Abstract
Streptococcus pneumoniae is a facultative anaerobic pathogen. Although it maintains fermentative metabolism, during aerobic growth pneumococci produce high levels of H2O2, which can have adverse effects on cell viability and DNA, and influence pneumococcal interaction with its host. The pneumococcus is unusual in its dealing with toxic reactive oxygen species (ROS) in that it neither has catalase nor the global regulators of peroxide stress resistance. Previously, we identified pneumococcal thiol peroxidase (TpxD) as the key enzyme for enzymatic removal of H2O2, and showed that TpxD synthesis is up-regulated upon exposure to H2O2. This study aimed to reveal the mechanism controlling TpxD expression under H2O2 stress. We hypothesize that H2O2 activates a transcription factor which in turn up-regulates tpxD expression. Microarray analysis revealed a pneumococcal global transcriptional response to H2O2. Mutation of tpxD abolished H2O2-mediated response to high H2O2 levels, signifying the need for an active TpxD under oxidative stress conditions. Bioinformatic tools, applied to search for a transcription factor modulating tpxD expression, pointed toward CodY as a potential candidate. Indeed, a putative 15-bp consensus CodY binding site was found in the proximal region of tpxD-coding sequence. Binding of CodY to this site was confirmed by EMSA, and genetic engineering techniques demonstrated that this site is essential for TpxD up-regulation under H2O2 stress. Furthermore, tpxD expression was reduced in a ΔcodY mutant. These data indicate that CodY is an activator of tpxD expression, triggering its up-regulation under H2O2 stress. In addition we show that H2O2 specifically oxidizes the 2 CodY cysteines. This oxidation may trigger a conformational change in CodY, resulting in enhanced binding to DNA. A schematic model illustrating the contribution of TpxD and CodY to pneumococcal global transcriptional response to H2O2 is proposed.
Keywords: Streptococcus pneumoniae, TpxD, CodY, global gene regulation, H2O2
Introduction
Streptococcus pneumoniae commonly causes severe infections such as pneumonia, meningitis, and septicemia. More than 2 million deaths per year are caused as a result of pneumococcal septicemia (O'Brien et al., 2009). The pneumococcus has the ability to colonize and invade into different tissue sites with distinct environmental conditions; however, the mechanisms enabling its survival in the host are poorly understood. One of the detriments of pneumococcal survival in the host is its exposure to toxic levels of reactive oxygen species (ROS) such as H2O2, produced by the host immune cells. Furthermore, during aerobic growth the pneumococcus produces extremely high levels of H2O2, up to 2 mM (Pericone et al., 2003; Taniai et al., 2008). H2O2 increases the mutation rate, leads to DNA damage and apoptosis in lung cells and has damaging effects on many macromolecular compounds (Pericone et al., 2002, 2003; Rai et al., 2015). On the other hand, peroxide production was suggested to provide a competitive advantage to the pneumococcus against other colonizing bacteria in the upper respiratory tract (Pericone et al., 2000). H2O2 plays a pivotal role in pneumococcal pathogenesis through its cytotoxicity to epithelial cells (Duane et al., 1993), and through induction of genes required for host inflammatory response (Loose et al., 2014). Therefore, it is important to understand how the pneumococcus responds to this toxic compound.
S. pneumoniae lacks the common proteins that have been shown to protect against oxidative stress in other bacterial species, such as the known H2O2 scavengers, catalase and NADH-peroxidase (Hoskins et al., 2001; Tettelin et al., 2001). Nonetheless, previous studies have shown that S. pneumoniae uses other key enzymes to defend itself against oxidative stress, such as superoxide dismutase (Yesilkaya et al., 2000), NADH oxidase (Auzat et al., 1999), and alkyl hydroperoxidase (Paterson et al., 2006).
We have previously shown that the pneumococcal surface adhesion D gene, psaD, encodes a functional thiol peroxidase, which scavenges directly H2O2, and hence renamed it as tpxD (Hajaj et al., 2012). tpxD expression and synthesis were significantly up-regulated under aerobic compared to anaerobic conditions, and in the presence of exogenously added (10–1,000 μM) H2O2. In addition, H2O2 removal by catalase resulted in a drop in tpxD expression and synthesis (Hajaj et al., 2012).
Studies in other bacteria have shown that the change in thiol peroxidase expression in response to H2O2 is regulated by two transcription factors (TFs), OxyR and PerR (Hoskins et al., 2001; Paget and Buttner, 2003; Veal et al., 2007; Chiang and Schellhorn, 2012; Mishra and Imlay, 2012), but S. pneumoniae lacks these two TFs. However, the pneumococcus has a regulator named CodY, which regulates various metabolic pathways and cellular processes. The DNA binding activity of CodY in S. pneumoniae is increased by branched chain amino acids [BCAA: isoleucine, leucine and valine (ILV)] (Brinsmade et al., 2010). Notably, pneumococcal CodY does not respond to GTP (Hendriksen et al., 2008), unlike Bacillus subtilis, Staphylococcus aureus and several other bacterial species (Han et al., 2016). Inactivation of codY in S. pneumoniae dramatically decreased adherence to nasopharyngeal cells, an essential stage for successful pneumococcal infection (Hendriksen et al., 2008). No significant differences in bacterial loads were observed with the pneumonia and sepsis model of infection (Hendriksen et al., 2008). Although CodY has also been implicated in regulation of pneumococcal oxidative stress resistance through its role in iron metabolism (Caymaris et al., 2010; Johnston et al., 2015), the extent of CodY-mediated regulation of oxidative resistance has not been studied in detail.
This study aimed to reveal the mechanism controlling TpxD expression under H2O2 stress. We hypothesize that H2O2 activates a transcription factor which in turn up-regulates tpxD expression. We present data showing that H2O2 induces a global transcriptional response in S. pneumoniae and that TpxD enables this response by limiting H2O2 levels. Furthermore, we identified a conserved CodY-binding site (AATCATTGGAAAATT) in the putative regulatory region of tpxD, and present mechanistic evidence that CodY serves as an activator of tpxD, thereby preventing toxic levels of H2O2.
Materials and methods
Bacterial strains and growth condition
S. pneumoniae serotype 2 strain, D39 (WT), and its isogenic mutants were used in this work. Routinely, pneumococci were grown anaerobically in closed, completely filled 20 ml test-tubes, in a water bath without shaking, at 37°C in Todd–Hewitt (TH) broth with 0.5% (w/v) yeast extract (THY) to an approximate OD620 of 0.25. Where specified, bacteria were grown in chemically defined medium (CDM) (Yesilkaya et al., 2009), with the exception that BCAA were omitted from the amino acids mixture, and added separately. D39 strains mutated in tpxD (ΔtpxD) and brnQ (ΔbrnQ), were grown in medium supplemented with 100 μg ml−1 spectinomycin. The 6 ΔtpxD complemented strains 1–6 (ΔtpxDcomp1-6; Figure 2), were grown in medium supplemented with 100 μg ml−1 spectinomycin and 250 μg ml−1 kanamycin. The ΔcodY mutant (kindly donated by Dr. Calum Johnston, Laboratoire de Microbiologie et Genetique Moleculaires, France) was grown in medium supplemented with 20 μg ml−1 trimethoprim.
Construction of genetically modified strains
The construction of ΔtpxD and its genetically complemented strain containing the entire tpxD-coding sequence (SPD1464) plus 83 bp upstream, was described previously (Hajaj et al., 2012). The brnQ gene disruption was achieved by allelic replacement mutagenesis as described previously (Song et al., 2005). Briefly, the upstream and downstream flanking regions of brnQ were amplified using the primer combinations LFSPD0546F/LFSPD0546R, for the upstream region, and RFSPD0546F/RFSPD0546R, for the downstream region, respectively (Table S1). Subsequently, using LFSPD0546F and RFSPD0546R primers, the flanking fragments were fused to the spectinomycin resistance marker, which had been amplified with specF and specR primers from pDL278 (Yesilkaya et al., 2000). The resulting fused fragments were purified and transformed into S. pneumoniae D39 as described before using competence stimulating peptide (Alloing et al., 1996). The transformants were selected on blood agar base (BAB) containing 5% (v/v) sheep blood supplemented with spectinomycin. The mutation was confirmed by PCR using different primer combinations, and DNA sequencing.
To construct the 6 genetically cis-complemented strains (ΔtpxDcomp1-6), the chromosomal region containing the entire tpxD-coding sequence preceded by a variable length of the proximal upstream sequence harboring different parts of tpxD putative regulatory elements, was amplified using the appropriate primers (Table S1), which were designed to include NcoI or BamHI sites. The amplicons were purified (Qiagen), digested and cloned into similarly digested pCEP (Guiral et al., 2006). An aliquot of ligation mixture was transformed into E. coli BL21 (DE3), and kanamycin resistant transformants were analyzed for the presence of recombinant plasmid using primers, malF and pCEPR, that anneal to either side of the cloning site. Then the purified recombinant plasmid was transformed into ΔtpxD as described previously (Alloing et al., 1996). The successful construction of modified strains was then confirmed by PCR using malF and pCEPR primers, and DNA sequencing.
Point mutation of TpxD catalytic cysteine58
The replacement of cys58 to serine in TpxD was done by splicing overlap extension (Song et al., 2005). For this, the region surrounding cys58 was amplified using malF and TPX-S1, and pCEPR and TPX-S2 primers (Table S1). The amplicons were then fused together using malF and pCEPR primers. The fused products were digested with BamHI and NcoI, and ligated into the same sites of pCEP. The replacement of nucleotide sequence was confirmed by sequencing using malF and pCEPR primers, and the recombinant plasmid was transformed into ΔtpxD as before. The resulting strain was designated as TpxDC58S.
Relative gene expression
Bacteria were grown anaerobically at 37°C in THY medium to OD620 = 0.25 and then challenged with 100, 250 or 1000 μM H2O2 for 40 min while maintaining anaerobic conditions. Control cultures were incubated with Double-distilled water instead of H2O2. Viability of WT and ΔtpxD bacteria, challenged with 1 mM H2O2, was confirmed by colony forming unit counts, as previously described (Hajaj et al., 2012). Experiments were repeated at least twice, each with two biological replicates. RNA was prepared by using MasterPureTM RNA purification kit (Epicentre®, USA). RNA was treated with DNase I according to the above kit protocol. cDNA was synthesized with VersoTM cDNA kit (ABgene, UK). Real-time PCR reactions contained AbsoluteTM blue QPCR SYBR mix ROX (ABgene, UK), and the expression of individual genes was determined using gene specific primers (Table S1). The transcription level of target genes was normalized to gyrA. The results were analyzed by the comparative CT method (Livak and Schmittgen, 2001).
DNA microarray analysis
D39 and its isogenic ΔtpxD were grown anaerobically at 37°C in THY medium to OD620 = 0.25 and then challenged with 1 mM H2O2 for 40 min while maintaining anaerobic conditions. Control cultures were incubated with Double-distilled water instead of H2O2. All other procedures regarding DNA microarray experiments (RNA isolation, RNA quality test, cDNA synthesis and labeling) were performed as described (Shafeeq et al., 2011). Microarray slide images were scanned using GenPix Pro 6.1 (MSD analytical technologies). Processing and normalization (LOWESS spotpin-based) of slides was done with the in-house developed MicroPrep software. DNA microarray data were obtained from three independent biological replicates hybridized to glass slides with a dye swap. Expression ratios were calculated from the measurements of at least seven spots. Differential expression tests were performed on expression ratios with a local copy of the Cyber-T implementation of a variant of the t-test. False discovery rates were calculated as described (van Hijum et al., 2005). A gene with p-value of < 0.005 and a fold change cut-off of 1.8 was considered differential expressed. We have used the Cyber-T server to analyze our Microarray data. Therefore, the p-values mentioned in this study (Supplementary Material) should be considered as Bayes corrected p-values.
Microarray data have been submitted to Gene Expression Omnibus (GEO) database under the accession number GSE65157.
Expression, purification, and antibody production of TpxD
The coding sequence of tpxD was amplified by PCR with tpx-for and tpx-rev primers (Table S1) from S. pneumoniae D39 and cloned into pRSETc vector (Invitrogen) between XhoI and KpnI sites. E. coli BL21 cells harboring the constructed plasmid were grown in Luria-Bertani broth supplemented with ampicillin (100 μg/ml) for 24 h. Cells were harvested by centrifugation and stored at −70 °C. The pellet was suspended in lysis buffer [50 mM Tris pH 8, 100 mM phenylmethylsulfonyl fluoride (PMSF)], disintegrated by sonication, and centrifuged at 4000 × g for 1 h. Proteins in the supernatant were loaded onto a Ni-NTA column (Adar biotec, Israel), and incubated for 1 h at 4°C. The column was then washed with 10 mM imidazole, and the recombinant protein eluted from the column with 100 mM imidazole. The eluted protein was run on SDS-PAGE and a band which was of the predicted molecular weight of TpxD was visualized. Polyclonal antibodies against TpxD were custom made by Sigma Aldrich, Israel, using the purified recombinant TpxD.
SDS-PAGE and western blotting
Pneumococci were lysed and subjected to SDS-PAGE. Separated proteins were electroblotted onto 0.45 μm nitrocellulose membrane (Bio-Rad), and probed with the polyclonal antibodies against TpxD. Antigen complexes were detected using Peroxidase Affinity Pure Goat anti rabbit IgG (Jackson ImmunoResearch) and visualized with SuperSignal West Pico Chemiluminescent substrate (Pierce). Densitometry was performed using ImageJ software (Schneider et al., 2012).
Computational analysis of tpxD promoter region and regulatory sites
The sequence of tpxD upstream region in D39 was extracted from the National Center for Biotechnology Information (NCBI) database using the following entries: tpxD sequence gene ID: 4441865. Streptococcus pneumoniae D39 strain sequence ID: NC_008533.1. Promoter elements were predicted using the Softberry BPROM algorithm of bacterial promoters (http://www.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb). Known CodY-regulated genes in S. pneumoniae were retrieved from the RegPrecise database (Novichkov et al., 2010), and their CodY binding site aligned with the upstream sequence of tpxD, using ClustalW in BioEdit 7.0.9 (Hall, 1999). CodY binding motif was visualized using WebLogo 3.2 (Lewis et al., 2000).
Cloning, expression and purification of CodY
The coding sequence of codY was amplified by PCR from Genomic DNA from D39 strain with CodYF and CodYR primers (Table S1). The amplicons were cloned into plasmid pLEIC using In-fusion Cloning kit (Clontech), which allows ligation independent cloning method based on the annealing of complementary ends of a cloning insert and linearized cloning vector. The recombinant construct was transformed into Fusion-Blue competent E. coli, and transformants were plated on LA containing 100 μg/ml ampicillin. A transformant was selected for sequencing to rule out any mutation. The construct DNA then was transformed into E. coli BL21(DE3) (Novagen, United Kingdom) for recombinant protein expression. The strain with the recombinant plasmid was grown in Luria-Bertani broth containing ampicillin (100 μg/ml) for 16 h at 25°C, and the expression was induced with 0.5 mM Isopropyl β-D-1-thiogalactopyranoside. The pellet was collected by centrifugation, resuspended in lysis buffer (50 mM Tris pH 8, 100 mM PMSF), and subjected to sonication. The protein purification used immobilized metal affinity chromatography (IMAC) resin and non-denaturing conditions as instructed by the manufacturer (Clontech). The column was washed with 20 mM imidazole and the recombinant protein was eluted using 400 mM imidazole. The identity of the protein was verified as previously described (Kazakov et al., 2007) by matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) mass spectrometric analysis of tryptic digests of the products by the Protein nucleic acid chemistry laboratory at the University of Leicester.
Electrophoretic mobility shift assay (EMSA)
Fluorescently labeled DNA probe representing the putative promoter region of tpxD was generated using Spd1464E1FAM and Spd1464E2 primers (Table S1), and S. pneumoniae D39 genomic DNA. After amplification, the probe DNA was gel purified. The binding reaction was set up by mixing a constant amount of DNA promoter probe (10 nM), and increasing amounts of purified and dialyzed His-tagged recombinant CodY (0–1 μM) in 5X binding buffer (20 mM Tris-HCl pH 7.5, 30 mM KCl, 1 mM dithiothreitol (DTT), 1 mM ethylenediaminetetraacetic acid (EDTA) pH 8.0 and 10% glycerol). When required, 2 mM H2O2 and BCAA (isoleucine 1.62 mM, leucine 3.48 mM, and valine 2.77 mM) were also added into the binding buffer 20 min after addition of the DNA probe. The binding reaction was incubated at room temperature for 30 min in a total volume of 20 μl. Then, the reaction mixture was subjected to non-denaturing PAGE (8%) for 40 min at 200 V. DNA-protein complexes were visualized using a Typhoon Trio+ scanner (GE Healthcare Life Sciences) with a 526 nm short-pass wavelength filter. The image was analyzed using LI-COR image analysis software.
Determination of CodY cysteines redox state by mass spectrometry
D39 were grown under anaerobic conditions to OD620 = 0.25 and then challenged with 1 mM H2O2 for 40 min while maintaining anaerobic conditions. Alkylation was performed as previously described (Hajaj et al., 2012). Briefly, in vivo reduced protein-thiols were blocked by resuspending phosphate buffered saline washed bacteria in urea buffer (0.1 M Tris pH 8.2, 1 mM EDTA, 8 M urea) containing 32.5 mM iodoacetic acid (IAA). Samples were then treated with DTT (3.5 mM) and incubated with iodoacetamide (IAM) (10 mM). Following alkylation, samples were resolved on 18% (w/v) SDS-PAGE, stained with Coomassie blue, and a band corresponding to ~29.5 kDa cut from the gel, reduced with 3 mM DTT (60°C for 30 min), and modified with 10 mM IAM in 100 mM ammonium bicarbonate (in the dark, room temperature for 30 min). The modified protein was then digested with modified trypsin (Promega) at a 1:10 enzyme-to-substrate ratio, overnight at 37°C. The resulting peptides were resolved by reverse-phase chromatography on 0.075 × 200-mm fused silica capillaries (J&W) packed with Reprosil reversed phase material (Dr Maisch GmbH, Germany). Mass spectrometry was performed by a Q-Exactive plus mass spectrometer (Thermo) in a positive mode using repetitively full MS scan followed by High energy Collision Dissociation (HCD) of the 10 most dominant ion selected from the first MS scan. The mass spectrometry data was analyzed using Proteome Discoverer 1.4 software Using Sequest (Thermo) algorithm searching against the Streptococcus pneumoniae D39 proteome from Uniprot and specific database. Semi quantitation was done by calculating the peak area of each peptide based its extracted ion currents (XICs). The area of the protein is the average of the three most intense peptides from each protein.
Results were filtered with 1% false discovery rate.
Statistical analysis
The significance of differences was determined by the unpaired t-test. p < 0.05 was considered significant.
Safety statement
For research involving biohazards, correct standard procedures have been carried out.
Results
TpxD is involved in the pneumococcal global response to H2O2
In a previous study (Hajaj et al., 2012), we showed that TpxD is an efficient H2O2 scavenger, and that its expression is up-regulated under aerobic compared to anaerobic growth conditions. In order to isolate the effect of H2O2 from other ROS formed under aerobic environment, D39 and ΔtpxD were challenged with sub-lethal H2O2 concentrations under anaerobic conditions. Similar viability, confirmed by colony forming unit counts, was measured in WT and the mutant cells challenged with 1 mM H2O2. Microarray profiling of D39 challenged with 1 mM H2O2 revealed that 217 genes were differentially expressed by a factor of 1.8 or more compared to unchallenged bacteria: 146 were down-regulated and 71 were up-regulated (Table S2). When the ΔtpxD mutant was challenged with 1 mM H2O2, global transcriptional response was not observed (Table S3). Classification of the genes affected by H2O2 in the WT into functional categories revealed that the most prominent differentially expressed genes included those associated with amino acid transport and metabolism (Table S4).
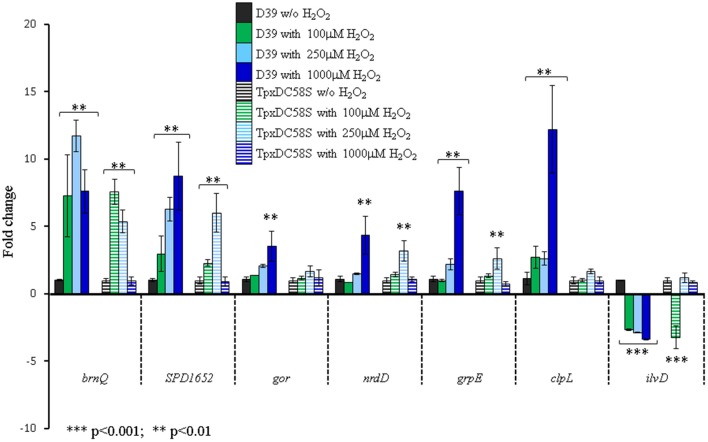
It was shown in eukaryotes that thiol peroxidases serve as transcriptional regulators of global gene expression in response to H2O2, in addition to enzymatic removal of H2O2 (Crooks et al., 2004). The finding that ΔtpxD response to H2O2 was significantly lower than that of the WT strain could originate from bacterial inability to respond to 1 mM H2O2 in the absence of TpxD scavenging activity. Alternatively, the low response could be due to TpxD role as a global transcriptional regulator in S. pneumoniae. Hence, we constructed a mutant in which the catalytic activity was disrupted by replacing the peroxidatic cys58 by serine (TpxDC58S). We selected 7 genes, whose expression was affected by 1 mM H2O2 in the WT but not in ΔtpxD mutant and checked their transcription levels in TpxDC58S. As shown in Figure 1, the expression levels of these genes were unaffected by 1 mM H2O2 in TpxDC58S, unlike the WT strain. However, when lower H2O2 concentrations (100 and 250 μM) were applied, TpxDC58S mutant could respond in a way similar to that of the WT strain (Figure 1). These data suggest that TpxD scavenging activity is crucial for pneumococcal global response to increased H2O2 levels. The fact that a transcriptional response to 100 and 250 μM H2O2 was induced in cells harboring inactive TpxD (TpxDC58S), rules out the possibility that TpxD serves as a transcriptional regulator.
Figure 1.
TpxD scavenging activity is crucial for pneumococcal global response to high H2O2 levels. Seven genes identified in the microarray to be affected by 1 mM H2O2 were selected for further examination by relative RT-PCR. D39 and TpxDC58S mutant were grown anaerobically to OD620 = 0.25 and then challenged with 100, 250, or 1000 μM H2O2 for 40 min. Control cultures were grown without (w/o) the addition of H2O2. Values are the mean of duplicate determinations of at least two independent experiments, and were normalized to the expression level of the relevant unchallenged strain (D39 or TpxDC58S). Bars indicate standard deviation. Significant alterations in gene expression compared to bacteria grown without H2O2 were determined using Student's t-test.
Identification of TpxD promoter elements
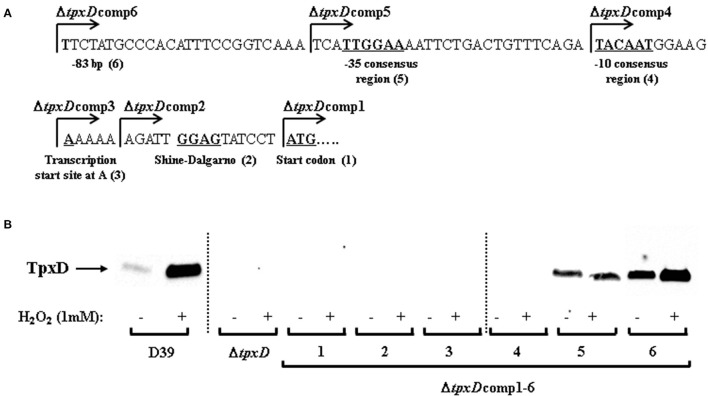
We have shown that TpxD expression and synthesis were significantly increased following a challenge with H2O2 (Hajaj et al., 2012). To discover the specific DNA-elements responsible for TpxD synthesis and regulation by H2O2, in silico tools were applied. BPROM algorithm predicted 5 core elements upstream of tpxD-coding sequence (Figure 2A). Based on these predictions, we constructed 6 genetically modified strains, in which ΔtpxD was cis-complemented with an intact copy of tpxD-coding sequence, preceded by a variable length of tpxD proximal upstream region: ΔtpxDcomp1 harboring only tpxD-coding sequence; ΔtpxDcomp2 harboring the sequence starting from the predicted Shine-Dalgarno element and up to and including tpxD-coding sequence; ΔtpxDcomp3 harboring the sequence starting from the predicted transcription start site and up to and including tpxD-coding sequence; ΔtpxDcomp4 and 5 harboring the sequence starting from the predicted −10 and −35 consensus regions of the promoter, respectively, and up to and including tpxD-coding sequence; ΔtpxDcomp6 harboring tpxD-coding sequence plus a region of 83 bp upstream of the gene (Figure 2A). Lysates of ΔtpxDcomp1-6 strains, grown anaerobically, were subjected to Western blotting using polyclonal antibodies against TpxD. No TpxD-band was seen in ΔtpxDcomp1-4 strains (Figure 2B). In ΔtpxDcomp5, basal TpxD protein level was detected, indicating that the Shine-Dalgarno, transcription start site, −10 and −35 consensus regions are essential for TpxD synthesis. However, ΔtpxDcomp5 did not show any up-regulation in response to 1 mM H2O2compared to the WT strain (Figure 2B). Only ΔtpxDcomp6 showed a significant increase in TpxD synthesis following a challenge with H2O2as measured by densitometry: 1.70 ± 0.13-fold, p < 0.05 (Figure 2B). These results point out that the regulatory elements essential for tpxD up-regulation by H2O2 are located within the immediate 83 bp upstream of tpxD-coding sequence.
Figure 2.
Identification of tpxD regulatory promoter elements. (A) In silico analysis of tpxD proximal promoter region. Identification of tpxD promoter elements using BPROM algorithm: ATG start codon (1); Shine Dalgarno (2); transcription start site at A (3); −10 and −35 elements of the σ70 promoter (4 and 5, respectively). The first nucleotide included in each of the ΔtpxDcomp mutants (1–6) is indicated by arrow. (B) Influence of tpxD putative regulatory elements on TpxD expression levels under H2O2 stress. TpxD levels, determined by Western blotting, were measured in D39, ΔtpxD and ΔtpxDcomp1-6 (described in Figure 2A). Bacteria were grown anaerobically to OD620 = 0.25 and then challenged with 1 mM H2O2 for 40 min. Separated proteins were electroblotted onto 0.45 μm nitrocellulose membrane, and stained with Ponceau S to ensure equal loading, before incubation with antibodies. Membranes were exposed for long time intervals to ensure the absence of bands in ΔtpxDcomp1-4 and are lined up, as indicated in the figure. Densitometry was performed using ImageJ software.
CodY binding motif in the proximal upstream region of tpxD
The next step was to identify a candidate TF responsible for tpxD up-regulation under H2O2 stress. To this end, we clustered the genes, shown in the microarray to be affected by H2O2, according to their specific regulator, by using RegTransBase database (Fomenko et al., 2011). Clustering was done solely to highlight a potential TF, being aware of the fact that H2O2 invokes a general stress response, involving many transcription factors and genes, making it very hard to distinguish direct from indirect effects of H2O2. This analysis yielded 14 TF-clusters covering 60/217 of the genes found by the microarray to be significantly affected by 1 mM H2O2 (Table 1). It should be noted that 16 of the 60 genes were classified in 2 TF-clusters (based on RegTransBase database) and that the regulon grouping of some genes is controversial.
Table 1.
Clustering of genes significantly affected by 1 mM H2O2, by at least a factor of 1.8, according to their specific TF.
| Transcription factor | D39 locus taga | No of genes |
|---|---|---|
| Transcriptional repressor, CodY | SPD_1412 | 21 |
| Catabolite control protein A, CcpA | SPD_1797 | 12 |
| DNA-binding response regulator, YycF | SPD_1085 | 10 |
| Transcriptional regulator, MarR family protein, FabT | SPD_0379 | 6 |
| DNA-binding response regulator | SPD_0344 | 5 |
| DNA-binding response regulator, CiaR | SPD_0701 | 5 |
| Transcriptional regulator, MerR family protein | SPD_0447 | 3 |
| Iron-dependent transcriptional regulator, PsaR | SPD_1450 | 3 |
| Heat-inducible transcription repressor, HrcA | SPD_0458 | 3 |
| Transcriptional regulator, CtsR | SPD_2023 | 2 |
| adc operon repressor, AdcR | SPD_2000 | 2 |
| Transcriptional regulator, NrdR | SPD_1523 | 2 |
| Arginine repressor, ArgR | SPD_1904 | 1 |
| Maltose operon transcriptional repressor, MalR | SPD_1938 | 1 |
Gene number refers to D39 locus tags.
We then used ClustalW to check whether tpxD upstream sequence contains a putative binding site for one of the TFs reported in Table 1. This analysis identified a CodY-binding site of 15 bp, between −26 and −40, upstream of tpxD-transcription start site (AATCATTGGAAAATT) which is highly homologous with CodY-binding motifs of pneumococcal genes known to be under CodY regulation (Figure 3). This putative binding site shares 73% homology with CodY-consensus sequence in Lactococcus lactis (den Hengst et al., 2005; Guedon et al., 2005).
Figure 3.
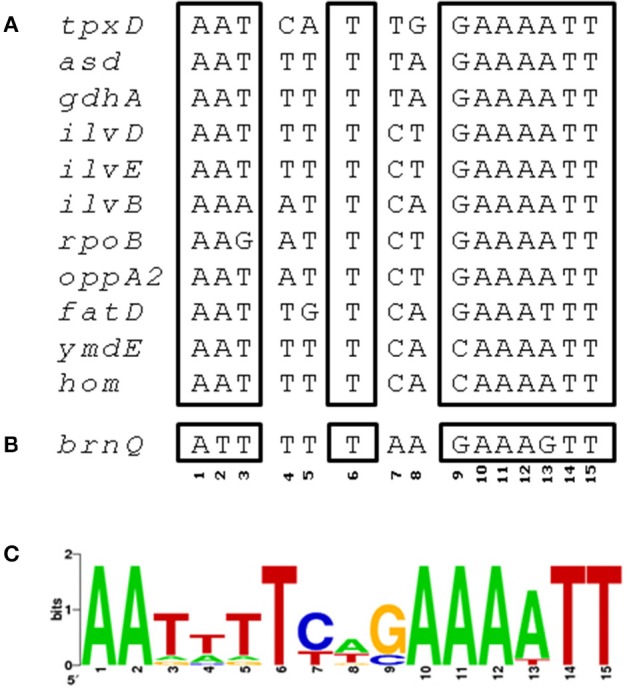
CodY binding site in tpxD promoter region. (A) ClustalW was used to align the 83 bp upstream to tpxD-coding sequence with several pneumococcal genes known to be regulated by CodY. A region of 15 bp, between −26 and −40, upstream to tpxD-transcription start site, with high homology to CodY binding motif in L. lactis, is presented. (B) Putative CodY binding site in the upstream region of brnQ. (C) Position specific weight matrix for S. pneumoniae CodY binding motif deduced from CodY-regulated, established and putative genes (Figure 3A), identified by bioinformatic analysis of the microarray data, using RegPrecise database. The matrix (created at http://weblogo.berkeley.edu/) shows the relative frequency of occurrence of each of the 4 nucleotides (A, T, C, G) at each of the 15 positions of the motif.
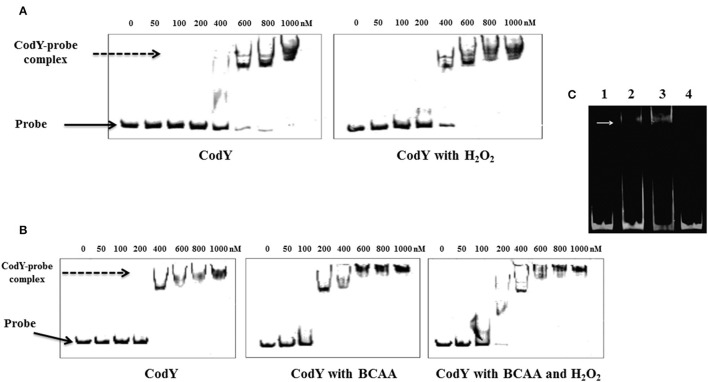
We then expressed and purified the pneumococcal CodY protein with the intent to test its in vitro binding to the motif upstream of tpxD-coding sequence. EMSA results showed indeed that CodY interacts with the DNA probe containing this region in a concentration dependent manner (Figure 4). CodY binding to tpxD upstream region was specific, first, as we demonstrated previously that CodY does not bind to the psaR promoter, which is known not to be regulated by CodY (Hendriksen et al., 2008). Second, we deleted the putative CodY binding sequence in the upstream region of tpxD, and found that the binding was abolished (Figure 4C). The effect of H2O2 on CodY binding to the motif upstream of tpxD-coding sequence was checked by the incubation of CodY with 2 mM H2O2 prior to the addition of target DNA. This experiment resulted in increased CodY affinity to its target: 400 nM were sufficient for CodY binding to DNA in the presence of H2O2 whereas 600 nM were needed without H2O2 (Figure 4A). CodY-DNA binding activity was shown to be increased by BCAA (Brinsmade et al., 2010). When the reaction mixture was supplemented with BCAA, DNA binding was observed already at 200 nM. In this case addition of H2O2 had no impact (Figure 4B). These data indicate that the 15 bp sequence serves as CodY binding site. Furthermore, H2O2 increases CodY binding to its DNA target under conditions where BCAA are not present.
Figure 4.
CodY interaction with the regulatory region of tpxD. EMSA assay was used to confirm the presence of CodY binding site in tpxD regulatory region: Effect of H2O2 (A) and BCAA (B). 10 nM of fluorescently labeled DNA probe was mixed with increasing concentration of recombinant CodY, and incubated for 30 min at room temperature. When required, 2 mM H2O2 and/or BCAA (isoleucine 1.62 mM, leucine 3.48 mM, and valine 2.77 mM) were added into the binding buffer. DNA-protein complexes were separated by electrophoresis on non-denaturing PAGE (8%). In all gels, Lane 1 contains DNA probe alone, indicated with a black arrow. DNA-protein interaction is seen as an upward shift of DNA probe, indicated with a dotted arrow. (C) Lane 1, 50 ng approximately of the 81 bp DNA fragment alone, lane 2 and 3, DNA plus 0.6- and 1-mM recombinant CodY, respectively. Lane 4, DNA probe, excluding 15 bp putative CodY binding site plus 1 mM recombinant CodY. White arrow indicates the position of DNA-protein complex. EMSA was performed using Molecular Probes fluorescence-based EMSA kit (Invitrogen). Briefly, 5X FY binding buffer (20 mM Tris-HCl pH 7.5, 30 mM KCl, 1 mM DTT, 1 mM EDTA pH 8.0 and 10% v/v glycerol) was prepared to incubate the promoter probe and protein. The binding reaction was set up by mixing a constant amount of target promoter probe (~30 ng), and increasing amounts (0.1–0.5 μM) of purified and dialyzed His-tagged protein. The binding reaction was incubated at room temperature for 20 min in a total volume of 20 μl, and then analyzed on an 8% w/v non-denaturing polyacrylamide gel. After electrophoresis, gels were stained with SYBR® Green EMSA gel stain (Invitrogen) and visualized using a Typhoon Trio+ scanner (GE Healthcare Life Sciences) with a 526 nm short-pass wavelength filter.
Effect of CodY-mutation on tpxD expression
Our attempts to inactivate codY were unsuccessful, probably due to its essentiality. Hence we used a codY-deletion mutant harboring suppressing mutations inactivating the fatC and amiC genes (Caymaris et al., 2010). To elucidate the influence of CodY on TpxD expression, bacteria were grown in CDM without BCAA. Under anaerobic conditions, tpxD expression was significantly reduced in ΔcodY compared to its wild type, 2.43 ± 0.25-fold reduction, indicating that CodY is an activator of tpxD, even without BCAA. A 4.00 ± 0.12-fold reduction in tpxD expression was measured when ΔcodY cells were challenged with 1 mM H2O2. Hence we conclude that CodY is required for tpxD up-regulation under high H2O2 levels.
Effect of BCAA on tpxD expression
To elucidate the role of BCAA in CodY regulation of TpxD expression and synthesis, D39 were grown in the absence of BCAA (Figure 5). This experiment revealed that BCAA had no effect on the basal level of TpxD under anaerobic growth conditions. In addition, TpxD up-regulation by H2O2 was less pronounced in bacteria grown without BCAA. These data indicate that BCAA are required for CodY-mediated, TpxD up-regulation by H2O2. We then checked whether BCAA itself affect tpxD expression in a mechanism independent of CodY. To this end, tpxD expression was assessed in ΔcodY grown with or without BCAA and challenged with H2O2. This comparison revealed that the presence of BCAA was accompanied by a 2.27 ± 0.20-fold reduction in tpxD expression in ΔcodY, suggesting that in the absence of CodY, BCAA interfere with tpxD expression in as yet unresolved mechanism.
Figure 5.
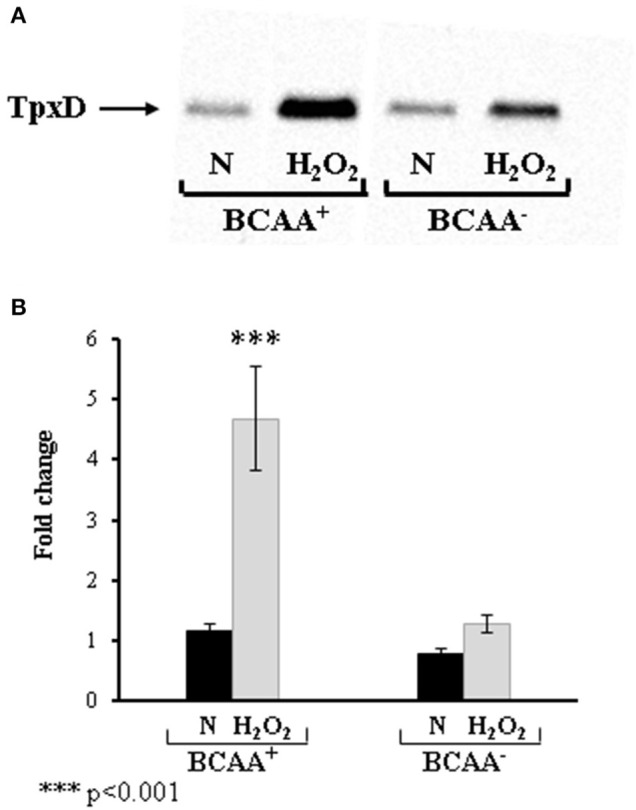
Effect of BCAA on TpxD expression in bacteria challenged with H2O2. TpxD synthesis and expression levels were determined by Western blotting (A) and by RT-PCR (B) in D39 grown anaerobically (N) in CDM with (BCAA+) or without (BCAA-) BCAA. Bacteria were grown to OD620 = 0.25 and then challenged with 1 mM H2O2 for 40 min. Control cultures were incubated without the addition of H2O2. Separated proteins were electroblotted onto 0.45 μm nitrocellulose membrane, and stained with Ponceau S to ensure equal loading, before incubation with antibodies. Values are the mean of duplicate determinations of at least two independent experiments, and were normalized to the expression level of D39 grown anaerobically in medium containing BCAA. Bars indicate standard deviation. Significant alterations in gene expression compared to bacteria grown without H2O2 were determined using Student's t-test.
The BCAA transporter brnQ is a putative CodY regulated gene
BCAA intracellular level is determined by their biosynthesis through the ilv operon (Brinsmade et al., 2010) and by their transport through specific transporters (Belitsky, 2015). The microarray data showed that the BCAA- biosynthetic enzymatic machinery (ilv) was down-regulated following a challenge with H2O2 (Table S2). Concomitantly, the expression of brnQ, a BCAA transport system II carrier protein, showed a remarkable increase, 7.6-fold, following a challenge with 1 mM H2O2 (Figure 1), probably to compensate for the decrease in BCAA biosynthesis. Noteworthy is the fact that a ΔbrnQ mutant could not overcome a challenge with 0.5–1.0 mM H2O2 (data not shown), indicating the importance of BCAA for oxidative stress resistance. ClustalW analysis revealed a putative CodY binding site in the upstream region of brnQ (Figure 3). Taken together the fact that CodY functions as a transcriptional repressor of the ilv operon (Hendriksen et al., 2008) on the one hand, and our the finding of a putative CodY binding site upstream of brnQ coding sequence on the other, implies that CodY is involved in maintaining homeostatic levels of BCAA in the cell.
H2O2 specifically oxidizes CodY cysteines
Microarray data showed that codY expression was increased by 1.8-fold in the presence of 1 mM H2O2 (Table S2). Validation by RT-PCR and Western blotting showed no significant change in CodY expression (Figure S1). This led us to conclude that CodY regulation by H2O2 is carried out by a different mechanism. Hence we checked the effect of H2O2 on the redox state of CodY-cysteines by a thiol double trapping method followed by mass spectrometry. Our data show that under anaerobic conditions, the 2 CodY-cysteines were always in the reduced state, as the pick area of the oxidized, IAM-alkylated cysteines, was zero. Following a challenge with H2O2, the pick area of the reduced, IAA-labeled cysteines, was 13E7 whereas that of the oxidized, IAM labeled cysteines, was 3.75E7. These data indicate that H2O2 triggers the oxidation of the 2 cysteines in about 1/4 of CodY molecules, which might influence CodY activity.
Discussion
Hydrogen peroxide is well known as an oxidant that can react with various cellular targets. At low levels, H2O2 functions as a signaling agent, whereas at high levels, H2O2 inhibits the growth of bacteria and induces cell death (Imlay and Linn, 1986; Johnston et al., 2015). Homeostatic control of this oxidant is maintained by several oxidant-scavenging enzymes. We have previously shown that tpxD encodes a functional thiol peroxidase, involved in H2O2 homeostasis in S. pneumoniae (Hajaj et al., 2012). In the current study we show that pneumococcal response to high H2O2 levels depends on TpxD, and that tpxD expression is regulated by the global transcription factor CodY.
Microarray analysis revealed a global transcriptional response to high H2O2 concentration in the WT but not in ΔtpxD-null cells. To verify whether this global response depends on TpxD activity, the peroxidatic cysteine58 was replaced by serine, TpxDC58S. Similar substitution of the peroxidatic Tpx-cysteine in E. coli resulted in an inactive enzyme (Baker and Poole, 2003), and in a peroxide-sensitive phenotype in Enterococcus faecalis (La Carbona et al., 2007). Based on the sequence similarity between S. pneumoniae TpxD and E. coli Tpx (56.9%) as well as between S. pneumoniae TpxD and E. faecalis Tpx (55.1%), and their established H2O2 scavenging activity, we surmise that the catalytic activity of TpxDC58S is ablated. The expression of 7 genes, shown to be affected by 1 mM H2O2 in the WT but not in ΔtpxD mutant, was checked in TpxDC58S. These genes showed no response to 1 mM H2O2, but were differentially expressed upon exposure to 100 and/or 250 μM H2O2. These findings indicate that TpxD scavenging activity is required for the bacterial response to increased H2O2 concentrations. Of note is the fact that H2O2 can be reduced to OH· by Fe+2 via the Fenton reaction under both aerobic and anaerobic conditions (Henle et al., 1996). Hence, some of the changes in gene expression could originate from the effect of OH· rather than H2O2. The effect of the Fenton reaction on global gene expression needs further investigation.
In a previous study (Hajaj et al., 2012) we found that TpxD expression is modulated in accordance with H2O2 levels: in a WT strain recovered from the nasopharynx of mice following intranasal infection, tpxD expression level was higher than that measured in bacteria recovered from the less aerated niche of the blood, in agreement with the in vitro expression levels in cells grown under aerobic vs. anaerobic conditions or challenged with H2O2. A recent publication (Lisher et al., 2017) demonstrated that tpxD level is increased by 6.8 following growth with limited aeration compared with growth under anaerobic condition. These authors show that TpxD plays a major role in preventing endogenous protein sulfenylation by H2O2, reinforcing our conclusion that TpxD is required for the bacterial response to H2O2.
Bioinformatic tools pointed toward CodY as a potential TF mediating the cellular response to H2O2, since expression of 21 genes known to be under its regulation were repressed/activated (Table S5). CodY is a highly conserved global regulator in many low G+C Gram positive bacteria. Targets of CodY include operons involved in amino acid metabolism, particularly BCAA, as well as in proteolysis and peptide uptake and degradation (Geiger and Wolz, 2014). CodY is also a repressor of the iron transport operon fatD–fatC–fecE–fatB and the Ami-Obl oligopeptide transporter (Hendriksen et al., 2008), and an activator of the essential iron storage-peroxide resistance Dpr protein (Pericone et al., 2003). Our attempts to inactivate codY without disrupting additional genes were unsuccessful, in line with the data published by Caymaris et al. (2010): codY inactivation results in derepression of iron uptake by the two mentioned transporters and depletion of iron storage by Dpr, leading to a severe oxidative stress (Hendriksen et al., 2008). Consequently, simultaneous mutations in fatC and amiC arises to prevent the accumulation of high levels of iron, and thereby limit the formation of reactive oxygen intermediates through the Fenton reaction (Caymaris et al., 2010; Johnston et al., 2015). We therefore used a codY-deletion mutant, possessing suppressing mutations in fatC and amiC, and found that CodY is an activator of tpxD. A putative CodY binding site was found in the proximal region of tpxD-coding sequence. Genetic engineering techniques and EMSA assays confirmed the presence of an active CodY binding site. This site overlaps the −35 consensus region of tpxD, suggesting that CodY belongs to Class II activators, which bind to sites that overlap the target promoter −35 region, and in most cases activate transcription by making a direct interaction with domain 4 of the RNA-polymerase subunit (Dove et al., 2003). H2O2 Increased CodY binding to tpxD regulatory sequence, indicating that H2O2 positively regulates tpxD. Streptococcus mutans tpx was also found to be regulated in a positive manner, although not by CodY but by the oxidative stress regulator SpxA1(Kajfasz et al., 2015).
BCAA are known to enhance CodY binding to DNA (Brinsmade et al., 2010). BCAA intracellular level is determined by their biosynthesis through the ilv operon (Brinsmade et al., 2010) and by their transport through specific transporters (Belitsky, 2015). The biosynthetic pathway can be affected by oxidative stress; in particular, the [4Fe-4S] prosthetic group of IlvD has been shown to react with H2O2 in E. coli (Flint et al., 1993). Henard et al. suggested that oxidation of the redox active center of IlvD in Salmonella is expected to decrease the biosynthesis of BCAA (Henard et al., 2010). If this is the case in S. pneumoniae, it may explain the down-regulation of ilvD as well as additional ilv genes by CodY, presumably to prevent the expression of genes belonging to a pathway that will subsequently undergo oxidation, and thus will be inactive. Bioinformatic analysis identified a putative CodY binding site in the upstream region of the BCAA transporter, brnQ, similar to that found in B. subtilis (Belitsky, 2015), suggesting that CodY controls both BCAA biosynthesis and transport, to ensure a steady supply of these essential amino acids. The reciprocity between CodY and BCAA constitutes an indirect auto-regulatory loop that allows the fine-tuning of BCAA level and CodY activity in S. pneumoniae, as published before for B. subtilis (Belitsky, 2015). These authors have shown that the basal level of BCAA achieved by intracellular synthesis supports a low level of CodY activity that is sufficient for partial regulation of some genes. However, maximal CodY activity is observed only in the presence of BCAA-containing amino acid mixtures. Thus, uptake of exogenous BCAA is critical for attaining conditions favoring CodY activation, and the efficiency of such uptake is likely to determine the level of CodY activation (Belitsky, 2015). However, in Listeria monocytogenes CodY was shown to be active even when bacteria were starved for BCAA (Lobel and Herskovits, 2016). These authors suggest that additional factors are involved in mediating CodY binding to its DNA targets. In line with this are our EMSA results showing that H2O2 enhances CodY binding to tpxD- target DNA in the absence of BCAA, resulting in tpxD up-regulation. CodY harbors a CXXC motif, thought to facilitate disulfide bond formation when exposed to an oxidizing environment (Kajfasz et al., 2015). We show that H2O2 specifically oxidizes these 2 adjacent cysteines and speculate that CodY-cysteines oxidation triggers a conformational change that ultimately facilitates CodY interaction with its DNA binding site. The finding that only 1/4 of CodY molecules undergo oxidation at high H2O2 levels suggests that tpxD expression is strictly controlled, to ensure homeostatic levels of H2O2 in the cell. Further research is needed to establish this mechanism.
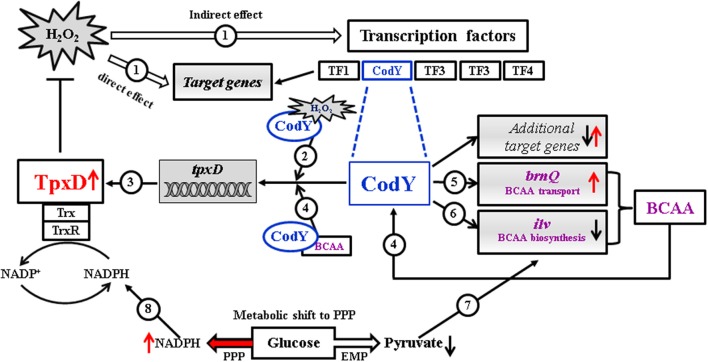
We present a schematic model (Figure 6) illustrating the contribution of TpxD and CodY to the pneumococcal global transcriptional response to H2O2. Under oxidative stress, H2O2 activates directly or indirectly a cascade of genes and transcription factors, among them CodY. We speculate that CodY activation by H2O2 is due to a conformational change originating from specific cysteine oxidation. As a result, CodY binding to tpxD regulatory sequence is enhanced, leading to TpxD up-regulation, thereby preventing the accumulation of high H2O2 levels. In addition, we show that under oxidative stress conditions, CodY modulates the intracellular level of BCAA (known to enhance CodY binding to DNA) through the up-regulation of the BCAA transporter, brnQ, and down-regulation of the BCAA biosynthetic machinery. Consequently, the need for pyruvate, which is the central precursor in BCAA biosynthesis, is expected to decrease. Hence, glucose catabolism will be shifted from the Embden-Meyerhof-Parnas (EMP) pathway toward the pentose phosphate pathway (PPP) resulting in NADPH formation, via the oxidative branch (Krüger et al., 2011). In line with this are the findings in E. coli (Sandoval et al., 2011) showing that high ROS levels lead to overexpression of glucose-6-phosphate dehydrogenase (zwf), the enzyme directing glucose to PPP. Increased NADPH levels are essential under high H2O2 levels, since TpxD activity relies on the recycling of its catalytic cysteine residues by the thioredoxin-thioredoxin reductase system.
Figure 6.
Proposed schematic model illustrating the contribution of TpxD and CodY to the pneumococcal global transcriptional response to H2O2. Under oxidative stress conditions H2O2 activates directly or indirectly a cascade of genes and transcription factors (TF), among them CodY (1). We speculate that CodY activation by H2O2 is due to a conformational change originating from specific cysteine oxidation (2). As a result, CodY binding to tpxD regulatory sequence is enhanced, leading to TpxD up-regulation (3), thereby preventing the accumulation of high H2O2 levels. BCAA are known to enhance CodY binding to DNA (4), thereby inducing tpxD expression (3). Under oxidative stress conditions, CodY modulates the intracellular level of BCAA through the up-regulation of the BCAA transporter, brnQ (5) and down-regulation of the BCAA biosynthetic machinery (ilv) (6). Consequently, the need for pyruvate, which is the central precursor in BCAA biosynthesis, is expected to decrease (7) leading to a shift in glucose catabolism toward the pentose phosphate pathway (PPP). This metabolic shift will result in increased NADPH formation (8), thus enhancing TpxD recycling by the thioredoxin-thioredoxin reductase system.
Results presented in this manuscript attribute a role for CodY in maintaining homeostatic levels of H2O2 in S. pneumoniae. This role is achieved by the regulation of tpxD, to enable bacterial survival under oxidative stress conditions. We also demonstrate that some of the genes involved in oxidative stress response, which are also known to be regulated by CodY, were not affected by high H2O2 levels in tpxD mutants ΔtpxD and TpxDC58S), signifying that active-TpxD is required for CodY function as a TF under oxidative stress conditions.
Author contributions
Substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data for the work: BH, HY, SS, XZ, RB, ST, OK, NP. Drafting the work or revising it critically for important intellectual content: BH, HY, SS, XZ, RB, ST, OK, NP. Final approval of the version to be published: BH, HY, SS, XZ, RB, ST, OK, NP. Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved: BH, HY, SS, XZ, RB, ST, OK, NP.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the Smoler Proteomics Center at Technion, Israel, for mass spectrometry analysis.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2017.00210/full#supplementary-material
References
- Alloing G., Granadel C., Morrison D. A., Claverys J. P. (1996). Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol. Microbiol. 21, 471–478. 10.1111/j.1365-2958.1996.tb02556.x [DOI] [PubMed] [Google Scholar]
- Auzat I., Chapuy-Regaud S., Le Bras G., Dos Santos D., Ogunniyi A. D., Le Thomas I., et al. (1999). The NADH oxidase of Streptococcus pneumoniae: its involvement in competence and virulence. Mol. Microbiol. 34, 1018–1028. 10.1046/j.1365-2958.1999.01663.x [DOI] [PubMed] [Google Scholar]
- Baker L., Poole L. B. (2003). Catalytic mechanism of thiol peroxidase from Escherichia coli: sulfenic acid formation and overoxidation of essential Cys61. J Biol. Chem. 278:9203. 10.1074/jbc.M209888200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky B. R. (2015). Role of branched-chain amino acid transport in Bacillus subtilis CodY activity. J. Bacteriol. 197, 1330–1338. 10.1128/JB.02563-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinsmade S. R., Kleijn R. J., Sauer U., Sonenshein A. L. (2010). Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools. J. Bacteriol. 192, 6357–6368. 10.1128/JB.00937-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caymaris S., Bootsma H. J., Martin B., Hermans P. W., Prudhomme M., Claverys J. (2010). The global nutritional regulator CodY is an essential protein in the human pathogen Streptococcus pneumoniae. Mol. Microbiol. 78, 344–360. 10.1111/j.1365-2958.2010.07339.x [DOI] [PubMed] [Google Scholar]
- Chiang S. M., Schellhorn H. E. (2012). Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch. Biochem. Biophys. 525, 161–169. 10.1016/j.abb.2012.02.007 [DOI] [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hengst C. D., van Hijum S. A., Geurts J. M., Nauta A., Kok J., Kuipers O. P. (2005). The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 280, 34332–34342. 10.1074/jbc.M502349200 [DOI] [PubMed] [Google Scholar]
- Dove S. L., Darst S. A., Hochschild A. (2003). Region 4 of σ as a target for transcription regulation. Mol. Microbiol. 48, 863–874. 10.1046/j.1365-2958.2003.03467.x [DOI] [PubMed] [Google Scholar]
- Duane P., Rubins J., Weisel H., Janoff E. (1993). Identification of hydrogen peroxide as a Streptococcus pneumoniae toxin for rat alveolar epithelial cells. Infect. Immun. 61, 4392–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint D. H., Tuminello J. F., Emptage M. H. (1993). The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol. Chem. 268, 22369–22376. [PubMed] [Google Scholar]
- Fomenko D. E., Koc A., Agisheva N., Jacobsen M., Kaya A., Malinouski M., et al. (2011). Thiol peroxidases mediate specific genome-wide regulation of gene expression in response to hydrogen peroxide. Proc. Natl. Acad. Sci. U.S.A. 108, 2729–2734. 10.1073/pnas.1010721108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T., Wolz C. (2014). Intersection of the stringent response and the CodY regulon in low GC Gram-positive bacteria. Int. J. Med. Microbiol. 304, 150–155. 10.1016/j.ijmm.2013.11.013 [DOI] [PubMed] [Google Scholar]
- Guedon E., Sperandio B., Pons N., Ehrlich S. D., Renault P. (2005). Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes. Microbiology 151(Pt 12), 3895–3909. 10.1099/mic.0.28186-0 [DOI] [PubMed] [Google Scholar]
- Guiral S., Henard V., Laaberki M. H., Granadel C., Prudhomme M., Martin B., et al. (2006). Construction and evaluation of a chromosomal expression platform (CEP) for ectopic, maltose-driven gene expression in Streptococcus pneumoniae. Microbiology 152(Pt 2), 343–349. 10.1099/mic.0.28433-0 [DOI] [PubMed] [Google Scholar]
- Hajaj B., Yesilkaya H., Benisty R., David M., Andrew P. W., Porat N. (2012). Thiol peroxidase is an important component of Streptococcus pneumoniae in oxygenated environments. Infect. Immun. 80, 4333–4343. 10.1128/IAI.00126-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A., Kang H., Son J., Kwon D. H., Kim S., Lee W. C., et al. (2016). The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism. Nucleic Acids Res. 44, 9483–9493. 10.1093/nar/gkw775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98. 10.12691/ajmr-3-2-1 [DOI] [Google Scholar]
- Henard C. A., Bourret T. J., Song M., Vazquez-Torres A. (2010). Control of redox balance by the stringent response regulatory protein promotes antioxidant defenses of Salmonella. J. Biol. Chem. 285, 36785–36793. 10.1074/jbc.m110.160960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriksen W. T., Bootsma H. J., Estevao S., Hoogenboezem T., de Jong A., de Groot R., et al. (2008). CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J. Bacteriol. 190, 590–601. 10.1128/JB.00917-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle E. S., Luo Y., Gassmann W., Linn S. (1996). Oxidative damage to DNA constituents by iron-mediated Fenton reactions. The deoxyguanosine family. J. Biol. Chem. 271, 21177–21186. [PubMed] [Google Scholar]
- Hoskins J., Alborn W. E., Jr., Arnold J., Blaszczak L. C., Burgett S., DeHoff B. S., et al. (2001). Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183, 5709–5717. 10.1128/JB.183.19.5709-5717.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J. A., Linn S. (1986). Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. J. Bacteriol. 166, 519–527. 10.1128/jb.166.2.519-527.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C., Bootsma H. J., Aldridge C., Manuse S., Gisch N., Schwudke D., et al. (2015). Co-Inactivation of GlnR and CodY regulators impacts pneumococcal cell wall physiology. PLoS ONE 10:e0123702. 10.1371/journal.pone.0123702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajfasz J. K., Rivera-Ramos I., Scott-Anne K., Gregoire S., Abranches J., Lemos J. A. (2015). Transcription of oxidative stress genes is directly activated by SpxA1 and, to a lesser extent, by SpxA2 in Streptococcus mutans. J. Bacteriol. 197, 2160–2170. 10.1128/JB.00118-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakov A. E., Cipriano M. J., Novichkov P. S., Minovitsky S., Vinogradov D. V., Arkin A., et al. (2007). RegTransBase-a database of regulatory sequences and interactions in a wide range of prokaryotic genomes. Nucleic Acids Res. 35, D407-412-12. 10.1093/nar/gkl865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger A., Grüning N., Wamelink M. M., Kerick M., Kirpy A., Parkhomchuk D., et al. (2011). The pentose phosphate pathway is a metabolic redox sensor and regulates transcription during the antioxidant response. Antioxid Redox Signal. 15, 311–324. 10.1089/ars.2010.3797 [DOI] [PubMed] [Google Scholar]
- La Carbona S., Sauvageot N., Giard J., Benachour A., Posteraro B., Auffray Y., et al. (2007). Comparative study of the physiological roles of three peroxidases (NADH peroxidase, Alkyl hydroperoxide reductase and Thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. Mol. Microbiol. 66, 1148–1163. 10.1111/j.1365-2958.2007.05987.x [DOI] [PubMed] [Google Scholar]
- Lewis J. K., Wei J., Siuzdak G. (2000). Matrix-Assisted Laser Desorption/Ionization mass spectrometry in peptide and protein analysis, in Encyclopedia of Analytical Chemistry, ed Meyers R. A. (Chichester: John Wiley & Sons, Ltd.), 5880–5894. [Google Scholar]
- Lisher J. P., Tsui H. T., Ramos-Monta-ez S., Hentchel K. L., Martin J. E., Trinidad J. C., et al. (2017). Biological and chemical adaptation to endogenous hydrogen peroxide production in Streptococcus pneumoniae D39. mSphere 2, e00291–e00216. 10.1128/mSphere.00291-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lobel L., Herskovits A. A. (2016). Systems level analyses reveal multiple regulatory activities of CodY controlling metabolism, motility and virulence in Listeria monocytogenes. PLoS Genet. 12:e1005870. 10.1371/journal.pgen.1005870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose M., Hudel M., Zimmer K. P., Garcia E., Hammerschmidt S., Lucas R., et al. (2014). Pneumococcal hydrogen peroxide-induced stress signaling regulates inflammatory genes. J. Infect. Dis. 211, 306–316. 10.1093/infdis/jiu428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S., Imlay J. (2012). Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch. Biochem. Biophys. 525, 145–160. 10.1016/j.abb.2012.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novichkov P. S., Rodionov D. A., Stavrovskaya E. D., Novichkova E. S., Kazakov A. E., Gelfand M. S., et al. (2010). RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res. 38, W299–W307. 10.1093/nar/gkq531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien K. L., Wolfson L. J., Watt J. P., Henkle E., Deloria-Knoll M., McCall N., et al. (2009). Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374, 893–902. 10.1016/S0140-6736(09)61204-6 [DOI] [PubMed] [Google Scholar]
- Paget M. S., Buttner M. J. (2003). Thiol-based regulatory switches. Annu. Rev. Genet. 37, 91–121. 10.1146/annurev.genet.37.110801.142538 [DOI] [PubMed] [Google Scholar]
- Paterson G. K., Blue C. E., Mitchell T. J. (2006). An operon in Streptococcus pneumoniae containing a putative alkylhydroperoxidase D homologue contributes to virulence and the response to oxidative stress. Microb. Pathog. 40, 152–160. 10.1016/j.micpath.2005.12.003 [DOI] [PubMed] [Google Scholar]
- Pericone C. D., Bae D., Shchepetov M., McCool T., Weiser J. N. (2002). Short-sequence tandem and nontandem DNA repeats and endogenous hydrogen peroxide production contribute to genetic instability of Streptococcus pneumoniae. J. Bacteriol. 184, 4392. 10.1128/jb.184.16.4392-4399.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericone C. D., Overweg K., Hermans P. W. M., Weiser J. N. (2000). Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect. Immun. 68, 3990–3997. 10.1128/IAI.68.7.3990-3997.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericone C. D., Park S., Imlay J. A., Weiser J. N. (2003). Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. J. Bacteriol. 185, 6815–6825. 10.1128/JB.185.23.6815-6825.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai P., Parrish M., Tay I. J., Li N., Ackerman S., He F., et al. (2015). Streptococcus pneumoniae secretes hydrogen peroxide leading to DNA damage and apoptosis in lung cells. Proc. Natl. Acad. Sci. U.S.A. 112, E3421–E3430. 10.1073/pnas.1424144112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval J. M., Arenas F. A., Vasquez C. C. (2011). Glucose-6-phosphate dehydrogenase protects Escherichia coli from tellurite-mediated oxidative stress. PLoS ONE 6:e25573. 10.1371/journal.pone.0025573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafeeq S., Yesilkaya H., Kloosterman T. G., Narayanan G., Wandel M., Andrew P. W., et al. (2011). The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol. Microbiol. 81, 1255–1270. 10.1111/j.1365-2958.2011.07758.x [DOI] [PubMed] [Google Scholar]
- Song J., Ko K. S., Lee J., Baek J., Oh W., Yoon H., et al. (2005). Identification of essential genes in Streptococcus pneumoniae by allelic replacement mutagenesis. Mol. Cells 19, 365–374. [PubMed] [Google Scholar]
- Taniai H., Iida K., Seki M., Saito M., Shiota S., Nakayama H., et al. (2008). Concerted action of lactate oxidase and pyruvate oxidase in aerobic growth of Streptococcus pneumoniae: role of lactate as an energy source. J. Bacteriol. 190, 3572. 10.1128/jb.01882-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H., Nelson K. E., Paulsen I. T., Eisen J. A., Read T. D., Peterson S., et al. (2001). Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293, 498–506. 10.1126/science.1061217 [DOI] [PubMed] [Google Scholar]
- van Hijum S. A., De Jong A., Baerends R. J., Karsens H. A., Kramer N. E., Larsen R., et al. (2005). A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77. 10.1186/1471-2164-6-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal E. A., Day A. M., Morgan B. A. (2007). Hydrogen peroxide sensing and signaling. Mol. Cell 26, 1–14. 10.1016/j.molcel.2007.03.016 [DOI] [PubMed] [Google Scholar]
- Yesilkaya H., Kadioglu A., Gingles N., Alexander J. E., Mitchell T. J., Andrew P. W. (2000). Role of manganese-containing superoxide dismutase in oxidative stress and virulence of Streptococcus pneumoniae. Infect. Immun. 68, 2819–2826. 10.1128/IAI.68.5.2819-2826.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesilkaya H., Spissu F., Carvalho S. M., Terra V. S., Homer K. A., Benisty R., et al. (2009). Pyruvate formate lyase is required for pneumococcal fermentative metabolism and virulence. Infect. Immun. 77, 5418–5427. 10.1128/IAI.00178-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.