithelium, making the posterior part of the epithelium incapable of assuming an appendage-producing fate in response to a later inductive signal (Fregoso Lomas et al., 2013). This posterior repression of the appendage cell fate is mediated by T-box transcription factors Midline (Mid) and H15, which are induced by the early phase of Grk signaling and which repress Br (Fregoso Lomas et al., 2013).
Figure 2.
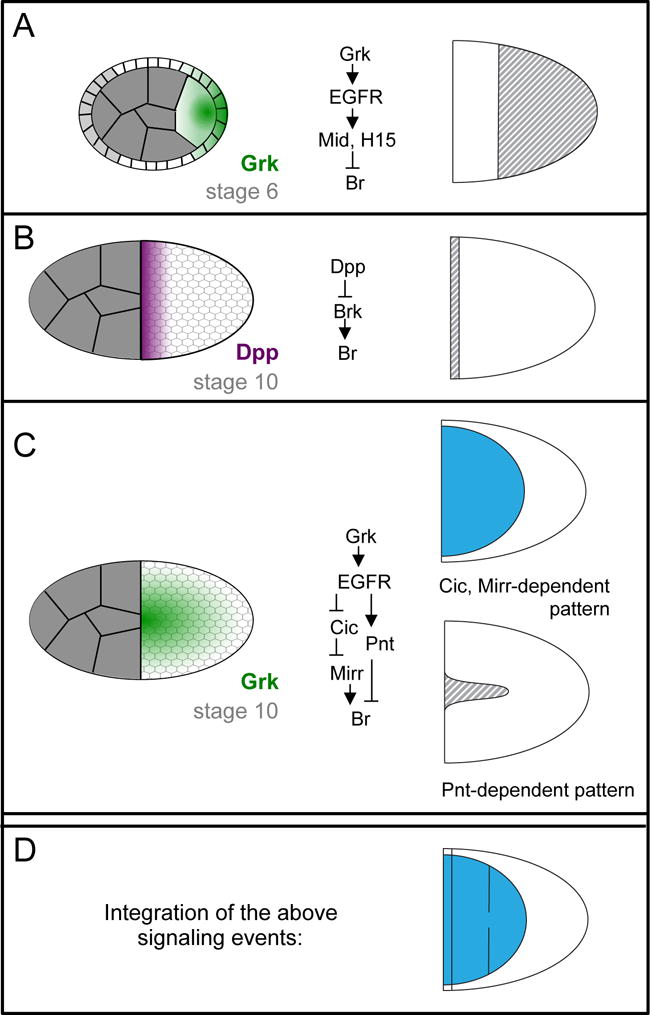
Several signaling gradients are integrated to specify the locations of two Br-expressing roof domains. A-C) Schematics of the signaling gradients that pattern the follicular epithelium are shown to the left. Simplified downstream signal-transduction networks are shown in the middle. The effect of these signals on Br expression during stage 10 is shown to the right; grey stripes indicate repression, blue indicates activation. Further details are provided in main text. Note that in (A), the light gray follicle cells in the schematized cross-section do not contribute to the columnar epithelium that surrounds the oocyte and produces the eggshell; instead, these cells become stretch and border cells. For more information on the developmental processes occurring between stages 6 and 10, see (Horne-Badovinac and Bilder, 2005). D) Due to the combined effect of all three signaling events, two patches of Br-expressing cells are formed (blue).
The second signaling event, which occurs at approximately stage 10, also involves repression of appendage-cell fate, this time by Decapentaplegic (Dpp) (Yakoby et al., 2008b). Dpp is produced by the stretch cells (Peri and Roth, 2000; Twombly et al., 1996), a population of squamous follicle cells that surround the nurse cells, creating an anterior-to-posterior gradient of Dpp signaling (Figure 2B). High levels of Dpp signaling in the anterior-most follicle cells repress brinker (brk), which encodes a transcriptional mediator of Dpp signaling that functions in multiple stages of fruit fly development (Chen and Schüpbach, 2006; Shravage et al., 2007). Brk is expressed in most follicle cells and is required for Br expression. Since brk expression is directly repressed by Dpp signaling in the anterior-most follicle cells, Br cannot be expressed in these cells (Charbonnier et al., 2015; Chen and Schüpbach, 2006; Shravage et al., 2007).
The third and most crucial signaling event occurs just prior to and during stage 10. By this stage, the oocyte nucleus has moved to the dorsal-anterior extreme of the oocyte, and so the Grk gradient is now highest there (Figure 2C) (Cheung et al., 2011). At this stage, high levels of Grk induce midline cell fates, intermediate levels induce Br-positive roof fates, and cells exposed to the lowest Grk levels maintain their default body-cell fate (Goentoro et al., 2006; Pai et al., 2000). In other words, ignoring the influence of the early Grk and Dpp signals discussed above, three domains are determined by distinct threshold levels of Grk. This observation poses two interesting questions; how is Br expressed specifically in response to intermediate levels of Grk at stage 10, and how can Grk signaling in the posterior at stage 6 repress appendage cell fate while Grk signaling in the dorsal-anterior at stage 10 promotes appendage cell fate? Regarding the latter question, one difference in these signaling events is that early processes require only low levels of Grk signaling, while later processes involve higher levels of ligand; that is, weak loss-of- function mutations do not affect posterior patterning, only dorsal patterning (Schüpbach, 1987). In addition, a recent study demonstrates that the different consequences of Grk signaling in the dorsal-anterior versus the posterior follicle cells is due to the cooperative action of Grk with two additional signaling pathways: the Dpp pathway, which is activated in the anterior as discussed above, and the JAK/STAT pathway, which is activated by the ligand Unpaired in the posterior (Fregoso Lomas et al., 2016). Signaling by Grk and Dpp together results in expression of Mirror (Mirr), a dorsal fate determinant discussed below. Signaling by Grk and JAK/STAT together results in expression of Mid and H15. Furthermore, Mid/H15 and Mirr mutually repress each other, potentially allowing for a switch-like response between the anterior and posterior interpretations of the Grk signal (Fregoso Lomas et al., 2016).
The expression of Br in response to intermediate levels of Grk is achieved through a network of transcription factors that form an incoherent feedforward loop. This term refers to a network motif in which the same input activates both a target gene and a repressor of that target gene. Grk-induced activation of Br expression occurs through the HMG-box repressor Capicua (Cic) and the Iroquois transcription factor Mirror (Mirr). Specifically, high and intermediate levels of Grk signaling result in the exclusion of Cic from nuclei of cells (Astigarraga et al., 2007). This removal relieves its transcriptional repression of mirr, which encodes a homeodomain protein that is essential for Br expression (Atkey et al., 2006). Grk- induced repression of Br expression occurs through the ETS-factor Pointed (Pnt). Pnt is expressed in response to the high levels of Grk signaling at the dorsal midline, where it then represses Br (Boisclair Lachance et al., 2009; Deng and Bownes, 1997; Morimoto et al., 1996). With Br repression occurring at high levels of Grk and activation at intermediate levels, this feedforward mechanism readily explains why Br is expressed in dorsolateral follicle cells. Furthermore, this mechanism accounts for the results of multiple studies involving genetic perturbations of the levels of EGFR signaling and manipulations of Cic, Mirr, and Pnt (Simakov et al., 2012; Yakoby et al., 2008b).
Currently, it is less clear how the floor-cell domain is patterned. This domain consists of a single line of cells that express rhomboid and that lie at the dorsal- anterior border of the roof domain (Dorman et al., 2004). Although rhomboid encodes a protease in the Drosophila EGF activation pathway (Urban et al., 2001), it is not required for appendage patterning (Boisclair Lachance et al., 2009) and can be viewed simply as a floor-cell marker. Importantly, in a wide variety of genetic backgrounds affecting the shape or number of roof domains, the rhomboid-positive floor domain maintains its single-cell width and tracks the Br domain, remaining at its dorsal-anterior border (Ward and Berg, 2005). The mechanisms responsible for the division of the appendage primordium into Br- and rhomboid-positive domains are still incompletely understood, though it is clear that Notch signaling is required (Ward et al., 2006). Furthermore, Br represses rhomboid, which explains why the rhomboid marker is excluded from the roof domain (Ward et al., 2006).
At this point, we have a solid framework for understanding patterning of the dorsal-appendage primordia. As alluded to earlier, one interesting feature of this framework is the use of one signal, EGF, to elicit multiple different responses. Another interesting feature is the use of combinatorial signaling (including EGFR, DPP, JAK/STAT and Notch) to establish distinct cell types. Within this framework, several questions remain. 1) What transcriptional regulators remain to be identified? 2) What is the function of negative feedback regulators of EGFR signaling, including Argos, Kekkon-1 and Sprouty, which modify the core Br regulatory pathway (Ghiglione et al., 1999; Wasserman and Freeman, 1998)? Do they affect interspecies variations in appendage patterning, as has been proposed? (Boisclair Lachance et al., 2009; Zartman et al., 2009; Zartman et al., 2011) 3) What is the role of developmental time in patterning? For example, it is unknown why patterns of gene expression continue to change quite dramatically even after distinct cell types are established (Peters et al., 2013; Yakoby et al., 2008a).
Morphogenesis
At its heart, tissue morphogenesis is a physical process, and in principal, it should be possible to describe this process simply in terms of temporal and spatial distributions of forces and material properties. Although a small number of studies have introduced techniques for directly measuring these properties in vivo (Campàs, 2016), most work on morphogenesis relies on more indirect techniques. Approaches to studying morphogenesis in a particular tissue begin with documenting and measuring cell- and tissue-level deformations. More detailed analyses include characterizing the localization of key molecular players, such as myosin II as a marker for force generation (Quintin et al., 2008); perturbing morphogenesis through genetic approaches or physical manipulations such as laser ablation (for example, Ducuing and Vincent, 2016; Kiehart et al., 2000); and computational modeling (Hashimoto et al., 2015; Wyczalkowski et al., 2012). As described below, many of these approaches have been applied to studying dorsal appendage morphogenesis.
Once the follicle cells have been patterned into different types, the appendage primordium begins its transformation from a flat patch of cells into a tube. The cell-shape changes mediating this transformation are fairly well characterized on the apical surface (Figure 3). The roof cells apically constrict as the tube first begins to form, buckling the roof patch into a small dome (Dorman et al., 2004). At the same time, the floor-cell domain twists underneath the roof domain, and the outer edges of the floor domain “zip” up to form a two-cell-wide floor of the tube (Osterfield et al., 2013). As the tube extends anteriorly, the roof cells re-expand apically, with the expansion biased in an anterior/posterior direction (Peters and Berg, 2016b), and undergo convergent extension (Ward and Berg, 2005), facilitating tube elongation (Figure 3). By the final stage of oogenesis, the floor of the distal “paddle” is two cells wide, while that of the proximal “stalk” is only one cell wide (Dorman et al., 2004), although the precise cellular changes involved in generating this distinctive shape remain unclear.
Figure 3.
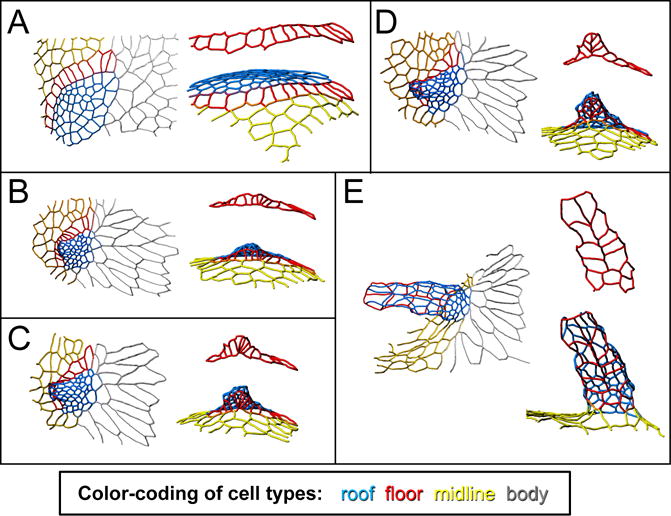
Changes in cell shape and neighbor relationships transform a flat appendage primordium into a dorsal-appendage tube. The tracings in these panels show the apical outlines of a patch of follicle cells, in their transformation from a flat primodium (A) to a fully formed, though not fully elongated, dorsal-appendage tube (E). Color-coding as in Figure 1: blue is roof, red is floor, yellow is midline (operculum), and gray is body. For each time point, the left image shows a dorsal view, while the right image is a view from below the appendage, i.e. a roughly ventral-anterior view. Of particular interest are the floor cells (also shown by themselves in red), which “zip” to form the two-cell wide appendage floor. This figure is adapted from (Osterfield et al., 2015).
What physical forces drive these morphogenetic changes? One proposal is that myosin localization along the apical surface generates patterns of tension that drive tube formation. Apically localized myosin is elevated throughout the roof-cell domain, and two myosin cables outline the floor-cell domain where it contacts midline and roof cells. The hypothesis that such a pattern of tension could drive morphogenesis has support from computational modeling. Specifically, implementing a qualitatively similar pattern of tension in a vertex-based model results in the model tissue buckling and zippering up its floor cells to form a tube (Osterfield et al., 2013). Interestingly, a similar process of myosin-mediated zippering appears to drive neural-tube closure in the chordate Ciona intestinalis (Hashimoto et al., 2015).
Although computational simulations suggest that apical patterns of tension, and particularly the myosin cable along the floor-midline boundary, may be sufficient for driving tube formation in simulations, this hypothesis has not yet been tested directly in vivo. Furthermore, other types of experimental observations suggest alternative mechanisms, including roof-cell constriction and/or convergent extension (Dorman et al., 2004), actin-cable formation along the roof-floor boundary controlled by Echinoid expression (Laplante and Nilson, 2006), and basal pulling. The latter two mechanisms are discussed further below.
Appendage tube elongation appears to be driven by forces at both the apical and basal surfaces. Tube elongation clearly requires the apical surfaces of the appendage cells to de-constrict or relax. This expansion occurs predominantly along the anterior-posterior axis (Peters and Berg, 2016b) and is controlled by the transcription factor Tramtrack69 (Boyle and Berg, 2009; French et al., 2003). At the same time, the basal sides of the appendage cells crawl forward into the space between the stretch cells (the squamous layer of follicle cells that surrounds the nurse-cell cluster) and the extracellular matrix. Several lines of genetic evidence indicate that this basal crawling is likely required for tube elongation. Paxillin, which encodes a focal adhesion protein, is upregulated in appendage primordia and is required for normal tube elongation (Peters et al., 2013). Additionally, disrupting the endocytosis regulator Dynamin impairs tube elongation. Defects in the subcellular localization of integrins in Dynamin mutants, combined with the observation that both knockdown and overexpression of integrins result in shorter dorsal appendages, suggest that Dynamin-mediated integrin turnover on the basal surface is involved in tube elongation (Peters and Berg, 2016b). Open questions include whether basal crawling plays any role in specifying appendage shape, and whether it might even drive appendage formation in the absence of patterned apical tension, as suggested by work in other species (Osterfield et al., 2015). It is worth noting that the elongation of epithelial tubes via cell crawling occurs in other systems, including Drosophila trachea and mouse mammary ducts (Andrew and Ewald, 2010).
So far we have considered dorsal-appendage morphogenesis in terms of physical forces. How does force relate to the patterns of gene expression discussed earlier? The myosin cables bordering the floor and roof domains have been explained in part by the expression pattern of Echinoid (Ed) (Laplante and Nilson, 2006), a homophilic immunoglobulin-family cell-adhesion molecule that is an important component of adherens junctions (Harris and Tepass, 2010). Contact between Ed-expressing and non-expressing cells results in formation of the myosin cables in the follicular epithelium, as well as in other cell types (Laplante and Nilson, 2006; Laplante and Nilson, 2011; Wei et al., 2005). Importantly, loss of Ed results in defective dorsal-appendage formation; however, since ed-mutant primordia can occasionally form tubes, it appears that redundant mechanisms, either for patterning the location of myosin cables or for forming appendages, must exist (Laplante and Nilson, 2006). Interestingly, myosin cables form at the boundaries between cells expressing different complements of a variety of other cell surface molecules, including Crumbs (Röper, 2012), Toll-family receptors (Paré et al., 2014), and E-cadherin (Wei et al., 2005).
Although most work has focused on the initial tube-forming process, some candidate genes have emerged as regulators of the apical relaxation of roof cells during tube extension. These candidates, which include Paxillin and Dynamin, were identified by screening for genes that are mis-expressed in a tramtracktwinpeaks genetic background (Peters et al., 2013). More generally, several dozen other genes are implicated in regulating some aspect of eggshell morphogenesis, based on their expression patterns in the follicular epithelium (Jordan et al., 2005; Yakoby et al., 2008a). We expect that future studies examining these candidates will shed further light on the mechanisms by which gene expression controls tissue shape.
Evolution
Several decades ago, systematic studies by entomologists established that eggshells of different Drosophilid species vary dramatically in the number, shape, and length of the respiratory appendages (Figure 4A) (Hinton, 1981; Patterson and Stone, 1952; Throckmorton, 1962). For example, eggshells from the Sophophora subgenus of Drosophila (represented in Figure 4A by D. melanogaster) generally have two appendages. In contrast, most species in the Drosophila subgenus produce eggshells with four appendages (for instance, see D. funebris and D. virilis in Figure 4A). The Drosophila genus is paraphyletic; that is, the genus excludes some species that share a common ancestor. For example, members of the Zaprionus genus are more closely related to members of the Drosophila subgenus than any of these species are related to D. melanogaster, yet Zaprionus species are classified into a separate genus. Species from genera whose ancestry groups them with members of the Drosophila subgenus also generally produce eggshells with four appendages (for example, Z. sepsoides in Figure 4A). However, from within this group of flies, several species have arisen that generate only two appendages (e.g., D. melanica and Z. davidi) or three respiratory appendages (e.g., D. phalerata). Interestingly, the more distantly related Drosophilid genera, Chymomyza and Scaptodrosophila, produce eggshells with larger numbers of appendages; furthermore, the appendage numbers vary within a species, and even amongst the eggs from a single female. Since key nodes in the phylogeny of Drosophilids remain unresolved (van der Linde et al., 2010), it is not yet clear which of these eggshell morphologies most closely resembles the ancestral type.
Figure 4.
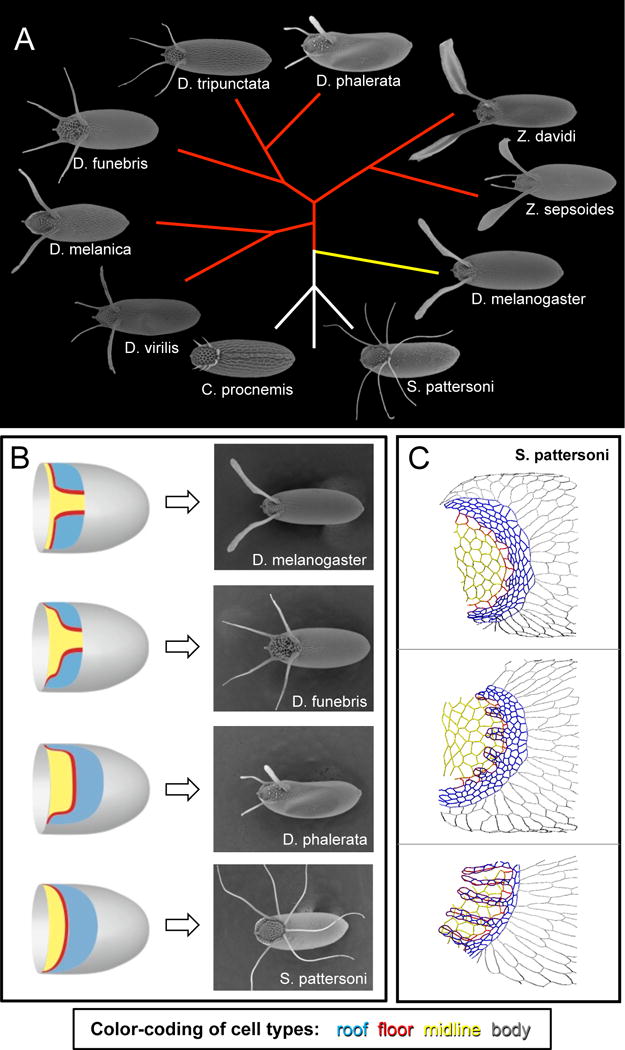
The eggshells of different Drosophilid species provide a rich system for testing models of patterning, morphogenesis, and adaptation. A) Eggshells of species from the genera Drosophila, Scaptodrosophila, Chymomyza, and Zaprionus. The eggshell appendages of various species can differ in both number and shape. This phylogenetic tree is based on (van der Linde et al., 2010) and shows inferred evolutionary relationships among the species, but it does not reflect evolutionary distances. Sizes of eggshells are not to scale. Yellow branch indicates the Sophophora subgenus. Red branches indicate species whose ancestry groups them with members of the Drosophila subgenus. B) The number of appendage primodia in a species (schematized on the left, with same color-coding as Figure 1) does not generally predict the number of eggshell appendages formed (right). See main text for further information. C) The tracings in these panels show apical outlines of S. pattersoni follicle cells; samples are developmentally ordered from top to bottom. In this species, a single appendage primordium is transformed into multiple dorsal appendages, not by the formation of new floor-floor boundaries, but rather by changes in floor-cell shape. Dorsal view. Color-coding as in Figure 1: blue is roof, red is floor, yellow is midline (operculum), and gray is body. Parts of figures B-C are adapted from (Osterfield et al., 2015)
Once the molecular mechanisms underlying appendage formation in D. melanogaster began to be identified, developmental biologists were attracted to the unexplained morphological diversity of Drosophilid eggshells, particularly the question of appendage number. Since two appendages are formed in D. melanogaster due to the earlier patterning of two separate appendage primordia, early proposals assumed that species with different numbers of eggshell appendages would have a corresponding number of appendage primordia. It soon became apparent, however, that this assumption is not true (Figure 4B). Most notably, D. virilis, which has four eggshell appendages, has only two appendage primordia in the egg chamber, as indicated by both early MAPK signaling and by high levels of Br expression (James and Berg, 2003; Nakamura and Matsuno, 2003). Similarly, some species with 3 appendages, such as D. guttifera or D. phalerata, initially have only one primordium (Kagesawa et al., 2008; Niepielko et al., 2011).
If the number of primordia that are patterned doesn’t dictate the number of appendages formed, then what does? In Drosophila species with two appendages, the dorsal-anterior boundary of each primordium is generally convex along its length, while in species with four appendages, this boundary is generally composed of two convex regions separated by a concave region (Figure 4B, top 2 panels). Thus one hypothesis is that the number of these convex regions or “corners” may dictate the number of appendages (Zartman et al., 2011). How exactly the primordium shape might control appendage number is unclear. On one hand, a purely mechanical model could be imagined, in which a two-lobed roof-cell domain might naturally form two buckles as the cells constrict. At the other end of the spectrum, one might imagine a patterning-driven model, in which two distinct regions of leading roof or floor cells could be specified, for example by a peak in the gradient of expression in some gene. Such proposals have not been tested, but provide interesting future directions for both computational modeling and experimental approaches.
At first sight, one might expect appendage formation in the eggshells of different species to use the same morphogenetic processes (i.e. roof-cell constriction and convergent extension, floor-cell zippering, etc.), but in different geometries based on gene patterning. This reasoning is in fact the viewpoint motivating the previous paragraph. It has recently become clear, however, that even the morphogenetic processes used to generate a tube can vary between species. This variation can be seen in vertebrate neurulation, where the “wrapping” mechanism of primary neurulation and the “cavitation” mechanism of secondary neurulation are used to different degrees and implemented differently in different species (Lowery and Sive, 2004).
Likewise, an examination of apical cell shapes in Scaptodrosophila pattersoni, which produces a large and variable number of dorsal appendages from a single appendage primordium (Figure 4B), reveals that this species generates appendages without the floor-floor zippering seen in D. melanogaster (Osterfield et al., 2015). Instead, the floor cells undergo an extreme cell-shape change involving the lengthening of alternate floor-floor edges (Figure 4C). Furthermore, the choice of edges to lengthen appears to involve an unexplained cell-polarity mechanism that localizes Bazooka (Par3) to only one edge per cell. Roof-cell behavior, including both apical constriction and convergent extension, is similar to that seen in other species, but it is not obvious whether these processes would be sufficient to drive formation of multiple, regularly spaced tubes. One proposed source for the physical force needed to drive appendage formation in this species is basal crawling; in other words, the basal pulling thought to drive appendage elongation in D. melanogaster might be used earlier in S. pattersoni for appendage formation (Osterfield et al., 2015).
One particularly interesting study involving cross-species comparisons of eggshells involves not the dorsal appendages, but the dorsal ridge, a dorsal midline structure that is missing from D. melanogaster eggs but is found in a variety of other species (e.g., C. procnemis in Figure 4A). Species with a dorsal ridge exhibit an unusual staining pattern at stage 10 for diphosphorylated ERK (dpERK), a marker for EGFR signaling. In many species, dpERK is confined to the dorsal anterior region, where the midline and appendage cell types will be specified, but in species with a dorsal ridge, dpERK extends more posteriorly, along the dorsal midline (Niepielko and Yakoby, 2014). The dorsal midline is located over the path the nucleus would be expected to traverse as it moves from the posterior of the oocyte, during early oogenesis, to the dorsal anterior, where it resides in mid and late oogenesis. Therefore, one hypothesis is that the dpERK signal on the dorsal midline is due to Grk released from the oocyte as the grk-containing ribonucleoprotein complexes move with the nucleus along this path. Indeed, grk knockdown by RNAi inhibits dorsal ridge formation in D. willistoni, a species that normally produces this structure. Strikingly, expressing the D. willistoni Grk protein (wGRK) in D. melanogaster not only rescues a grk mutant, but it can also cause the formation of a partial dorsal ridge (Niepielko and Yakoby, 2014). These exciting findings raise the question of how differences in Grk between D. willistoni and D. melanogaster control this aspect of morphology; possibilities may include differences in signal strength, or in ligand anchoring or stability (Niepielko and Yakoby, 2014). This observation also raises very interesting questions about the early follicular patterning summarized in Figure 2. Are Midline and H15 patterned differently in response to wGrk? If not, how does wGrk overcome the later inhibition to patterning that these transcription factors normally seem to confer? More generally, what patterning mechanisms can allow for the formation of a dorsal ridge along the dorsal midline without simultaneously preventing the splitting of the appendage forming primordium into two distinct domains? These and other intriguing questions reveal the need for continued investigation of these evolutionary developmental processes.
Computational modeling of pattern formation and morphogenesis
One outstanding feature of this model system is that quantitative models have been developed to study nearly every step of dorsal-appendage formation. These models describe the formation of the Grk and Dpp patterning gradients, their transcriptional interpretation by gene-regulatory networks, and subsequent epithelial morphogenesis. Each of these models has a distinct mathematical structure and addresses a specific set of observations and questions.
Models of the Grk and Dpp gradients are based on reaction-diffusion partial differential equations (PDEs), with the main variables corresponding to the spatial distributions of secreted ligands and their complexes with cell-surface receptors (Goentoro et al., 2006; Lembong et al., 2008). These models can readily explain how the distributions of inductive signals are affected by changes in the levels of ligand production or changes in the expression patterns of surface receptors. In addition to describing the Grk and Dpp gradients in Drosophila melanogaster, these models have facilitated mechanistic interpretation of quantitative changes in the spatial patterns of Grk and Dpp observed in related fly species (Niepielko et al., 2012; Zartman et al., 2011). For instance, within the framework of reaction-diffusion models, a dramatically elongated pattern of Grk observed in D. willistoni can be explained by quantitative changes in the diffusion and endocytic uptake rates of secreted Grk (Niepielko and Yakoby, 2014).
Solutions of these PDE-based models describing the Grk and Dpp signals have in turn been used as inputs to quantitative models of gene expression. In these models, each follicle cell is equipped with the same regulatory network but senses a different Grk or Dpp input depending on the position of the cell. These models implement a switch-like regulation of gene expression, and many of the key parameters in these models correspond to the threshold concentration at which a given transcription factor regulates one of its downstream targets. The first of these models demonstrated that a feedforward loop in the EGFR pathway (Figure 2C) is largely sufficient for Br patterning, and it furthermore integrated the effects of Dpp signaling (Figure 2D) (Lembong et al., 2009). One subsequent version extended this model to a 2D description of patterning and demonstrated that incorporating an early pre-patterning event by Grk (Figure 2A) could account for several additional mutant phenotypes (Zartman et al., 2011). Another extension incorporated additional important molecular features, including the role of two enhancers, in the primary feedforward loop regulating br (Figure 2C) (Cheung et al., 2013).
These computational models of patterning have not only been useful in testing the feasibility of proposed mechanisms of gene regulation, but have also generated ideas that remain to be tested. For example, based on results from the 2D model, the authors proposed that changes in negative feedback regulators such as Sprouty might account for different shapes of appendage primordia in different species (Zartman et al., 2011). Direct tests of this and related predictions could include cross-species analysis of enhancers of br and its regulators. Additionally, a model for the regulation of rhomboid expression suggests a relatively simple gene- regulatory network that could explain key features of the floor-cell domain; namely, that it is one cell wide and is located just anterior to the roof domain in every mutant yet examined (Simakov et al., 2012). This model is speculative, requiring the action of one as-yet-unidentified gene, but still makes testable predictions; for example, it predicts that br should be initially expressed in all appendage cell types before down-regulation in the floor cells.
Morphogenesis of the dorsal appendages has also been analyzed computationally, specifically using vertex models. In these models, cells are represented as polygons, and an energy function is assigned to the model epithelium based on cell geometry. Free parameters correspond to cellular properties (i.e. edge tension), and are assigned in a spatial pattern to reflect the proposed pattern of cellular properties (Fletcher et al., 2014). One form of vertex model, using a two-dimensional network of polygons that is free to move in three dimensions, showed that patterns of tensions within the apical surface of the follicular epithelium may be sufficient to explain the tissue buckling and cell-neighbor rearrangements seen during dorsal-appendage formation (Osterfield et al., 2013). Interestingly, this computational model requires a non-uniform pattern of tension along the floor- midline boundary, with a peak at the site of floor-floor zippering. The floor-midline boundary does exhibit a peak of myosin at this location (Osterfield et al., 2013), and a combination of laser ablation and genetic mosaic techniques have shown that the neighboring population of “leading” roof cells, which form the distal tip of the appendage roof, are specifically required for normal appendage formation, (Boyle et al., 2010). However, experiments directly testing the role of myosin peaks in dorsal- appendage formation, in D. melanogaster or in any other species, have not been reported and remain an important test of this model.
Comparison to other systems
The developmental processes underlying Drosophila eggshell patterning and morphogenesis exhibit many similarities with those found in other tissues. As a result, the techniques developed for and insights derived from this system should be applicable to a host of other systems.
Dorsal-appendage formation (Figure 5A) occurs through a type of tubulogenesis termed “wrapping” (Lubarsky and Krasnow, 2003). During wrapping, part of an epithelial sheet bends and seals itself off from the rest of the sheet, forming a tube parallel to the original epithelial sheet (Figure 5B). Two other highly studied wrapping processes, ventral-furrow formation in Drosophila (Figure 5C) (Sweeton et al., 1991) and primary neurulation in vertebrates (Figure 5D) (Massarwa et al., 2014), have much in common with dorsal-appendage formation. Patterning in both systems begins with morphogen gradients (BMP and Tollpathway in the fly; BMP and Wnt in vertebrates) that are translated into a stereotyped spatial array of different cell types through complex gene-regulatory networks (Betancur et al., 2010; Groves and LaBonne, 2014; Reeves and Stathopoulos, 2009). This patterning specifies the cells that will form the bulk of the tube (blue in Figure 5C,D), as well as a distinct cell type along the seam where the tube separates from its parental sheet (red in Figure 5C,D). Morphogenesis in both systems is largely driven by apically localized myosin, which modulates actin and adhesive junctions and enables closure of the tube (Colas and Schoenwolf, 2001; Dietz et al., 2006; Haigo et al., 2003; Hildebrand and Soriano, 1999; Martin et al., 2010; Nishimura and Takeichi, 2008). Although few to no cell rearrangements occur during ventral furrow formation, convergent extension, driven by canonical Planar Cell Polarity (PCP) proteins, is required for proper neural tube closure (Massarwa et al., 2014). It is currently unknown whether any type of planar polarity is important for dorsal-appendage formation or extension, but it may be informative to compare how these two systems coordinate cell rearrangements with tissue bending.
Figure 5.
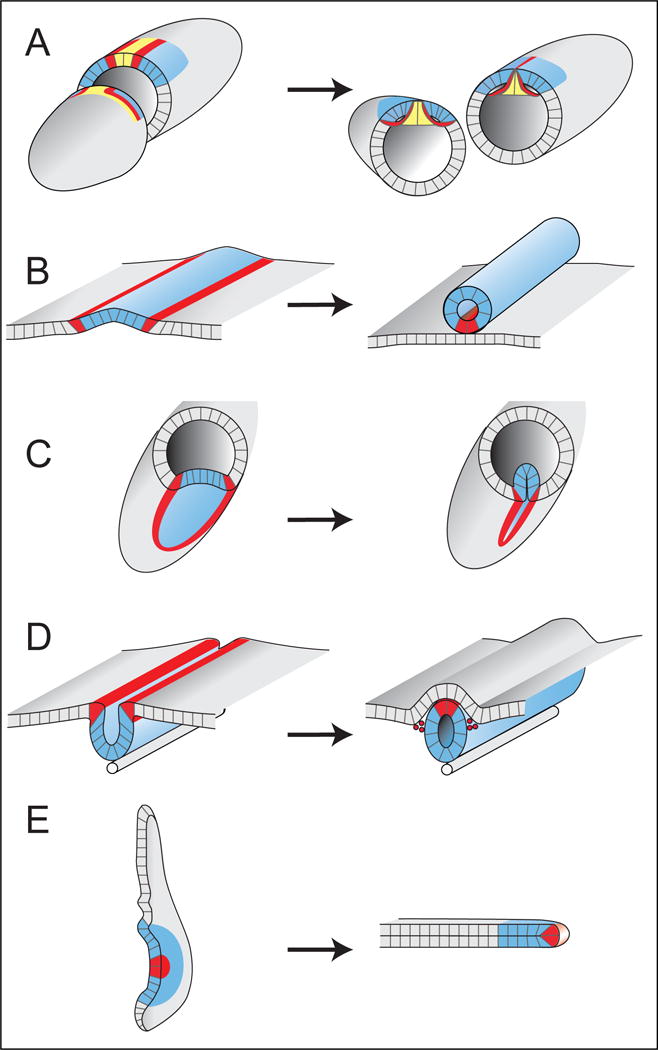
Dorsal-appendage tube formation shares features with other developmental processes. Left panels show early developmental time points; right panels show transitions into tubes. A) Drosophila egg chamber: Left. At S10B, patterning markers define future roof (blue, Broad) and floor (red, rhomboid) cells of the DA tubes. Midline cells (yellow) will form the operculum. Right. At the end of S11, the DA tubes have formed by wrapping the floor cells underneath the roof cells; midline cells constrict basally but still separate the two tubes. B) A generalized scheme for how a wrapping process creates a tube of cells parallel to the original epithelial sheet. The bulk of the tube (blue) is usually formed from a distinct cell type. Often, cells of a different type (red) that border the blue cells will form a seam either within the tube (shown), or within the original sheet. C) Drosophila embryo: Ventral-furrow formation internalizes future mesodermal cells by creating a transient tube. The midline cells (red) remain in the original sheet where they form the CNS. The mesodermal cells (blue) will eventually dissociate and migrate dorsally to create muscle. D) Primary neural-tube formation in vertebrate embryos; neural crest cells (red) seal the tube, then delaminate and migrate to form peripheral nervous system and other structures. E) Drosophila leg imaginal disc: Cutaway shows concentric rings of the leg disc; centrally located cells (red) become the most distal tip of the leg. The main text includes a brief comparison of dorsal-appendage formation to leg disc elongation and other tissue extension processes.
More generally, it may be interesting to compare dorsal-appendage elongation to other tissue-elongation processes. Dorsal-appendage elongation involves a combination of biased apical expansion, filopodia-associated basal pulling, and cell-neighbor exchanges. In contrast, germ-band extension in Drosophila, a major model for elongation, is caused mostly by cell rearrangements, which are in turn driven by planar-polarized myosin along the apical surface (Blankenship et al., 2006). On the other extreme, ascidian notochord elongation involves cell-shape changes and intercalation that are driven by basal crawling (Munro and Odell, 2002). There are also other elongation processes, however, that appear to involve multiple cellular mechanisms. For example, in Drosophila leg-disc elongation (Fig. 5E), myosin is required for the cell-shape changes and rearrangements that help transform the single-layered disc epithelium into an elongated tube (Condic et al., 1991; Edwards and Kiehart, 1996; Fristrom and Fristrom, 1993; Taylor and Adler, 2008), but additional mechanisms, including cell division, tissue spreading, and invasion, may also contribute to shaping the tissue (Fristrom and Chihara, 1978; Pastor-Pareja et al., 2004; Taylor and Adler, 2008).
Conclusion
Significant progress has been made towards understanding the patterning and morphogenesis underlying Drosophila eggshell formation, while opening up new questions. Recent studies highlight how examining differences between species can reveal surprises that may help deepen our understanding of basic developmental mechanisms. Future work in this system should yield further insight not only into developmental processes, but also into the mechanisms underpinning morphological change across evolution.
Table 1.
Gene names and symbols used in the main text.
| Gene | Symbol |
|---|---|
| argos | aos |
| brinker | brk |
| broad | br |
| capicua | cic |
| decapentaplegic (BMP) | dpp |
| echinoid | ed |
| Epidermal growth factor receptor | EGFR |
| gurken (EGF) | grk |
| H15 | H15 |
| kekkon-1 | kek1 |
| midline | mid |
| mirror | mirr |
| Paxillin | Pax |
| pointed | pnt |
| rhomboid | rho |
| rolled (ERK) | rl |
| shibire (Dynamin) | shi |
| sprouty | sty |
| tramtrack | ttk |
Acknowledgments
S.Y.S. and M.O. acknowledge support from the 1R01GM107103 grant from NIGMS. C.A.B. acknowledges support from grant NIH R01 GM079433. We thank Anne Sustar for the drawings in Figure 5.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiyama T, Gibson M. Decapentaplegic and Growth Control in the Developing Drosophila Wing. Nature. 2015;527:375–378. doi: 10.1038/nature15730. [DOI] [PubMed] [Google Scholar]
- Andrew DJ, Ewald AJ. Morphogenesis of epithelial tubes: Insights into tube formation, elongation and elaboration. Dev Biol. 2010;341:34–55. doi: 10.1016/j.ydbio.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astigarraga S, Grossman R, Díaz-Delfín J, Caelles C, Paroush Z, Jiménez G. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J. 2007;26:668–677. doi: 10.1038/sj.emboj.7601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkey MR, Lachance J-FB, Walczak M, Rebello T, Nilson LA. Capicua regulates follicle cell fate in the Drosophila ovary through repression of mirror. Development. 2006;133:2115–2123. doi: 10.1242/dev.02369. [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling Neural Crest Regulatory Circuits into a Gene Regulatory Network. Annu Rev Cell Dev Biol. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship JT, Backovic ST, Sanny JS, Weitz O, Zallen JA. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev Cell. 2006;11:459–470. doi: 10.1016/j.devcel.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Boisclair Lachance J-F, Fregoso Lomas M, Eleiche A, Bouchard Kerr P, Nilson LA. Graded Egfr activity patterns the Drosophila eggshell independently of autocrine feedback. Development. 2009;136:2893–2902. doi: 10.1242/dev.036103. [DOI] [PubMed] [Google Scholar]
- Boyle MJ, Berg CA. Control in time and space: Tramtrack69 cooperates with Notch and Ecdysone to repress ectopic fate and shape changes during Drosophila egg chamber maturation. Development. 2009;136:4187–4197. doi: 10.1242/dev.042770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle MJ, French RL, Cosand KA, Dorman JB, Kiehart DP, Berg CA. Division of labor: Subsets of dorsal-appendage-forming cells control the shape of the entire tube. Dev Biol. 2010;346:68–79. doi: 10.1016/j.ydbio.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Small S. Morphogen rules: design principles of gradient-mediated embryo patterning. Development. 2015;142:3996–4009. doi: 10.1242/dev.129452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campàs O. A toolbox to explore the mechanics of living embryonic tissues. Semin Cell Dev Biol. 2016;55:119–130. doi: 10.1016/j.semcdb.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetera M, Ramirez-San Juan G, Oakes P, Lewellyn L, Fairchild M, Tanetzapf G, Gardel M, Horne-Badovinac S. Epithelial rotation promotes the global alignment of contractile actin bundles during Drosophila egg chamber elongation. Nat Commun. 2014;5:5511. doi: 10.1038/ncomms6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnier E, Fuchs A, Cheung LS, Chayengia M, Shvartsman SY, Pyrowolakis G. BMP-dependent gene repression cascade in Drosophila eggshell patterning. Development. 2015;400:258–265. doi: 10.1016/j.ydbio.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Schüpbach T. The role of brinker in eggshell patterning. Mech Dev. 2006;123:395–406. doi: 10.1016/j.mod.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Cheung LS, Schüpbach T, Shvartsman SY. Pattern formation by receptor tyrosine kinases: Analysis of the Gurken gradient in Drosophila oogenesis. Curr Opin Genet Dev. 2011;21:719–725. doi: 10.1016/j.gde.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung LS, Simakov DSA, Fuchs A, Pyrowolakis G, Shvartsman SY. Dynamic model for the coordination of two enhancers of broad by EGFR signaling. Proc Natl Acad Sci U S A. 2013;110:17939–17944. doi: 10.1073/pnas.1304753110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe A, Poukkula M, Rørth P. Culturing Drosophila egg chambers and imaging border cell migration. Protoc Exch. 2007 doi: 10.1038/nprot.2007.289. [DOI] [Google Scholar]
- Colas JF, Schoenwolf GC. Towards a cellular and molecular understanding of neurulation. Dev Dyn. 2001;221:117–145. doi: 10.1002/dvdy.1144. [DOI] [PubMed] [Google Scholar]
- Condic ML, Fristrom D, Fristrom JW. Apical cell shape changes during Drosophila imaginal leg disc elongation: a novel morphogenetic mechanism. Development. 1991;111:23–33. doi: 10.1242/dev.111.1.23. [DOI] [PubMed] [Google Scholar]
- Cornell RA, Kimelman D. Combinatorial signaling in development. Bioessays. 1994;16:577–581. doi: 10.1002/bies.950160811. [DOI] [PubMed] [Google Scholar]
- Deng WM, Bownes M. Two signalling pathways specify localised expression of the Broad-Complex in Drosophila eggshell patterning and morphogenesis. Development. 1997;124:4639–4647. doi: 10.1242/dev.124.22.4639. [DOI] [PubMed] [Google Scholar]
- Dietz ML, Bernaciak TM, Vendetti F, Kielec JM, Hildebrand JD. Differential actin-dependent localization modulates the evolutionarily conserved activity of Shroom family proteins. J Biol Chem. 2006;281:20542–20554. doi: 10.1074/jbc.M512463200. [DOI] [PubMed] [Google Scholar]
- Dorman JB, James KE, Fraser SE, Kiehart DP, Berg CA. bullwinkle is required for epithelial morphogenesis during Drosophila oogenesis. Dev Biol. 2004;267:320–341. doi: 10.1016/j.ydbio.2003.10.020. [DOI] [PubMed] [Google Scholar]
- Ducuing A, Vincent S. The actin cable is dispensable in directing dorsal closure dynamics but neutralizes mechanical stress to prevent scarring in the Drosophila embryo. Nat Cell Biol. 2016;18:1149–1160. doi: 10.1038/ncb3421. [DOI] [PubMed] [Google Scholar]
- Edwards KA, Kiehart DP. Drosophila nonmuscle myosin II has multiple essential roles in imaginal disc and egg chamber morphogenesis. Development. 1996;122:1499–1511. doi: 10.1242/dev.122.5.1499. [DOI] [PubMed] [Google Scholar]
- Fletcher AG, Osterfield M, Baker RE, Shvartsman SY. Vertex models of epithelial morphogenesis. 2014;106:2291–2304. doi: 10.1016/j.bpj.2013.11.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores G, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- Fregoso Lomas M, Hails F, Lachance JFB, Nilson LA. Response to the dorsal anterior gradient of EGFR signaling in drosophila oogenesis is prepatterned by earlier posterior EGFR activation. Cell Rep. 2013;4:791–802. doi: 10.1016/j.celrep.2013.07.038. [DOI] [PubMed] [Google Scholar]
- Fregoso Lomas M, De Vito S, Boisclair Lachance J, Houde J, Nilson L. Determination of EGFR Signaling Output by Opposing Gradients of BMP and JAK/STAT Activity. Curr Biol. 2016;26:2572–2582. doi: 10.1016/j.cub.2016.07.073. [DOI] [PubMed] [Google Scholar]
- French RL, Cosand KA, Berg CA. The Drosophila female sterile mutation twin peaks is a novel allele of tramtrack and reveals a requirement for Ttk69 in epithelial morphogenesis. Dev Biol. 2003;253:18–35. doi: 10.1006/dbio.2002.0856. [DOI] [PubMed] [Google Scholar]
- Fristrom D, Chihara C. The mechanism of evagination of imaginal discs of Drosophila melanogaster. V. Evagination of disc fragments. Dev Biol. 1978;66:564–570. doi: 10.1016/0012-1606(78)90261-0. [DOI] [PubMed] [Google Scholar]
- Fristrom D, Fristrom JW. The metamorphic development of the adult epidermis. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila Melanogaster. Cold Spring Harbor; New York: 1993. pp. 843–897. [Google Scholar]
- Gates J. Drosophila egg chamber elongation: insights into how tissues and organs are shaped. Fly. 2012;6:213–227. doi: 10.4161/fly.21969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart J. 1998 Warkany Lecture: Signaling Pathways in Development. Teratology. 1999;60:226–239. doi: 10.1002/(SICI)1096-9926(199910)60:4<226::AID-TERA7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Ghiglione C, Carraway KL, Amundadottir LT, Boswell RE, Perrimon N, Duffy JB. The Transmembrane Molecule Kekkon 1 Acts in a Feedback Loop to Negatively Regulate the Activity of the Drosophila EGF Receptor During Oogenesis. Cell. 1999;96:847–856. doi: 10.1016/s0092-8674(00)80594-2. [DOI] [PubMed] [Google Scholar]
- Goentoro LA, Reeves GT, Kowal CP, Martinelli L, Schüpbach T, Shvartsman SY. Quantifying the Gurken Morphogen Gradient in Drosophila Oogenesis. Dev Cell. 2006;11:263–272. doi: 10.1016/j.devcel.2006.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK, LaBonne C. Setting appropriate boundaries: Fate, patterning and competence at the neural plate border. Dev Biol. 2014;389:2–12. doi: 10.1016/j.ydbio.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigo S, Bilder D. Global tissue revolutions in a morphogenetic movement controlling elongation. Science (80−) 2011;331:1071–1074. doi: 10.1126/science.1199424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigo SL, Hildebrand JD, Harland RM, Wallingford JB. Shroom induces apical constriction and is required for hingepoint formation during neural tube closure. Curr Biol. 2003;13:2125–2137. doi: 10.1016/j.cub.2003.11.054. [DOI] [PubMed] [Google Scholar]
- Harris TJ, Tepass U. Adherens junctions: from molecules to morphogenesis. Nat Rev Mol Cell Biol. 2010;11:502–514. doi: 10.1038/nrm2927. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Robin FB, Sherrard KM, Munroe EM. Sequential contraction and exchange of apical junctions drives zippering and neural tube closure in a simple chordate. Dev Cell. 2015;32:241–255. doi: 10.1016/j.devcel.2014.12.017. [DOI] [PubMed] [Google Scholar]
- He L, Wang X, Tang H, Montell D. Tissue elongation requires oscillating contractions of a basal actomyosin network. Nat Cell Biol. 2010;12:1133–1142. doi: 10.1038/ncb2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand JD, Soriano P. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell. 1999;99:485–497. doi: 10.1016/s0092-8674(00)81537-8. [DOI] [PubMed] [Google Scholar]
- Hinton HE. The structure and function of the respiratory horns of the eggs of some flies. Phil Trans R Soc L B. 1960;243:45–73. [Google Scholar]
- Hinton HE. Biology of Insect Eggs. Pergamon Press; 1981. [Google Scholar]
- Horne-Badovinac S. The Drosophila egg chamber-a new spin on how tissues elongate. Integr Comp Biol. 2014;54:667–676. doi: 10.1093/icb/icu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne-Badovinac S, Bilder D. Mass transit: Epithelial morphogenesis in the Drosophila egg chamber. Dev Dyn. 2005;232:559–574. doi: 10.1002/dvdy.20286. [DOI] [PubMed] [Google Scholar]
- Hudson AM, Cooley L. Methods for studying oogenesis. Methods. 2014;68:207–217. doi: 10.1016/j.ymeth.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabella A, Horne-Badovinac S. Dynamic regulation of basement membrane protein levels promotes egg chamber elongation in Drosophila. Dev Biol. 2015;406:212–221. doi: 10.1016/j.ydbio.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James KE, Berg CA. Temporal comparison of Broad-Complex expression during eggshell-appendage patterning and morphogenesis in two Drosophila species with different eggshell-appendage numbers. Gene Expr Patterns. 2003;3:629–634. doi: 10.1016/s1567-133x(03)00136-4. [DOI] [PubMed] [Google Scholar]
- Jordan KC, Hatfield SD, Tworoger M, Ward EJ, Fischer KA, Bowers S, Ruohola-Baker H. Genome wide analysis of transcript levels after perturbation of the EGFR pathway in the Drosophila ovary. Dev Dyn. 2005;232:709–724. doi: 10.1002/dvdy.20318. [DOI] [PubMed] [Google Scholar]
- Kagesawa T, Nakamura Y, Nishikawa M, Akiyama Y, Kajiwara M, Matsuno K. Distinct activation patterns of EGF receptor signaling in the homoplastic evolution of eggshell morphology in genus Drosophila. Mech Dev. 2008;125:1020–1032. doi: 10.1016/j.mod.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Kiehart DP, Galbraith CG, Edwards KA, Rickoll WL, Montague RA. Multiple Forces Contribute to Cell Sheet Morphogenesis for Dorsal Closure in Drosophila. J Cell Biol. 2000;149:471–490. doi: 10.1083/jcb.149.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante C, Nilson LA. Differential expression of the adhesion molecule Echinoid drives epithelial morphogenesis in Drosophila. Development. 2006;133:3255–3264. doi: 10.1242/dev.02492. [DOI] [PubMed] [Google Scholar]
- Laplante C, Nilson LA. Asymmetric distribution of Echinoid defines the epidermal leading edge during Drosophila dorsal closure. J Cell Biol. 2011;192:335–348. doi: 10.1083/jcb.201009022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembong J, Yakoby N, Shvartsman SY. Spatial regulation of BMP signaling by patterned receptor expression. Tissue Eng Part A. 2008;14:1469–1477. doi: 10.1089/ten.tea.2008.0098. [DOI] [PubMed] [Google Scholar]
- Lembong J, Yakoby N, Shvartsman SY. Pattern formation by dynamically interacting network motifs. Proc Natl Acad Sci U S A. 2009;106:3213–3218. doi: 10.1073/pnas.0810728106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner D, McCoy D, Isabella A, Mahowald A, Gerlach G, Chaudry T, Horne-Badovinac S. A Rab10-dependent mechanism for polarized basement membrane secretion during organ morphogenesis. Dev Cell. 2013;24:159–168. doi: 10.1016/j.devcel.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Hackney JF, Bergen A, Dobens L, Truesdale A, Dobens L. Opposing interactions between Drosophila Cut and the C/EBP encoded by Slow Border Cells direct apical constriction and epithelial invagination. Dev Biol. 2010;344:196–209. doi: 10.1016/j.ydbio.2010.04.030. [DOI] [PubMed] [Google Scholar]
- Lowery LA, Sive H. Strategies of vertebrate neurulation and a reevaluation of teleost neural tube formation. Mech Dev. 2004;121:1189–1197. doi: 10.1016/j.mod.2004.04.022. [DOI] [PubMed] [Google Scholar]
- Lubarsky B, Krasnow MA. Tube morphogenesis: Making and shaping biological tubes. Cell. 2003;112:19–28. doi: 10.1016/s0092-8674(02)01283-7. [DOI] [PubMed] [Google Scholar]
- Mahajan-Miklos S, Cooley L. Intercellular cytoplasm transport dring Drosophila oogenesis. Dev Biol. 1994;165:336–351. doi: 10.1006/dbio.1994.1257. [DOI] [PubMed] [Google Scholar]
- Manning L, Starz-Gaiano M. Culturing Drosophila Egg Chambers and Investigating Developmental Processes Through Live Imaging. In: Bratu DP, McNeil GP, editors. Drosophila Oogenesis: Methods and Protocols, Methods in Molecular Biology, vol 1328. New York: Springer Science+Business Media; 2015. pp. 73–88. [DOI] [PubMed] [Google Scholar]
- Martin AC, Gelbart M, Fernandez-Gonzalez R, Kaschube M, Wieschaus EF. Integration of contractile forces during tissue invagination. J Cell Biol. 2010;188:735–749. doi: 10.1083/jcb.200910099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massarwa R, Ray HJ, Niswander L. Morphogenetic movements in the neural plate and neural tube: mouse. WIREs Dev Biol. 2014;3:59–68. doi: 10.1002/wdev.120. [DOI] [PubMed] [Google Scholar]
- Montell DJ, Yoon WH, Starz-Gaiano M. Group choreography: mechanisms orchestrating the collective movement of border cells. Nat Rev Mol Cell Biol. 2012;13:631–645. doi: 10.1038/nrm3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales D, Kania A. Cooperation and Crosstalk in Axon Guidance Cue Integration: Additivity, Synergy, and Fine-Tuning in Combinatorial Signaling. Dev Neurobiol. 2016 doi: 10.1002/dneu.22463. [DOI] [PubMed] [Google Scholar]
- Morimoto AM, Jordan KC, Tietze K, Britton JS, O’Neill EM, Ruohola-Baker H. Pointed, an ETS domain transcription factor, negatively regulates the EGF receptor pathway in Drosophila oogenesis. Development. 1996;122:3745–3754. doi: 10.1242/dev.122.12.3745. [DOI] [PubMed] [Google Scholar]
- Munro EM, Odell GM. Polarized basolateral cell motility underlies invagination and convergent extension of the ascidian notochord. Development. 2002;129:13–24. doi: 10.1242/dev.129.1.13. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Matsuno K. Species-specific activation of EGF receptor signaling underlies evolutionary diversity in the dorsal appendage number of the genus Drosophila eggshells. Mech Dev. 2003;120:897–907. doi: 10.1016/s0925-4773(03)00164-3. [DOI] [PubMed] [Google Scholar]
- Niepielko MG, Yakoby N. Evolutionary changes in TGF distribution underlie morphological diversity in eggshells from Drosophila species. Development. 2014;141:4710–4715. doi: 10.1242/dev.111898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepielko MG, Hernáiz-Hernández Y, Yakoby N. BMP signaling dynamics in the follicle cells of multiple Drosophila species. Dev Biol. 2011;354:151–159. doi: 10.1016/j.ydbio.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Niepielko MG, Ip K, Kanodia JS, Lun DS, Yakoby N. Evolution of BMP signaling in Drosophila oogenesis: A receptor-based mechanism. Biophys J. 2012;102:1722–1730. doi: 10.1016/j.bpj.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Takeichi M. Shroom3-mediated recruitment of Rho kinases to the apical cell junctions regulates epithelial and neuroepithelial planar remodeling. Development. 2008;135:1493–1502. doi: 10.1242/dev.019646. [DOI] [PubMed] [Google Scholar]
- Osterfield M, Du X, Schüpbach T, Wieschaus E, Shvartsman SY. Three-Dimensional Epithelial Morphogenesis in the Developing Drosophila Egg. Dev Cell. 2013;24:400–410. doi: 10.1016/j.devcel.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterfield M, Schüpbach T, Wieschaus E, Shvartsman SY. Diversity of epithelial morphogenesis during eggshell formation in drosophilids. Development. 2015;142:1971–1977. doi: 10.1242/dev.119404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai LM, Barcelo G, Schüpbach T. D-cbl, a negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell. 2000;103:51–61. doi: 10.1016/s0092-8674(00)00104-5. [DOI] [PubMed] [Google Scholar]
- Paré AC, Vichas A, Fincher CT, Mirman Z, Farrell DL, Mainieri A, Zallen JA. A positional Toll receptor code directs convergent extension in Drosophila. Nature. 2014;515:523–527. doi: 10.1038/nature13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Pareja JC, Grawe F, Martín-Blanco E, García-Bellido A. Invasive cell behavior during Drosophila imaginal disc eversion is mediated by the JNK signaling cascade. Dev Cell. 2004;7:387–399. doi: 10.1016/j.devcel.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Patterson JT, Stone WS. Evolution in the genus Drosophila. New York: The Macmillan Company; 1952. [Google Scholar]
- Peri F, Roth S. Combined activities of Gurken and Decapentaplegic specify dorsal chorion structures of the Drosophila egg. Development. 2000;127:841–850. doi: 10.1242/dev.127.4.841. [DOI] [PubMed] [Google Scholar]
- Peters NC, Berg CA. In vitro culturing and live imaging of Drosophila egg chambers: A history and adaptable method. In: Nezis IP, editor. Oogenesis: Methods and Protocols, Methods in Molecular Biology, vol 1457. New York: Springer Science+Business Media; 2016a. pp. 35–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters NC, Berg CA. Dynamin-mediated endocytosis is required for tube closure, cell intercalation, and biased apical expansion during epithelial tubulogenesis in the Drosophila ovary. Dev Biol. 2016b;409:39–54. doi: 10.1016/j.ydbio.2015.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters NC, Thayer NH, Kerr SA, Tompa M, Berg CA. Following the “tracks”: Tramtrack69 regulates epithelial tube expansion in the Drosophila ovary through Paxillin, Dynamin, and the homeobox protein Mirror. Dev Biol. 2013;378:154–169. doi: 10.1016/j.ydbio.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires-daSilva A, Sommer RJ. The evolution of signalling pathways in animal development. Nat Rev Genet. 2003;4:39–49. doi: 10.1038/nrg977. [DOI] [PubMed] [Google Scholar]
- Prasad M, Jang AC-C, Starz-Gaiano M, Melani M, Montell DJ. A protocol for culturing Drosophila melanogaster stage 9 egg chambers for live imaging. 2007;2:2467–2473. doi: 10.1038/nprot.2007.363. [DOI] [PubMed] [Google Scholar]
- Quintin S, Gally C, Labouesse M. Epithelial morphogenesis in embryos: asymmetries, motors and brakes. Trends Genet. 2008;24:221–230. doi: 10.1016/j.tig.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Reeves GT, Stathopoulos A. Graded Dorsal and differential gene regulation in the Drosophila embryo. Cold Spring Harb Perspect Biol. 2009;1:a000836. doi: 10.1101/cshperspect.a000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röper K. Anisotropy of Crumbs and aPKC drives myosin cable assembly during tube formation. Dev Cell. 2012;23:939–953. doi: 10.1016/j.devcel.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T. Germ Line and Soma Cooperate During Oogenesis to Establish the Dorsoventral Pattern of Egg Shell and Embryo in Drosophila Melanogaster. Cell. 1987;49:699–707. doi: 10.1016/0092-8674(87)90546-0. [DOI] [PubMed] [Google Scholar]
- Shravage BV, Altmann G, Technau M, Roth S. The role of Dpp and its inhibitors during eggshell patterning in Drosophila. Development. 2007;134:2261–2271. doi: 10.1242/dev.02856. [DOI] [PubMed] [Google Scholar]
- Simakov DSA, Cheung LS, Pismen LM, Shvartsman SY. EGFR-dependent network interactions that pattern Drosophila eggshell appendages. Development. 2012;139:2814–2820. doi: 10.1242/dev.077669. [DOI] [PubMed] [Google Scholar]
- Spradling AC. Developmental genetics of oogenesis. In: Bate M, Martinez Arias A, editors. The Development of Drosophila Melanogaster. Cold Spring Harbor Laboratory Press; 1993. pp. 1–70. [Google Scholar]
- Swarup S, Verheyen E. Wnt/Wingless Signalig in Drosophila. Cold Spring Harb Perspect Biol. 2012;4:a007930. doi: 10.1101/cshperspect.a007930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeton D, Parks S, Costa M, Wieschaus E. Gastrulation in Drosophila: the formation of the ventral furrow and posterior midgut invaginations. Development. 1991;112:775–789. doi: 10.1242/dev.112.3.775. [DOI] [PubMed] [Google Scholar]
- Taylor J, Adler PN. Cell rearrangement and cell division during the tissue level morphogenesis of evaginating Drosophila imaginal discs. Dev Biol. 2008;313:739–751. doi: 10.1016/j.ydbio.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throckmorton LH. The problem of phylogeny in the genus Drosophila. In: Wheeler MR, editor. Studies in Genetics, vol. II. Austin, TX: University of Texas Publication; 1962. pp. 207–343. [Google Scholar]
- Twombly V, Blackman RK, Jin H, Graff JM, Padgett RW, Gelbart WM. The TGF-beta signaling pathway is essential for Drosophila oogenesis. Development. 1996;122:1555–1565. doi: 10.1242/dev.122.5.1555. [DOI] [PubMed] [Google Scholar]
- Tzolovsky G, Deng W, Schlitt T, Bownes M. The Function of the Broad-Complex During Drosophila melanogaster Oogenesis. Genetics. 1999;153:1371–1383. doi: 10.1093/genetics/153.3.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S, Lee J, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- van der Linde K, Houle D, Spicer GS, Steppan SJ. A supermatrix-based molecular phylogeny of the family Drosophilidae. Genet Res (Camb) 2010;92:25–38. doi: 10.1017/S001667231000008X. [DOI] [PubMed] [Google Scholar]
- Ward EJ, Berg CA. Juxtaposition between two cell types is necessary for dorsal appendage tube formation. Mech Dev. 2005;122:241–255. doi: 10.1016/j.mod.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Ward EJ, Zhou X, Riddiford LM, Berg CA, Ruohola-Baker H. Border of Notch activity establishes a boundary between the two dorsal appendage tube cell types. Dev Biol. 2006;297:461–470. doi: 10.1016/j.ydbio.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Waring GL. Morphogenesis of the Eggshell in Drosophila. Int Rev Cytol. 2000;198:67–108. doi: 10.1016/s0074-7696(00)98003-3. [DOI] [PubMed] [Google Scholar]
- Wasserman JD, Freeman M. An autoregulatory cascade of EGF receptor signaling patterns the Drosophila egg. Cell. 1998;95:355–364. doi: 10.1016/s0092-8674(00)81767-5. [DOI] [PubMed] [Google Scholar]
- Wei SY, Escudero LM, Yu F, Chang LH, Chen LY, Ho YH, Lin CM, Chou CS, Chia W, Modolell J, et al. Echinoid Is a Component of Adherens Junctions That Cooperates with DE-Cadherin to Mediate Cell Adhesion. Dev Cell. 2005;8:493–504. doi: 10.1016/j.devcel.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Wu X, Tanwar PS, Raftery LA. Drosophila follicle cells: morphogenesis in an eggshell. Semin Cell Dev Biol. 2008;19:271–282. doi: 10.1016/j.semcdb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyczalkowski MA, Chen Z, Filas BA, Varner VD, Taber L. Computational Models for Mechanics of Morphogenesis. Birth Defects Res C Embryo Today. 2012;96:132–152. doi: 10.1002/bdrc.21013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakoby N, Bristow CA, Gong D, Schafer X, Lembong J, Zartman JJ, Halfon MS, Schüpbach T, Shvartsman SY. A Combinatorial Code for Pattern Formation in Drosophila Oogenesis. Dev Cell. 2008a;15:725–737. doi: 10.1016/j.devcel.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakoby N, Lembong J, Schüpbach T, Shvartsman SY. Drosophila eggshell is patterned by sequential action of feedforward and feedback loops. Development. 2008b;135:343–351. doi: 10.1242/dev.008920. [DOI] [PubMed] [Google Scholar]
- Zartman JJ, Shvartsman SY. Unit Operations of Tissue Development: Epithelial Folding. Annu Rev Chem Biomol Eng. 2010;1:231–246. doi: 10.1146/annurev-chembioeng-073009-100919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zartman JJ, Kanodia JS, Cheung LS, Shvartsman SY. Feedback control of the EGFR signaling gradient: superposition of domain-splitting events in Drosophila oogenesis. Development. 2009;136:2903–2911. doi: 10.1242/dev.039545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zartman JJ, Cheung LS, Niepielko MG, Bonini C, Haley B, Yakoby N, Shvartsman SY. Pattern formation by a moving morphogen source. Phys Biol. 2011;8:045003. doi: 10.1088/1478-3975/8/4/045003. [DOI] [PMC free article] [PubMed] [Google Scholar]


