Abstract
Bacteria use small diffusible molecules to exchange information in a process called quorum sensing. An important class of autoinducers used by Gram-negative bacteria is the family of N-acylhomoserine lactones. Here, we report the discovery of a previously undescribed nonenzymatically formed product from N-(3-oxododecanoyl)-L-homoserine lactone; both the N-acylhomoserine and its novel tetramic acid degradation product, 3-(1-hydroxydecylidene)-5-(2-hydroxyethyl)pyrrolidine-2,4-dione, are potent antibacterial agents. Bactericidal activity was observed against all tested Gram-positive bacterial strains, whereas no toxicity was seen against Gram-negative bacteria. We propose that Pseudomonas aeruginosa utilizes this tetramic acid as an interference strategy to preclude encroachment by competing bacteria. Additionally, we have discovered that this tetramic acid binds iron with comparable affinity to known bacterial siderophores, possibly providing an unrecognized mechanism for iron solubilization. These findings merit new attention such that other previously identified autoinducers be reevaluated for additional biological functions.
Keywords: tetramic acid, bactericidal agents, evolution
The term “quorum sensing” has been coined to describe the ability of a population of unicellular bacteria to act as a multicellular organism in a cell-density-dependent manner; that is, a way to sense “how many are out there” (1-3). Bacteria use small diffusible molecules to exchange information among themselves (2). An important class of “quormones,” or autoinducers, is the family of N-acylhomoserine lactones (AHLs) used by Gram-negative bacteria. Variation in N-acyl chain length and the oxidation state of AHLs provide for bacterial strain specificity in the signaling process and subsequent synchronization of gene expression. Depending on their acylation pattern, most AHLs diffuse freely across the bacterial cell membrane. Upon reaching a critical threshold concentration, they bind to their cognate receptor proteins, triggering the expression of target genes; for example, in the case of Vibrio fischeri, genes located on the lux operon are transcribed that are responsible for the production of bioluminescence (4). Indeed, this account of V. fischeri was the first description of a bacterial population acting in a concerted fashion to achieve a common goal: the simultaneous expression of a particular set of genes.
Pseudomonas aeruginosa is a common environmental microorganism that has acquired the ability to take advantage of weaknesses in the host immune system to become an opportunistic pathogen in humans (5). Most prominent is the role of P. aeruginosa in patients suffering from cystic fibrosis because lung-defense functions are severely impaired. As a result, infections with P. aeruginosa and the damage caused by the inflammatory infection process increase the mortality rate in cystic fibrosis patients. Additionally, nosocomial infections by P. aeruginosa, particularly in burn victims, cause serious complications because of the wide variety of virulence factors and inherent antibiotic resistance. Significant percentages of burn patients become infected with P. aeruginosa, and, of those, the majority die of septicemia. These infections are especially troublesome because P. aeruginosa continues to grow more resistant to many antibiotics.
Over the last 10 years, significant progress has been made in elucidating the molecular mechanisms underlying P. aeruginosa pathogenicity. Two different AHLs, N-(3-oxododecanoyl) homoserine lactone 1, synthesized by LasI, and N-butyrylhomoserine lactone 2, synthesized by RhlI, have been identified as the main quorum-sensing signaling molecules in P. aeruginosa (Fig. 1) (6). Genes regulated by this mechanism encode enzymes such as elastases A and B, catalase, and superoxide dismutase. Furthermore, quorum sensing also has been demonstrated to control the expression of other virulence factors as well as the formation of structures known as biofilms (7). Here, we demonstrate that N-(3-oxododecanoyl) homoserine lactone 1 performs a previously unrecognized role: The autoinducer itself and a corresponding degradation product derived from an unusual Claisen-like condensation reaction function as innate bactericidal agents. Furthermore, the AHL degradation product tightly binds essential metals such as iron, possibly providing a previously unrecognized primordial siderophore.
Fig. 1.
AHLs used by P. aeruginosa in quorum sensing.
Materials and Methods
General Synthetic Methods. Unless otherwise stated, all reactions were performed under an inert atmosphere with dry reagents and solvents and flame-dried glassware. Analytical TLC was performed by using 0.25-mm precoated silica gel Kieselgel 60 F254 plates. Visualization of the chromatogram was by UV absorbance, iodine, dinitrophenylhydrazine, ceric ammonium molybdate, ninhydrin, or potassium permanganate as appropriate. All 1H NMR spectra were recorded on either a Bruker AMX-500 or DRX-600 spectrometer (Billerica, MA) at 500 and 600 MHz, respectively. All 13C NMR spectra were recorded on a Bruker AMX-500 spectrometer at 125 MHz. Optical rotations were determined at 598 nm in a conventional 10-cm cell by using a PerkinElmer 241 MC polarimeter. MALDI-Fourier transform-MS experiments were performed on an IonSpec-Fourier transform mass spectrometer (Lake Forest, CA). Electrospray ionization-MS experiments were performed on an API 100 PerkinElmer Sciex single-quadrupole mass spectrometer. Analytical reverse-phase HPLC was performed on a Hitachi L-5000 series instrument equipped with a Vydac-C18 analytical column, a UV detector at 254 nm, and mobile phases composed of mixtures of acetonitrile/water (0.1% trifluoroacetic acid).
General Procedure for Synthesis of Tetramic Acids. As a representative example, the synthesis of (S)-3-(1-hydroxydecylidene)-5-(2-hydroxyethyl)pyrrolidine-2,4-dione 4 is described. To a vial containing (S)-3-oxo-C12-AHL 1 (111 mg, 0.374 mmol) in dry MeOH (1 ml), NaOMe in MeOH (0.5 M, 0.747 ml, 0.374 mmol) was added at room temperature under argon. After stirring at 55°C for 3 h, the reaction mixture was passed through acidic ion-exchange resin (Dowex 50WX2-200, ≈2 cm3) and further eluted with MeOH (20 ml). The combined filtrate was concentrated under reduced pressure, and the resulting residue was recrystallized (EtOAc/hexanes) to give an off-white small crystalline solid (80 mg). This product then was purified by preparative HPLC to give tetramic acid 4 as white fluffy solid. Tetramic acids were purified further by reverse-phase HPLC using a Vydac 214TP101522 column (Hesperia, CA) at a flow rate of 10 ml/min with detection at 230 nm on a dual-pump Rainin Dynamax HPLC system (Rainin Instruments). In each preparative separation, recrystallized material (40 mg) was dissolved in 5 ml of 45:45:10 acetic acid/water/DMSO, filtered through a 0.45-μm poly(vinylidene difluoride) filter, and purified by using a gradient of 40-60% solvent B (0.085% trifluoroacetic acid in acetonitrile) in solvent A (0.1% trifluoroacetic acid in water) over 30 min. Pure fractions were identified by electrospray ionization-MS on a Sciex API-150EX single quadrupole mass spectrometer (DP 10 V, FP 50 V) operated in multichannel analysis mode, pooled, and lyophilized. In all cases, the homogeneity of pure pooled products was verified by analytical reverse-phase HPLC using a C4-functionalized Vydac 218TP5415 column running a gradient of 20-80% B over 30 min; 1H NMR (600 MHz, CD3OD): δ 0.90 (t, J = 7.0 Hz, 3 H), 1.23-1.43 (m, 12 H), 1.66 (qt, J = 7.4 Hz, 2 H), 1.71-1.82 (m, 1 H), 1.96-2.08 (m, 1 H), 2.72-2.94 (m, 2 H), 3.62-3.78 (m, 2 H), 3.97 (br s, 1H); 13C NMR (125 MHz, CDCl3): δ 14.1, 22.6, 25.9, 29.2, 29.4, 31.8, 32.9, 34.0, 61.3, 62.0, 100.5, 175.0, 190.0, 195.3. MALDI-Fourier transform-MS for C16H28NO4 (M + H+) calculated 298.2017, found 298.2013. [α]25d = +14.0 (c = 7.20, MeOH).
Detection of 3-(1-Hydroxydecylidene)-5-(2-Hydroxyethyl)Pyrrolidine-2,4-Dione 4 from P. aeruginosa Culture. P. aeruginosa PAO1 cells were grown for 24 h in Luria-Bertani broth (2 liters) containing 50 mM Mops (pH 7.4). The cells were harvested by centrifugation and resuspended in BugBuster (50 ml; Novagen). After incubation for 1 h at room temperature, the cells were removed by centrifugation, and an aliquot of the supernatant (20 ml) was poured into a separatory funnel containing brine (1 liter) and CHCl3 (1 liter). The layers then were separated, and the aqueous layer was acidified to pH 2 with 6 M HCl. The aqueous mixture then was extracted with CHCl3 (2 × 1 liter), and the organic layers were combined and concentrated. The resulting residue was dissolved in hot acetonitrile (100 ml) and filtered through a 0.45-μm poly(vinylidene difluoride) filter. The filtrate then was concentrated, redissolved in hot acetonitrile (6 ml), and filtered again. Upon cooling, the solution was brown and turbid. Analysis of the extract was performed by using the described reverse-phase HPLC system (see above). The gradient used was as follows: t = 0, 50% solvent B (0.1% trifluoroacetic acid in acetonitrile) in solvent A (0.1% trifluoroacetic acid in water); t = 5, min 50% solvent B; t = 25 min, 80% solvent B; t = 35 min, 100% solvent B; t = 50 min, 100% solvent B. Fractions were collected every minute and analyzed by electrospray ionization-MS. Peaks with mass corresponding to tetramic acid 4 were found to elute at a retention time of 14.7 min and were confirmed by coinjection with authentic standard samples.
Kinetic Assays for the Formation of 3-(1-Hydroxydecylidene)-5-(2-Hydroxyethyl)Pyrrolidine-2,4-Dione 4. The assay was initiated by the addition of a solution of AHL (5 mM in DMSO) to phosphate buffer at 25°C (200 mM, pH 7.4/10% DMSO cosolvent; 1 ml total volume). Continuous monitoring of the reaction was performed over 2,000 s by using a Hewlett Packard 8452A UV-visible spectrophotometer. Tetramic acid product formation was determined spectrophotometrically by using an extinction coefficient of 13,900 M-1·cm-1 at λ = 278 nm. The acquired data were fit to a two-state model in which competing reactions were assumed to be nonequilibrating. Data analysis was performed by nonlinear curve-fitting algorithms by using the program kaleidagraph 3.6.2 (Synergy Software, Reading, PA).
Antibacterial Assays. The following bacterial strains were purchased from the American Type Culture Collection (ATCC) unless otherwise stated: Bacillus cereus ATCC 11778, B. cereus ATCC 13061, B. cereus ATCC 14579, B. cereus ATCC 27348, Bacillus licheniformis 5A36 (from Bacillus Genetic Stock Center, Columbus, OH), Bacillus mycoides ATCC 6462, Bacillus subtilis ATCC 6051, Enterococcus faecalis ATCC 29212, Escherichia coli ATCC 35150, Listeria monocytogenes ATCC 43251, P. aeruginosa PAO1 (generous gift from B. Iglewski, University of Rochester, Rochester, NY), Staphylococcus aureus ATCC 25923, Staphylococcus epidermidis ATCC 12228, Streptococcus pyogenes ATCC 49399, and Salmonella typhimurium ATCC 13311.
A bacterial colony was picked and grown overnight in the growth medium and at the temperature recommended by ATCC. On the next day, the culture was diluted 1:1,000 in fresh growth medium and grown until an OD650 of 0.1 was reached. The culture then was diluted 1:200 in fresh growth medium. Aliquots (198 μl) were added into a 96-well microtiter plate containing the test compound dissolved in 2 μl of DMSO (Sigma) at 100× the desired concentration. Control experiments contained only DMSO. The microtiter plate was sealed (BreatheEasy, Research Products International) and incubated on a shaker at the appropriate temperature overnight. The next day, OD650 was measured by using a ThermoMax plate reader (Molecular Devices). EC50 values then were determined by nonlinear curve-fitting using kaleidagraph 3.6.2 (Synergy Software). Reported values are the average of a minimum of three replicates.
Electron Microscopy of Gram-Positive Bacterial Cells Exposed to Tetramic Acid 4. B. cereus (ATCC 11778) was grown overnight at 30°C. The next day, the cells were diluted 1:500 and grown at 30°C. After 3 h, tetramic acid 4 or AHL 1 was added to a final concentration of 100 mM (1% DMSO). Control cultures contained only 1% DMSO. After further incubation (5 h) on a shaker at 30°C, the B. cereus samples were fixed in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.12 M cacodylate buffer (pH 7.3) and pelleted by using a tabletop microfuge, and the resulting pellets were postfixed in cacodylate-buffered 1% osmium tetroxide. After dehydration in graded ethanol series followed by propylene oxide, the samples were embedded in EMbed 812/Araldite 502 (Electron Microscopy Sciences, Fort Washington, PA). Thin sections were cut transversely, mounted on parlodion-coated copper slot grids, and stained with uranyl acetate and lead citrate; images were documented on a Philips CM100 electron microscope (FEI, Hillsborough, OR) using Kodak SO163 film. Negatives were scanned at 600 dots per inch by using a Fuji FineScan 2750 and converted to TIFF format for subsequent handling in photoshop (Adobe Systems, San Jose, CA).
Iron Binding to Tetramic Acid 4. Stock solutions of ferric chloride (10 mM) and tetramic acid 4 (10 mM) were prepared in spectroscopic-grade methanol. Aliquots of both the ferric chloride and tetramic acid 4 stocks then were diluted to 150 and 500 μM, respectively, with 4:1 MeOH/0.1 M NaOAc buffer solution (pH 7.4), with and without EDTA (100 μM). The absorbances of the formed complexes were measured at 440 nm in triplicate in the presence and absence of EDTA. Samples containing varying ligand and ferric chloride concentrations in methanol also were analyzed by MS as described above.
Results and Discussion
Our studies were initiated to examine the lifetime of AHL 1 in an aqueous environment. Incubation of compound 1 in water produced an undocumented compound in addition to the expected hydrolysis product 3. Structural characterization of this anomalous molecule revealed it to be 3-(1-hydroxydecylidene)-5-(2-hydroxyethyl)pyrrolidine-2,4-dione 4, a compound belonging to a class of antibacterial compounds known as tetramic acids (Fig. 2) (8). The mechanism of formation is a Claisen-like intramolecular alkylation of the β-ketoamide moiety (Fig. 3). Specifically, the α-carbon between the ketone and amide moieties is deprotonated, leading to an anion that cyclizes intramolecularly on the lactone generating the tetramic acid motif. Tetramic acid 4 was found to be very stable, with no decomposition or detectable reversion to compound 1; most importantly, compound 4 also was detected in P. aeruginosa culture. To assess the generality of this phenomenon, a variety of AHLs with varying acyl chain lengths were prepared and incubated in buffer. This chemical reaction was not limited to compound 1 because all tested 3-oxo-AHLs with varying chain lengths also underwent this intramolecular rearrangement (Table 1). Surprisingly, although previous studies have attempted to measure the kinetics of AHL hydrolysis, the formation of 4 has never been reported, to our knowledge (9).
Fig. 2.
Representative natural products containing a tetramic acid motif. Interestingly, various tetramic acids have been demonstrated to possess mycotoxic, antibacterial, antiviral, and antioxidant activities.
Fig. 3.
Reaction of 3-oxo-AHL 1 to generate lactone hydrolysis product 3 (path a) and 3-(1-hydroxydecylidene)-5-(2-hydroxyethyl)pyrrolidine-2,4-dione 4 (path b).
Table 1. Summary of kinetic data for the decomposition of AHL 1.
| Compound | khydrolysis, S-1 | ktetramic acid, S-1 |
|---|---|---|
| 3-Oxo-C4-AHL | 8.06 × 10-6 | 5.97 × 10-6 |
| 3-Oxo-C6-AHL | 3.07 × 10-5 | 2.29 × 10-6 |
| 3-Oxo-C8-AHL | 1.48 × 10-5 | 1.28 × 10-6 |
| 3-Oxo-C12-AHL 1 | 4.77 × 10-6 | 1.49 × 10-6 |
In each entry, Cn refers to the number of carbon atoms present in the N-acyl chain.
AHL 1 and Tetramic Acid 4 Are Innate Bactericidal Agents. Given that P. aeruginosa employs AHL 1 as the principal autoinducer and the known bactericidal activity of tetramic acids, we hypothesized that compound 4 could have a biological function in the context of bacterial viability. We were guided by the observation that lipid-substituted antibiotics have been shown to localize near the cell membrane of Gram-positive bacteria (10). Indeed, significant antibacterial activity was observed against all tested Gram-positive bacterial strains in the presence of 4 after overnight incubation (Table 2), whereas no toxicity was observed against P. aeruginosa or other tested Gram-negative bacteria (Fig. 4). The effective concentration of 4 is biologically relevant, because high concentrations of compound 1 (>600 μM) have been detected previously in P. aeruginosa biofilms (11). Furthermore, 4 has comparable activity (average EC50 = 23 μM or ≈60 ng/ml) to other known antibacterial compounds.
Table 2. Cytotoxicity of tetramic acid 4 and AHL 1 in various bacterial cell lines.
| Bacteria* | 4 EC50, μM | 1 EC50, μM |
|---|---|---|
| Gram-positive | ||
| B. cereus (11778) | 8.3 | 22.1 |
| B. cereus (14579) | 14.8 | >100 |
| B. cereus (13061) | 12.5 | >100 |
| B. cereus (27348) | 18.3 | >100 |
| B. licheniformis† | 36.1 | n.d. |
| B. mycoides (6462) | 13.8 | n.d. |
| B. subtilis (6051) | 13.9 | 31.3 |
| E. faecalis (29212) | 33.7 | n.d. |
| L. monocytogenes (43251) | 34.1 | n.d. |
| S. aureus (25923) | 26.7 | 80.2 |
| S. epidermidis (12228) | 15.8 | n.d. |
| S. pyogenes (49399) | 55.3 | 40.8 |
| Gram-negative | ||
| E. coli (35150) | >100 | >100 |
| P. aeruginosa‡ | >100 | >100 |
| S. typhimurium (13311) | >100 | >100 |
n.d., not determined.
ATCC numbers are given in parentheses
Strain 5A36 from Bacillus Genetic Stock Center
PAO1 from B. Iglewski
Fig. 4.
Electron micrographs of B. cereus ATCC 11778 after 5 h of incubation with DMSO (A), 100 μM tetramic acid 4 (B), or 100 μM AHL 1 (C). Cells in both B and C show significant morphological changes.
Similar to previously described tetramic acids and related analogs, sensitivity to compound 4 was apparent only in Gram-positive cells with little variation present in EC50 values across different species and genera (12). Analogous observations have been made in the case of the natural product reutericyclin, a tetramic acid isolated from the sourdough isolate Lactobacillus reuteri LTH2584. This compound has been demonstrated to be bactericidal in Gram-positive bacteria by acting as a protonionophore, thereby dissipating the transmembrane change in pH and leading to cell lysis (13). The efficiency of this mechanism is a function of the high hydrophobicity of reutericyclin, thereby favoring partitioning into the cytoplasmic membrane. Based on the similarities between this compound and 4, including the long hydrophobic carbon chain and tetramic acid functionality, it is plausible that 4 also would operate by means of a similar mechanism.
Notably, compound 1 also displayed cytotoxicity (Table 2) suggesting a dual role for 1 in P. aeruginosa communities as both quorum-sensing molecules and as an interference mechanism against bacterial competitors. Here, cytotoxicity cannot be attributed simply to tetramic acid because the length at which the experiment was conducted was such that tetramic acid formation is expected to be minimal (see below). Indeed, although significant cell death was observed after overnight incubation with AHL 1 Fig. 4C), the bacteria recovered after additional incubation, suggesting that although AHL 1 is bactericidal, upon hydrolysis, the resulting compound 3 is tolerated by bacteria. Interestingly, resistant B. cereus strains known to express a penicillinase or cephalosporinase in conjunction with a penicillinase (ATCC 13061 and 27348, respectively) were not affected by exposure to 1, yet displayed comparable sensitivity to tetramic acid 4 as other tested B. cereus strains (Table 2). We acknowledge that the observed antibacterial activity of 4 is relatively modest in comparison with other known more potent bactericidal agents; however, the fact that this compound is nonenzymatically produced from a molecule designed to play a role in cell-cell signaling is significant. Furthermore, even in the instance that only a small percentage of the competitor population is killed by 4, this characteristic then should provide a competitive advantage for P. aeruginosa, thereby insuring the survival of the colony or biofilm.
The identification of AHL-hydrolyzing enzymes, or lactonases, in B. cereus 240B1 led to the hypothesis that certain bacteria have evolved quorum-sensing signaling interference strategies, or so-called “quorum quenching” (14, 15). The hydrolysis of AHLs not only prevents cell density-dependent signaling events from occurring, but also presumably could mitigate the cytotoxic effect of AHLs and furthermore prevent the formation of tetramic acid 4. Indeed, B. cereus ATCC 14579 cells, previously shown to express lactonase (16), do not show sensitivity to compound 1, yet are still affected by compound 4 (EC50 = 13.4 ± 1.6 μM). This finding advocates two distinct implications: the first is that, as previously hypothesized, lactonases may have evolved in certain Gram-positive bacteria to interrupt quorum-sensing signaling. However, our findings also suggest that these enzymes have a previously unrecognized function: to abrogate the toxic effects of 1 and prevent the formation 4. This hypothesis is concurrent with recent discussions about a more complex role of lactonases in vivo (17). Second, this finding raises the enticing evolutionary prospect that 3-oxo-AHLs and their corresponding tetramic acids were selected for as a result of their fortuitous cytotoxic abilities and resistance to lactonase degradation, respectively.
3-(1-Hydroxydecylidene)-5-(2-Hydroxyethyl)Pyrrolidine-2,4-Dione 4 Binds Iron with High Affinity. Iron plays an essential role in physiological processes and the pathogenesis of bacteria (18, 19). Many bacteria are known to produce siderophores to sequester iron, an element that, although essential for their growth, has poor solubility under physiological conditions (20). P. aeruginosa synthesizes two siderophores, pyoverdin and pyochelin, and has been shown to be able to use a variety of heterologous siderophores of microbial origin. The P. aeruginosa genome contains many homologues of iron-siderophore receptor genes, reflecting the enormous flexibility of this organism to use various iron carriers. The ability to use such a wide variety of siderophores underscores the importance of iron for bacterial growth and survival as well as the need to compete with other microorganisms within the environments they inhabit for this essential metal (21, 22).
The 3-acetyl-pyrrolidine-2,4-dione heterocycle found in compound 4 and in many other naturally occurring tetramic acids has been shown to efficiently chelate a variety of metal cations including iron (23). The role of metal binding in the bactericidal activity of tetramic acids is unclear, with some tetramic acids displaying increased toxicity as a metal complex, whereas in others, the toxicity is attenuated by chelation of a metal ion (24). Given this disparity, we investigated the ability of AHL-derived tetramic acid 4 to complex metals to evaluate the potential bioactivity of these complexes. Bearing in mind the crucial role that iron plays in bacterial physiology (see above), and the demonstrated potential of tetramic acids to bind this metal, we focused our investigations on the chelation of iron by 4.
Lebrun et al. (25) studied the complexation of the fungal tetramic acid metabolite tenuazonic acid (Fig. 2) with iron(III) and observed the formation an octahedral complex with a 3:1 stoichiometry. Analogous to these studies, we have envisioned a similar mode of complexation for the compound 4-Fe3+ complex (Fig. 5). To verify our hypothesis, we performed studies by using varying concentrations of ferric chloride and tetramic acid and observed the formation of a 4-Fe3+ complex by spectrophotometric analysis at 440 nm; the corresponding 3-oxo-C12-AHL did not show significant absorption at this wavelength upon FeCl3 addition. The complex was further characterized by using electrospray ionization-MS, confirming the formation of a 3:1 4-Fe3+ complex ([M + H]+ m/z = 945.5).
Fig. 5.
Proposed structure of the compound 4-Fe3+ complex demonstrating a 3:1 ratio of tetramic acid/metal.
Additional support for our hypothesis was given through the measurement of the apparent binding constant (Kd,app) for iron(III) with compound 4. These experiments were performed by using a previously described protocol based on the competition between tetramic acid 4 and EDTA for iron and the facile detection of 4-Fe3+ complexes by their characteristic absorption (26). The loss of 4-Fe3+ absorbance at 440 nm upon addition of EDTA was used to calculate the equilibrium constant, presuming the formation of a 3:1 tetramic acid-Fe complex, according to the following equation:
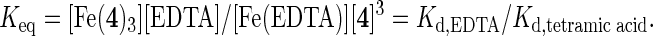 |
[1] |
By using the known affinity of EDTA for Fe3+ (Kd = 5 × 10-23 M), we were able to determine the relative affinity (Kd,app) of compound 4 for Fe3+ to be 1.6 × 10-29 M3. A direct comparison of iron affinity between this presumed bidentate chelator and known hexadentate chelators such as desferrioxamine, EDTA, and pyoverdin is not possible because the affinity of hexadenate chelators are measured in units of molarity, whereas bidentate chelators are measured in units of (molarity)3. However, a parameter termed “pM” has been used previously as a method of standardization and is given by
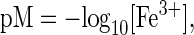 |
[2] |
where [Fe3+] is equal to the concentration of iron in the presence of 1 mM total iron chelator and 1 μM total Fe3+ at pH 7.4 (25). By using these values (Table 3), compound 4 was deduced to have a roughly three times stronger affinity for iron(III) than EDTA, but weaker than pyoverdin. Interestingly, the reported Kd for the secondary siderophore, pyochelin (10-5 M), is quite poor, suggesting that 4 could compete for available iron in solution and provide an additional method for iron solubilization.
Table 3. Affinity constants for iron (III) chelators.
In summary, we propose that P. aeruginosa utilizes compound 4 as an interference strategy to preclude encroachment by competing bacteria. In essence, this behavior parallels other known quorum-controlled processes in that an individual cell cannot produce sufficient quantities of tetramic acid to affect other bacteria; the cytotoxic properties of 4 only emerge when a population of cells synthesizes 3-oxo-AHLs. However, the autoinducer not only functions as a transcriptional regulator but also serves as a bactericidal agent. Although the complexation of critical metals such as iron may play a role in this process, further study is required into the mechanism and scope of the observed bactericidal activity and the potential of 4 to act as a primordial siderophore. The compilation of our data recommends that other autoinducers be reevaluated for the potential to perform additional biological functions.
Acknowledgments
We thank Professor Barbara Iglewski for generously providing P. aeruginosa PAO1 and Malcolm Wood and Theresa Fassel for expert assistance with electron microscopy experiments. This work was supported by National Institutes of Health Grant AI055781 and by The Skaggs Institute for Chemical Biology.
Author contributions: G.F.K., M.M.M., T.J.D., and K.D.J. designed research; G.F.K., R.S., S.-H.L., C.J.R., M.M.M., J.A.M., B.C., A.P.B., and T.J.D. performed research; G.F.K., R.S., S.-H.L., C.J.R., M.M.M., J.A.M., B.C., A.P.B., and T.J.D. analyzed data; S.-H.L. and A.P.B. contributed new reagents/analytic tools; and G.F.K., T.J.D., and K.D.J. wrote the paper.
Abbreviation: AHL, N-acylhomoserine lactone.
References
- 1.Schauder, S. & Bassler, B. L. (2001) Genes Dev. 15, 1468-1480. [DOI] [PubMed] [Google Scholar]
- 2.Miller, M. B. & Bassler, B. L. (2001) Annu. Rev. Microbiol. 55, 165-199. [DOI] [PubMed] [Google Scholar]
- 3.Fuqua, C., Parsek, M. R. & Greenberg, E. P. (2001) Annu. Rev. Genet. 35, 439-468. [DOI] [PubMed] [Google Scholar]
- 4.Dunlap, P. V. (1999) J. Mol. Microbiol. Biotechnol. 1, 5-12. [PubMed] [Google Scholar]
- 5.Lyczak, J. B., Cannon, C. L. & Pier, G. B. (2000) Microbes Infection 2, 1051-1060. [DOI] [PubMed] [Google Scholar]
- 6.Pearson, J. P., Passador, L., Iglewski, B. H. & Greenberg, E. P. (1995) Proc. Natl. Acad. Sci. USA 92, 1490-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith, R. S. & Iglewski, B. H. (2003) J. Clin. Invest. 112, 1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghisalberti, E. L. (2003) Studies Nat. Products Chem. 28, 109-163. [Google Scholar]
- 9.Yates, E. A., Philipp, B., Buckley, C., Atkinson, S., Chhabra, S. R., Sockett, R. E., Goldner, M., Dessaux, Y., Camara, M., Smith, H., et al. (2002) Infect. Immun. 70, 5635-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, S. D., Oberthur, M., Losey, H. C., Anderson, J. W., Eggert, U. S., Peczuh, M. W., Walsh, C. T. & Kahne, D. (2002) J. Am. Chem. Soc. 124, 9064-9065. [DOI] [PubMed] [Google Scholar]
- 11.Charlton, T. S., de Nys, R., Netting, A., Kumar, N., Hentzer, M., Givskov, M. & Kjelleberg, S. (2000) Environ. Microbiol. 2, 530-541. [DOI] [PubMed] [Google Scholar]
- 12.Gänzle, M. G., Höltzel, A., Walter, J., Jung, G. & Hammes, W. P. (2000) Appl. Environ. Microbiol. 66, 4325-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gänzle, M. G. & Vogel, R. F. (2003) Appl. Environ. Microbiol. 69, 1305-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong, Y. H., Xu, J. L., Li, X. Z. & Zhang, L. H. (2000) Proc. Natl. Acad. Sci. USA 97, 3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong, Y. H., Wang, L. H., Xu, J. L., Zhang, H. B., Zhang, X. F. & Zhang, L. H. (2001) Nature 411, 813-817. [DOI] [PubMed] [Google Scholar]
- 16.Dong, Y. H., Gusti, A. R., Zhang, Q., Xu, J. L. & Zhang, L. H. (2002) Appl. Environ. Microbiol. 68, 1754-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roche, D. M., Byers, J. T., Smith, D. S., Glansdorp, F. G., Spring, D. R. & Welch, M. (2004) Microbiology 150, 2023-2028. [DOI] [PubMed] [Google Scholar]
- 18.Cox, C. D. & Adams, P. (1985) Infect. Immun. 48, 130-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gensberg, K., Hughes, K. & Smith, A. W. (1992) J. Gen. Microbiol. 138, 2381-2387. [DOI] [PubMed] [Google Scholar]
- 20.Payne, S. M. (1994) Methods Enzymol. 235, 329-344. [DOI] [PubMed] [Google Scholar]
- 21.Poole, K. & McKay, G. A. (2003) Front. Biosci. 8, d661-d686. [DOI] [PubMed] [Google Scholar]
- 22.Takase, H., Nitanai, H., Hoshino, K. & Otani, T. (2000) Infect. Immun. 68, 4498-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henning, H.-G. & Gelbin, A. (1993) Adv. Heterocycl. Chem. 57, 139-185. [Google Scholar]
- 24.Gandhi, N. M., Nazareth, J., Divekar, P. V., Kohl, H. & de Souza, N. J. (1973) J. Antibiot. 797-798. [DOI] [PubMed]
- 25.Lebrun, M. H., Nicolas, L., Boutar, M., Gaudemer, F., Ranomenjanahary, S. & Gaudemer, A. (1988) Phytochemistry 27, 77-84. [Google Scholar]
- 26.Wang, J., Buss, J. L., Chen, G., Ponka, P. & Pantopoulos, K. (2002) FEBS Lett. 529, 309-312. [DOI] [PubMed] [Google Scholar]
- 27.Martell, A. E. (1977) Critical Stability Constants (Plenum, New York).
- 28.Ratledge C. & Dover, L. G. (2000) Annu. Rev. Microbiol. 54, 881-941. [DOI] [PubMed] [Google Scholar]







