Abstract
AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) receptors mediate fast excitatory synaptic transmission in brain and underlie aspects of synaptic plasticity. Numerous AMPA receptor-binding proteins have been implicated in AMPA receptor trafficking and anchoring. However, the relative contributions of these proteins to the composition of native AMPA receptor complexes in brain remain uncertain. Here, we use blue native gel electrophoresis to analyze the composition of native AMPA receptor complexes in cerebellar extracts. We identify two receptor populations: a functional form that contains the transmembrane AMPA receptor-regulatory protein stargazin and an apo-form that lacks stargazin. Limited proteolysis confirms assembly of stargazin with a large proportion of native AMPA receptors. In contrast, other AMPA receptor-interacting proteins, such as synapse-associated protein 97, glutamate receptor-interacting protein 1, protein kinase Cα binding protein, N-ethylmaleimide-sensitive fusion protein, AP2, and protein 4.1N, do not show significant association with AMPA receptor complexes on native gels. These data identify stargazin as an auxiliary subunit for a neurotransmitter-gated ion channel.
Keywords: synapse, plasticity, glutamate, assembly, trafficking
AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) receptors are glutamate-gated ion channels that mediate fast excitatory synaptic transmission in the mammalian central nervous system. AMPA receptors are tetrameric proteins composed of subunits GluR1-4 (1, 2). These receptors cluster at the postsynaptic density (PSD), which poises them to respond to synaptically released glutamate (3). Interestingly, AMPA receptors rapidly cycle into and out of the synaptic membrane (4). These changes in synaptic AMPA receptor density provide a key mechanism for activity-dependent modulation of synaptic strength, a phenomenon thought to underlie aspects of learning and memory (5). Understanding the mechanisms that traffic AMPA receptors to the plasma membrane and the synapse is, therefore, of fundamental importance.
Trafficking and synaptic anchoring of AMPA receptors are regulated by numerous binding partners (3). Many of these partners are cytosolic PDZ domain-containing proteins that directly interact with PDZ binding sites in the cytoplasmic tails of AMPA receptor subunits. For example, glutamate receptor-interacting proteins (GRIP1/ABP) and protein kinase Cα binding protein (PICK1) bind to GluR2 and GluR3 (6-8), and synapse-associated protein 97 (SAP97) interacts with GluR1 (9). AMPA receptor tails also engage in non-PDZ domain interactions, as exemplified by the binding of GluR1 and GluR4 to the cytoskeletal protein 4.1N (10, 11) and the associations of GluR2 and GluR3 with N-ethylmaleimide-sensitive fusion (NSF) protein and the clathrin adaptor AP2 (12-15).
Genetic studies identified stargazin as the first transmembrane protein to interact with AMPA receptors (16). Stargazin is the tetraspanin absent in stargazer mice (17), which suffer from epilepsy, abnormal head movements, and ataxia. Stargazer cerebellar granule neurons contain intracellular AMPA receptor protein but lack AMPA receptors on the cell surface, establishing a requirement for stargazin in AMPA receptor trafficking (16, 18). Stargazin directly binds all four AMPA receptor subunits through both intra- and extracellular domains (16, 19). Stargazin is one of four homologous transmembrane AMPA receptor regulatory proteins (TARPs), which have differential, partially overlapping distributions in brain (20), and this can explain why the AMPA receptor defect in stargazer mice appears restricted to cerebellar granule cells.
Although there are numerous AMPA receptor-binding proteins, their potential cooperation and relative contributions to native AMPA receptor complexes remain uncertain. Whether these proteins bind receptors simultaneously or sequentially or interact with mutually exclusive receptor populations is not clear. Also, some interactions may be transient and others more stable. Here, we use blue native (BN)-PAGE and limited proteolysis to characterize native AMPA receptor complexes from cerebellum. We find that stargazin assembles quantitatively with functional AMPA receptors. In contrast, other reported AMPA receptor-interacting proteins do not show detectable association with AMPA receptors. These findings identify stargazin as an auxiliary subunit of a neurotransmitter-gated ion channel.
Methods
Antibodies. Rabbit anti-GluR2/3 and anti-GluR1, mouse anti-GluR2, and guinea pig anti-VGLUT1 were from Chemicon; mouse anti-GRIP, anti-β-adaptin (recognizing AP2), and anti-4.1N were from BD Transduction Laboratories; rabbit anti-GRIP1 was from Upstate Biotechnology; mouse anti-SAP97 was from Stressgen; and rabbit anti-PICK1 and anti-NSF were from Santa Cruz Biotechnology. Mouse monoclonal anti-NSF and rabbit polyclonal anti-GRIP were kind gifts from S. Whiteheart (University of Kentucky, Lexington) and R. Huganir (Johns Hopkins University, Baltimore), respectively. Rabbit anti-stargazin and sheep anti-PSD-95 have been described (20, 21).
BN-PAGE. For BN-PAGE of crude lysate, cerebellum from 4- to 8-week-old mice was homogenized in 20 volumes of 20 mM Tris·HCl, pH 7.4. After centrifugation at 20,000 × g for 2 min, the pellet was solubilized in 14 volumes of 1% Triton X-100/20 mM Tris·HCl (pH 7.4) or 0.5% n-dodecyl maltoside/20% glycerol/25 mM Bis-Tris·HCl, pH 7.0 for 30 min at 4°C. These low ionic strength buffers were used to maximally preserve AMPA receptor interactions (22) and because salts can lead to precipitation of Coomassie dye and stained proteins on BN gels (23). After centrifugation at 20,000 × g for 5 min, 120 μg of solubilized protein was mixed in a 10:1 vol/vol ratio with loading buffer (5% wt/vol Coomassie Blue G 250/200 mM Bis-Tris·HCl/1 M 6-aminocaproic acid/30% wt/vol sucrose, pH 7.0) and loaded on polyacrylamide gels in parallel with native markers (Amersham Pharmacia). Gradient (4-12%) gels were prepared with a gradient maker and a peristaltic pump (Minipuls 3, Gilson) and overlaid with a 3% stacking gel. Gel, cathode, and anode buffers were as described (23). BN-PAGE was performed at 4°C for 4-5 h. Cathode buffer was replaced by buffer without Coomassie 1.5 h before the end of PAGE. After PAGE, gels were washed for 10 min at room temperature with transfer buffer (20% methanol/0.1% wt/vol SDS/50 mM TrisBase/385 mM glycine) and blotted onto Immobilon membranes (Millipore). Blots were destained for 1 h in 10% acetic acid/30% methanol/H2O and incubated with blocking solution and antibodies. Immunoreaction was visualized with ECL (Pierce). Scanned signals were analyzed by using National Institutes of Health software. Before reprobing with antibody from the same species, the blot was stripped for 15 min at 55°C in 62 mM Tris, pH 6.7/120 mM 2-mercaptoethanol/2% SDS, washed five times for 5 min at room temperature in 10 mM TrisBase/100 mM NaCl/0.1% Tween, pH 7.5, and incubated in blocking solution.
To obtain better resolution of protein bands, cerebellar homogenates were centrifuged at 200 × g for 5 min at 4°C, followed by an additional 5 min spin at 350 × g. After centrifugation of the supernatants at 20,000 × g for 2 min, pellets were solubilized for 30 min in 10 volumes of 1% Triton X-100/20 mM Tris, pH 7.4. After centrifugation at 20,000 × g for 5 min, 0.5-0.7 mg of protein extract was layered onto 10-50% glycerol gradients (24). Gradients were centrifuged in a SW-55 rotor (Beckman) at 4°C for 18 h at 165,000 × gav (maximum acceleration and deceleration). Twelve fractions of 450 μl were collected manually from the top, and 60 μl of each fraction was loaded on BN gels.
To determine presence of AMPA receptors and binding proteins in the same complex, 40-μl aliquots taken from the 450-μl glycerol gradient fractions were mixed with 30 μl of 0.1% Triton X-100/20 mM Tris·HCl (pH 7.4) containing 4 μg of rabbit anti-stargazin, anti-GRIP1, anti-PICK1, and anti-NSF or IgG (Chemicon). After incubation at 4°C for 90 min, 60 μl of sample was loaded on BN gels.
Limited Proteolysis. Cerebella were homogenized and incubated in 1% Triton X-100 as described above. After centrifugation at 20,000 × g for 5 min at 4°C, the pellet was resuspended in 20 mM Tris·HCl (pH 7.4). Samples containing 25-30 μg of protein were incubated with trypsin at room temperature. After 10 min, samples were mixed with a 100-fold excess of trypsin inhibitor and put on ice. Protein was then denatured by mixing with SDS sample buffer and urea (final concentration, 1.6 M), followed by heating to 67°C for 10 min, SDS/PAGE, and Western blotting.
Genotyping. Stargazer mice were bred from heterozygous parents (The Jackson Laboratory). For genotyping of tail samples, we used PCR primers γ2F (5′-CATTTCCTGTCTCATCCTTT-3′), γ2R (5′-ACTGTCACTCTATCTGGAATC-3′), and γ2KO (5′-GAGCAAGCAGGTTTCAGGC-3′). PCRs with γ2F and γ2R and with γ2KO and γ2R were performed on each sample to amplify the wild-type stargazin and knockout alleles, respectively.
Statistics. Statistical significance of differences between more than two groups was analyzed with one-way ANOVA and the Student-Newman-Keuls test. Differences were considered significant at P < 0.05. Values represent mean ± SEM.
Results
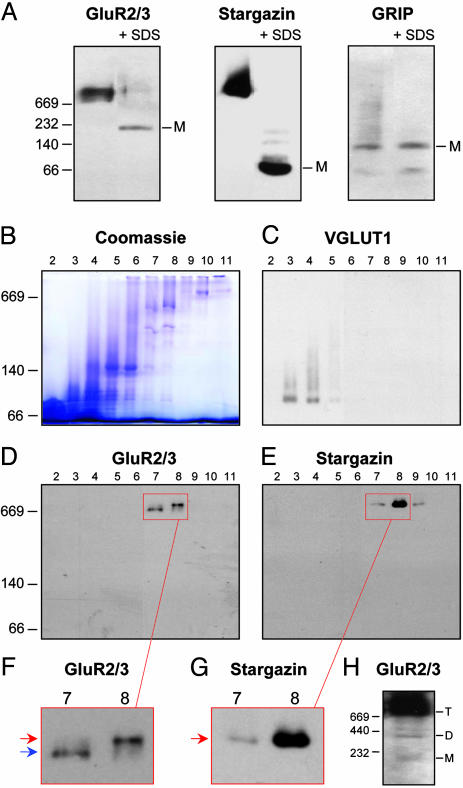
Two Major Populations of Native AMPA Receptors. BN-PAGE permits analysis of multimeric protein complexes (23) and has been used to monitor oligomerization of AMPA receptors in cultured neurons (24). We used this method on crude cerebellar lysates solubilized with 1% Triton X-100 (Fig. 1). Solubilization with 0.5% n-dodecyl maltoside produced similar results (data not shown). After electrophoresis, we performed immunoblotting to compare the migration of AMPA receptors and known interacting proteins. Native AMPA receptors migrated as a broad band of high apparent molecular mass (Fig. 1 A). Parallel BN-PAGE and SDS/PAGE of Triton X-100 extracts followed by immunoblotting onto the same membrane resulted in similar amounts of AMPA receptor immunoreactivity (data not shown), indicating that solubilized AMPA receptor complexes fully entered the BN gels. Importantly, stargazin comigrated with AMPA receptors on BN gels; by contrast, GRIP did not (Fig. 1 A). Addition of SDS before BN-PAGE disassembles protein complexes (24, 25), and we found this yielded monomeric GluR2/3, stargazin, and GRIP species with apparent molecular masses of ≈200, 60, and 130 kDa, respectively (Fig. 1 A). The apparent molecular masses of monomeric GluR2/3 and stargazin were larger than the predicted values (≈105 and 36 kDa, respectively). Similar overestimations of the molecular masses of transmembrane proteins have been observed in previous BN-PAGE studies (24-26). Remarkably, monomeric stargazin was undetectable in cerebellar extracts (Fig. 1 A).
Fig. 1.
Two populations of native AMPA receptor complexes. (A) Crude cerebellar lysate was solubilized in 1% Triton X-100 and analyzed by BN-PAGE and Western blotting with antibodies against GluR2/3, stargazin, and GRIP. To identify monomeric (M) protein species, some samples were treated with 1.5% SDS for 10 min before loading (+ SDS). (B-H) Cerebellar extracts were separated on glycerol gradients, and 12 fractions (numbered from top to bottom) were analyzed by BN-PAGE, followed by Coomassie blue staining (B) or Western blotting (C-H). GluR2/3 (D) and stargazin (E) are concentrated in fractions 7-9, in contrast to the vesicular glutamate transporter VGLUT1 (C). (F and G) Higher magnification of the boxed areas in D and E. GluR2/3 (F) migrates as two distinct bands (red and blue arrows), and stargazin (G) comigrates exclusively with the upper band (red arrow). (H) Prolonged film exposure of fraction 8 reveals two minor GluR2/3 bands, most likely representing AMPA receptor monomers (M) and dimers (D), in addition to the major, presumably tetrameric (T) AMPA receptor complexes. Numbers to the left of A, B, D, and H indicate native molecular masses of the markers thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), lactate dehydrogenase (140 kDa), and BSA (66 kDa).
To obtain better resolution of the broad AMPA receptor band, we fractionated solubilized cerebellar extracts on 10-50% glycerol gradients (24) and analyzed the fractions by BN-PAGE (Fig. 1 B-H). AMPA receptors (Fig. 1D) and stargazin (Fig. 1E) were both concentrated in gradient fractions 7-9, in contrast to the vesicular glutamate transporter VGLUT1 (Fig. 1C), a synaptic transmembrane protein of 58 kDa (27). Intriguingly, GluR2/3 migrated as two discrete bands (Fig. 1 D and F), with the upper band comprising 52.0 ± 6.6% (n = 7) of total GluR2/3. Stargazin comigrated exclusively with the upper band (Fig. 1 E and G). Similarly, GluR2/3 from cerebral cortex migrated as two distinct bands, and only the upper band colocalized with stargazin (data not shown). After prolonged film exposure, two additional minor GluR2/3 bands could be observed, probably representing small amounts of AMPA receptor monomers and dimers (24) (Fig. 1H). There was no detectable colocalization of stargazin with these monomeric and dimeric bands.
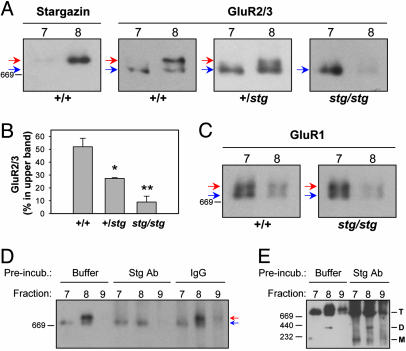
AMPA Receptors and Stargazin Are Present in the Same Complex. To determine whether the upper GluR2/3 band represented a complex of AMPA receptors with stargazin, we analyzed cerebellar extracts from stargazer mice (Fig. 2 A and B). Strikingly, the upper GluR2/3 band was absent in stargazer (Fig. 2 A). This finding has two important implications. First, the upper GluR2/3 band likely consists of AMPA receptor/stargazin complexes. Second, because stargazer granule cells lack AMPA receptors on the cell surface (16, 18), the lower and upper GluR2/3 bands reflect intracellular and functional receptor pools, respectively. There was no detectable increase in mono- or dimeric AMPA receptor species in stargazer cerebellum (data not shown), suggesting that stargazin is not required for AMPA receptor subunit oligomerization. Interestingly, heterozygous mice showed an intermediate upper to lower band ratio for GluR2/3 (Fig. 2 A and B), indicating that stargazin levels are limiting in formation of the AMPA receptor/stargazin complex. Trace amounts of residual upper band GluR2/3 in stargazer extracts (Fig. 2B) were probably derived from cell types such as Purkinje neurons, which express TARPs in addition to stargazin (20).
Fig. 2.
Demonstration of AMPA receptor/stargazin complexes. (A-C) Glycerol gradient fractions 7 and 8 of cerebellar extracts from wild-type (+/+), heterozygous (+/stg), and stargazer (stg/stg) mice were analyzed by BN-PAGE and Western blotting. (A) GluR2/3 from wild type migrates as two discrete bands (red and blue arrows). In contrast, only the lower GluR2/3 band (blue arrow) is found in stargazer extract. Heterozygotes show an intermediate amount of the upper band. (B) Quantification of the GluR2/3 upper band, expressed as percentage of total GluR2/3, in wild-type (n = 7), heterozygous (n = 3), and stargazer (n = 3) extracts. Asterisks and double asterisks denote significant difference (P < 0.05 and P < 0.01, respectively) from wild-type. (C) GluR1 migrates as two discrete bands (red and blue arrows) in cerebellar extracts from both wild type and stargazer. (D) Gradient fractions 7-9 from wild type were incubated with 4 μg of rabbit anti-stargazin (Stg Ab), control IgG, or buffer alone before BN-PAGE and Western blotting with mouse anti-GluR2. Preincubation with anti-stargazin completely and selectively eliminates the upper GluR2 band (red arrow). (E) Longer film exposure of the blot in D shows that preincubation with anti-stargazin has no effect on the migration of AMPA receptor monomers (M) and dimers (D). T, tetrameric AMPA receptors.
As a control we analyzed the migration of GluR1, which is not expressed in granule cells in adult cerebellum (28). Instead, most GluR1 in cerebellar extracts comes from Bergmann glia, which express the stargazin homologue γ-4 (20). GluR1 from wild-type migrated as two distinct bands (Fig. 2C) that colocalized with GluR2/3. However, there was no difference in GluR1 migration between wild-type and stargazer (Fig. 2C), consistent with the granule cell-specific AMPA receptor defect in stargazer (18, 20).
To determine directly whether stargazin and AMPA receptors occur in the same complex, we incubated glycerol gradient fractions of cerebellar extracts with anti-stargazin antibody before BN-PAGE (Fig. 2D). Antibody binding should change the migration of stargazin-containing protein complexes, or crosslink the complexes and prevent them from entering the gel. Importantly, preincubation with anti-stargazin completely removed the upper GluR2 band, but did not affect the lower band (Fig. 2D). Longer film exposures revealed no effect of preincubation with anti-stargazin on AMPA receptor monomers or dimers (Fig. 2E), suggesting that stargazin exclusively associates with AMPA receptor tetramers.
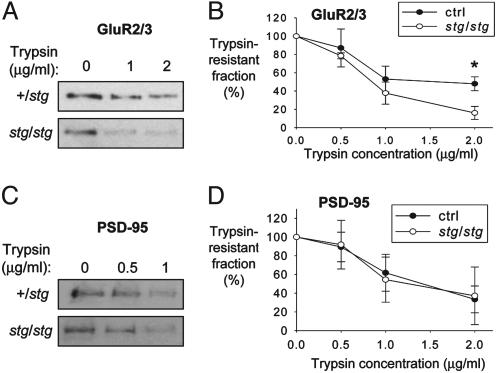
Incompletely assembled multisubunit protein complexes often show increased susceptibility to limited proteolysis (29). Interestingly, GluR2/3 was more efficiently digested by trypsin in extracts from stargazer than from control littermates (Fig. 3 A and B), in contrast with the postsynaptic protein PSD-95 (Fig. 3 C and D). This finding is consistent with the notion that stargazin stably assembles with a large proportion of cerebellar AMPA receptors and reduces trypsin access.
Fig. 3.
Stargazin makes AMPA receptors more resistant to protease. (A and C) Cerebellar proteins from stargazer (stg/stg) and heterozygous (+/stg) littermates were digested with trypsin for 10 min. Undigested proteins were detected by SDS/PAGE and Western blotting with anti-GluR2/3 (A) or anti-PSD-95 (C). (B and D) Quantification of trypsin-resistant GluR2/3 (B) and PSD-95 (D) in stargazer and control (ctrl) cerebella (n = 3 in B; n = 4 in D). Asterisk in B indicates significant difference from control.
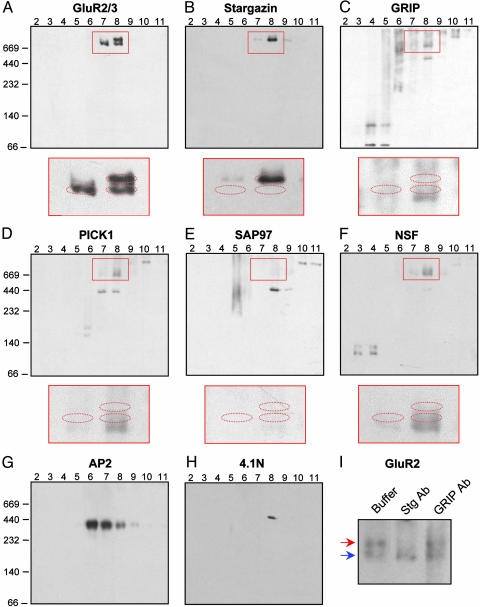
Stargazin Is Unique Among AMPA Receptor-Binding Proteins. To evaluate other AMPA receptor-binding proteins, we performed BN-PAGE and immunoblotting with antibodies against GRIP, PICK1, SAP97, NSF, AP2, and 4.1N (Fig. 4 C-H). These proteins migrated in characteristic, reproducible band patterns that clearly differed from those of AMPA receptors (Fig. 4A) or stargazin (Fig. 4B). SAP97 (Fig. 4E), AP2 (Fig. 4G), and 4.1N (Fig. 4H) did not show detectable immunoreactivity in the vicinity of the AMPA receptor bands. For GRIP (Fig. 4C), PICK1 (Fig. 4D), and NSF (Fig. 4F), there was faint immunoreactivity partially overlapping with some of the GluR2/3 bands. To test whether this minimal overlap reflected association with AMPA receptors, we incubated samples with antibodies against GRIP1, PICK1, and NSF before BN-PAGE. However, these antibody preincubations did not affect GluR2 migration (Fig. 4I and data not shown). This finding indicated that only stargazin assembles with a significant proportion of AMPA receptors in these extracts.
Fig. 4.
Among AMPA receptor-interacting proteins, only stargazin associates quantitatively with the receptor. (A-H) Cerebellar extracts separated by glycerol gradients were analyzed by BN-PAGE and Western blotting for the protein indicated. Magnifications of the red boxed areas are shown below A-F. The red dashed ellipses indicate the positions of upper and lower GluR2/3 bands, as determined by reprobing each blot with anti-GluR2/3. (I) Glycerol gradient fraction 8 was incubated with 4 μg of rabbit anti-GRIP1 (GRIP Ab), rabbit anti-stargazin (Stg Ab), or buffer alone, and analyzed by BN-PAGE and Western blotting with mouse anti-GluR2. Preincubation with anti-GRIP did not affect AMPA receptor complexes (red and blue arrows).
Discussion
The present study provides a comprehensive analysis of the molecular composition of AMPA receptor complexes from cerebellum. We find that stargazin is a prominent component of these complexes, whereas associations with other AMPA receptor-interacting proteins are not detected. These data identify stargazin as an auxiliary AMPA receptor subunit, and suggest functions for stargazin that are distinct from the roles of other AMPA receptor-binding partners.
Stargazin Is the First Auxiliary Subunit of a Neurotransmitter-Gated Ion Channel. Stargazin interacts directly with all four AMPA receptor subunits and drastically enhances the efficiency of AMPA receptor surface trafficking (16, 19). The present data reveal that stargazin forms a stable complex with AMPA receptors in cerebellum, and no free stargazin is detectable. Based on these properties, we designate stargazin an auxiliary AMPA receptor subunit. Most voltage-gated ion channels comprise assemblies of principal subunits, which form the channel pore, together with auxiliary subunits (30-32). Like stargazin, the auxiliary subunits directly bind to the principal subunits, are stable components of the functional channel complex, and typically enhance channel trafficking to the surface. Stargazin is an example of an auxiliary subunit of a neurotransmitter-gated ion channel. Our findings suggest that as yet unidentified auxiliary subunits may control trafficking of other ionotropic neurotransmitter receptors. Recent elegant work has shown that the transmembrane protein SOL-1 may function as an auxiliary subunit for the glutamate-gated channel GLR-1 in Caenorhabditis elegans (33), although it remains to be established whether SOL-1 directly binds to GLR-1.
How many stargazin molecules associate with each tetramer of principal AMPA receptor subunits is not yet clear. Molecular weight determinations from the upper portions of BN gradient gels are not accurate enough to determine the stoichiometry of this complex. Our data suggest that stargazin associates exclusively with AMPA receptor tetramers and not with monomers or dimers. This could mean that the AMPA receptor dimer-dimer interface forms the binding site for stargazin and could fit with a stoichiometry of two stargazin molecules per AMPA receptor tetramer.
A previous study showed that prolonged exposure of cortical neuronal cultures to AMPA receptor agonists can dissociate TARPs from AMPA receptors (19). In the present study, we did not detect monomeric stargazin in cerebellar extracts, suggesting that similar dissociation does not occur during basal cerebellar transmission in vivo.
Our BN-PAGE analysis did not detect AMPA receptor interactions with SAP97, PICK1, GRIP, NSF, AP2, or protein 4.1N, even though the AMPA receptor-binding properties of these proteins have been thoroughly established by a number of laboratories (6-15). Based on the previous studies, these AMPA receptor interactions are not disrupted by 1% Triton X-100. A more plausible explanation for our failure to detect these associations is that they may be transient or involve only a small percentage of the receptors. For example, NSF, a protein involved in vesicle fusion events, and AP2, a component of the endocytic machinery, could be evanescent AMPA receptor partners. Another explanation could be that some of these interactions may be restricted to receptor subpopulations that were underrepresented in our extracts. Although Triton X-100 effectively solubilizes synaptic AMPA receptors compared with NMDA receptors (9), a proportion of synaptic AMPA receptors are poorly extracted in this detergent (34). AMPA receptors from the most detergent-resistant portions of the PSD may also be the most tightly anchored to proteins like GRIP or the cytoskeletal protein 4.1N. Thus, associations of the core AMPA receptor/stargazin complex with other binding partners may be transient or highly localized.
Functional Implications of AMPA Receptor Assembly with an Auxiliary Subunit. How could assembly with an auxiliary subunit enhance AMPA receptor expression on the cell surface? AMPA receptors from stargazer cerebellum show an immature, endoplasmic reticulum (ER)-type glycosylation pattern (20), indicating that stargazin facilitates AMPA receptor export from the ER. A stringent quality control system in the ER prevents incompletely folded or assembled proteins from exiting to the Golgi (35). Because there was no increase in AMPA receptor monomers or dimers in stargazer extracts, it seems unlikely that stargazin mediates oligomerization of GluR subunits. Instead, tetrameric AMPA receptors unassembled with stargazin (“apo-AMPA receptors”) may expose ER retention signals that are masked by stargazin. This would be analogous to the regulated trafficking of ATP-sensitive K+ channel complexes (36). Alternatively, stargazin may promote concentrative sorting of AMPA receptors into ER export vesicles by coupling them to COPII subunits (37).
What could be the advantage of AMPA receptor assembly with TARPs? The catalytic subunits of many enzymes are controlled by regulatory subunits, which bind allosteric effectors or are reversibly phosphorylated. Analogously, assembly of AMPA receptors with a modifiable regulatory subunit may permit more subtle and flexible receptor regulation. Interestingly, stargazin can interact with synaptic PDZ proteins, and this interaction is regulated by stargazin phosphorylation (38), raising the possibility that TARPs could participate in rapid activity-dependent trafficking of AMPA receptors in synaptic plasticity. Intriguingly, invertebrates lack close homologs of TARPs, which may have evolved to mediate the complex regulation of AMPA receptor trafficking and synaptic plasticity that occurs in higher organisms.
The absence of monomeric stargazin in cerebellar extracts, the presence of a large pool of apo-AMPA receptors, and the intermediate amount of fully assembled AMPA receptors in stargazer heterozygotes all indicate that stargazin levels limit the formation of AMPA receptor/stargazin complexes. We previously showed that stargazin overexpression in wild-type hippocampal neurons increased the total surface levels of AMPA receptors (39), suggesting that TARP levels are limiting for functional AMPA receptor expression in hippocampus as well. Whether TARP levels are dynamically regulated by neuronal activity remains uncertain, and represents an exciting avenue of future exploration.
Acknowledgments
We thank Drs. S. Whiteheart and R. Huganir for antibodies and Dr. R. Edwards for reading the manuscript. W.V. is supported by a postdoctoral fellowship of the Fund for Scientific Research, Flanders; D.S.B. is supported by the National Institutes of Health (NIH), the Christopher Reeve Paralysis Foundation, the Human Frontier Science Program, and the American Heart Association; and R.A.N. is supported by the NIH. R.A.N. is a member of the Keck Center for Integrative Neuroscience and the Silvio Conte Center for Neuroscience Research.
Author contributions: R.A.N. and D.S.B. designed research; W.V. performed research; and W.V. wrote the paper.
Abbreviations: AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid; PSD, postsynaptic density; GRIP, glutamate receptor-interacting protein; SAP, synapse-associated protein; NSF, N-ethylmaleimide-sensitive fusion; TARP, transmembrane AMPA receptor regulatory protein; BN, blue native; PICK, protein kinase Cα binding protein.
References
- 1.Hollmann, M. & Heinemann, S. (1994) Annu. Rev. Neurosci. 17, 31-108. [DOI] [PubMed] [Google Scholar]
- 2.Rosenmund, C., Stern-Bach, Y. & Stevens, C. F. (1998) Science 280, 1596-1599. [DOI] [PubMed] [Google Scholar]
- 3.Scannevin, R. H. & Huganir, R. L. (2000) Nat. Rev. Neurosci. 1, 133-141. [DOI] [PubMed] [Google Scholar]
- 4.Lüscher, C., Xia, H., Beattie, E. C., Carroll, R. C., von Zastrow, M., Malenka, R. C. & Nicoll, R. A. (1999) Neuron 24, 649-658. [DOI] [PubMed] [Google Scholar]
- 5.Malinow, R. & Malenka, R. C. (2002) Annu. Rev. Neurosci. 25, 103-126. [DOI] [PubMed] [Google Scholar]
- 6.Dong, H., O'Brien, R. J., Fung, E. T., Lanahan, A. A., Worley, P. F. & Huganir, R. L. (1997) Nature 386, 279-284. [DOI] [PubMed] [Google Scholar]
- 7.Srivastava, S., Osten, P., Vilim, F. S., Khatri, L., Inman, G., States, B., Daly, C., DeSouza, S., Abagyan, R., Valtschanoff, J. G., et al. (1998) Neuron 21, 581-591. [DOI] [PubMed] [Google Scholar]
- 8.Xia, J., Zhang, X., Staudinger, J. & Huganir, R. L. (1999) Neuron 22, 179-187. [DOI] [PubMed] [Google Scholar]
- 9.Leonard, A. S., Davare, M. A., Horne, M. C., Garner, C. C. & Hell, J. W. (1998) J. Biol. Chem. 273, 19518-19524. [DOI] [PubMed] [Google Scholar]
- 10.Shen, L., Liang, F., Walensky, L. D. & Huganir, R. L. (2000) J. Neurosci. 20, 7932-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coleman, S. K., Cai, C., Mottershead, D. G., Haapalahti, J. P. & Keinänen, K. (2003) J. Neurosci. 23, 798-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishimune, A., Isaac, J. T., Molnar, E., Noel, J., Nash, S. R., Tagaya, M., Collingridge, G. L., Nakanishi, S. & Henley, J. M. (1998) Neuron 21, 87-97. [DOI] [PubMed] [Google Scholar]
- 13.Osten, P., Srivastava, S., Inman, G. J., Vilim, F. S., Khatri, L., Lee, L. M., States, B. A., Einheber, S., Milner, T. A., Hanson, P. I., et al. (1998) Neuron 21, 99-110. [DOI] [PubMed] [Google Scholar]
- 14.Song, I., Kamboj, S., Xia, J., Dong, H., Liao, D. & Huganir, R. L. (1998) Neuron 21, 393-400. [DOI] [PubMed] [Google Scholar]
- 15.Lee, S. H., Liu, L., Wang, Y. T. & Sheng, M. (2002) Neuron 36, 661-674. [DOI] [PubMed] [Google Scholar]
- 16.Chen, L., Chetkovich, D. M., Petralia, R. S., Sweeney, N. T., Kawasaki, Y., Wenthold, R. J., Bredt, D. S. & Nicoll, R. A. (2000) Nature 408, 936-943. [DOI] [PubMed] [Google Scholar]
- 17.Letts, V. A., Felix, R., Biddlecome, G. H., Arikkath, J., Mahaffey, C. L., Valenzuela, A., Bartlett, F. S., Jr., Mori, Y., Campbell, K. P. & Frankel, W. N. (1998) Nat. Genet. 19, 340-347. [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto, K., Fukaya, M., Qiao, X., Sakimura, K., Watanabe, M. & Kano, M. (1999) J. Neurosci. 19, 6027-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomita, S., Fukata, M., Nicoll, R. A. & Bredt, D. S. (2004) Science 303, 1508-1511. [DOI] [PubMed] [Google Scholar]
- 20.Tomita, S., Chen, L., Kawasaki, Y., Petralia, R. S., Wenthold, R. J., Nicoll, R. A. & Bredt, D. S. (2003) J. Cell Biol. 161, 805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGee, A. W. & Bredt, D. S. (1999) J. Biol. Chem. 274, 17431-17436. [DOI] [PubMed] [Google Scholar]
- 22.Rubio, M. E. & Wenthold, R. J. (1999) J. Neurochem. 73, 942-948. [DOI] [PubMed] [Google Scholar]
- 23.Schägger, H., Cramer, W. A. & von Jagow, G. (1994) Anal. Biochem. 217, 220-230. [DOI] [PubMed] [Google Scholar]
- 24.Greger, I. H., Khatri, L., Kong, X. & Ziff, E. B. (2003) Neuron 40, 763-774. [DOI] [PubMed] [Google Scholar]
- 25.Lee, M.C., Hamamoto, S. & Schekman, R. (2002) J. Biol. Chem. 277, 22395-22401. [DOI] [PubMed] [Google Scholar]
- 26.Meddows, E., Le Bourdelles, B., Grimwood, S., Wafford, K., Sandhu, S., Whiting, P. & McIlhinney, R. A. (2001) J. Biol. Chem. 276, 18795-18803. [DOI] [PubMed] [Google Scholar]
- 27.Fremeau, R. T., Jr., Troyer, M. D., Pahner, I., Nygaard, G. O., Tran, C. H., Reimer, R. J., Bellocchio, E. E., Fortin, D., Storm-Mathisen, J. & Edwards, R. H. (2001) Neuron 31, 247-260. [DOI] [PubMed] [Google Scholar]
- 28.Huh, K. H. & Wenthold, R. J. (1999) J. Biol. Chem. 274, 151-157. [DOI] [PubMed] [Google Scholar]
- 29.Rahmatullah, M. & Robishaw, J. D. (1994) J. Biol. Chem. 269, 3574-3580. [PubMed] [Google Scholar]
- 30.Isom, L. L., DeJongh, K. S. & Catterall, W. A. (1994) Neuron 12, 1183-1194. [DOI] [PubMed] [Google Scholar]
- 31.Trimmer, J. S. (1998) Curr. Opin. Neurobiol. 8, 370-374. [DOI] [PubMed] [Google Scholar]
- 32.Hille, B. (2001) Ion Channels of Excitable Membranes (Sinauer, Sunderland, MA).
- 33.Zheng, Y., Mellem, J. E., Brockie, P. J., Madsen, D. M. & Maricq, A. V. (2004) Nature 427, 451-457. [DOI] [PubMed] [Google Scholar]
- 34.Allison, D. W., Gelfand, V. I., Spector, I. & Craig, A. M. (1998) J. Neurosci. 18, 2423-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellgaard, L. & Helenius, A. (2003) Nat. Rev. Mol. Cell. Biol. 4, 181-191. [DOI] [PubMed] [Google Scholar]
- 36.Zerangue, N., Schwappach, B., Jan, Y. N. & Jan, L. Y. (1999) Neuron 22, 537-548. [DOI] [PubMed] [Google Scholar]
- 37.Mu, Y., Otsuka, T., Horton, A. C., Scott, D. B. & Ehlers, M. D. (2003) Neuron 40, 581-594. [DOI] [PubMed] [Google Scholar]
- 38.Chetkovich, D. M., Chen, L., Stocker, T. J., Nicoll, R. A. & Bredt, D. S. (2002) J. Neurosci. 22, 5791-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnell, E., Sizemore, M., Karimzadegan, S., Chen, L., Bredt, D. S. & Nicoll, R. A. (2002) Proc. Natl. Acad. Sci. USA 99, 13902-13907. [DOI] [PMC free article] [PubMed] [Google Scholar]