Abstract
The present study attempted to identify potential key genes and pathways of peri-implantitis, and to investigate the possible mechanisms associated with it. An array data of GSE57631 was downloaded, including six samples of peri-implantitis tissue and two samples of normal tissue from the Gene Expression Omnibus (GEO) database. The differentially expressed genes (DEGs) in the peri-implantitis samples compared with normal ones were analyzed with the limma package. Moreover, Gene Ontology annotation and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses for DEGs were performed by DAVID. A protein-protein interaction (PPI) network was established using Cytoscape software, and significant modules were analyzed using Molecular Complex Detection. A total of 819 DEGs (759 upregulated and 60 downregulated) were identified in the peri-implantitis samples compared with normal ones. Moreover, the PPI network was constructed with 413 nodes and 1,114 protein pairs. Heat shock protein HSP90AA1 (90 kDa α, member 1), a hub node with higher node degrees in module 4, was significantly enriched in antigen processing, in the presentation pathway and nucleotide-binding oligomerization domain (NOD)-like receptor-signaling pathway. In addition, nuclear factor-κ-B1 (NFKB1) was enriched in the NOD-like receptor-signaling pathway in KEGG pathway enrichment analysis for upregulated genes. The proteasome is the most significant pathway in module 1 with the highest P-value. Therefore, the results of the present study suggested that HSP90AA1 and NFKB1 may be potential key genes, and the NOD-like receptor signaling pathway and proteasome may be potential pathways associated with peri-implantitis development.
Keywords: differentially expressed genes, Kyoto Encyclopedia of Genes and Genomes pathway, modules, peri-implantitis, protein-protein interaction network
Introduction
Peri-implantitis is a destructive inflammatory disease that affects the tissues surrounding dental implants (1,2). It has been demonstrated that peri-implantitis is a crucial element in implant failure. In total, ~30% of patients that receive dental implants develop peri-implantitis (3). However, there are currently no effective therapeutic strategies against peri-implantitis. Moreover, the use of dental implants is constantly increasing, therefore an effective therapy to treat peri-implantitis is required. Thus, further investigations into the molecular pathophysiology of peri-implantitis are necessary in order to provide novel options for effective treatment (1).
Recently, a number of studies have investigated the pathological mechanisms underlying peri-implantitis progression. Becker et al (4) indicated that serglycin (SRGN) expression was significantly upregulated in peri-implantitis when compared with healthy individuals. It has been suggested that this gene may inhibit bone mineralization in vitro (5). Another study indicated that concentrations of the nuclear factor-κB (NF-B), soluble RANK ligand (sRANKL), osteoprotegerin (OPG) and sclerostin are significantly increased in patients with peri-implantitis (6). A number of typical bone matrix molecules, including collagen, type IX, α 1 (COL9A1), bone gamma-carboxyglutamate (Gla) protein (BGLAP) and secreted phosphoprotein 1 (SPP1) are decreased in the peri-implantitis tissues (1). Furthermore, it has been identified that fibroblasts are involved in the pathogenesis of peri-implantitis (7). The regulation of inflammatory mediators and matrix metalloproteinases (MMPs) in peri-implantitis fibroblasts function in the pathogenesis of the disease (8), and levels of the anti-inflammatory cytokine interleukin (IL)-10 are decreased in peri-implantitis (9). Furthermore, peroxisome proliferator-activated receptor γ (PPARγ) that can inhibit inflammation and promote osteoblast function is downregulated in the peri-implantitis tissues (10). However, other mechanisms associated with peri-implantitis have not been identified. Therefore, further research should focus on elucidating other potential mechanisms and investigate target genes for the treatment of peri-implantitis.
The microarray data of GSE57631 was used to confirm the similarities and differences of inflamed peri-implantitis tissues vs. normal peri-implantitis tissues at the mRNA level (1). In contrast to results from a previous study (1), the array data of GSE57631 was downloaded and the differentially expressed genes (DEGs) associated with peri-implantitis were analyzed using a biological informatics approach. In addition, functional enrichment analyses were performed for DEGs. In addition, a protein-protein interaction (PPI) network was established and four significant modules were analyzed. The present study aimed to identify the key genes and pathways of peri-implantitis, and identify possible significant mechanisms associated with it.
Materials and methods
Affymetrix microarray data
The array data of GSE57631 was downloaded from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database, which was deposited by Schminke et al (1). Six samples of peri-implantitis tissues and two samples of normal tissues were included in the present study. The raw data and annotation files were downloaded for subsequent analysis, based on the GPL15034 platform (Affymetrix Human Gene 1.0 ST Array [HuGene10stv1_Hs_ENTREZ, Brainarray v14]; Affymetrix, Inc., Santa Clara, CA, USA).
Data preprocessing
The raw expression data was preprocessed using the robust multiarray average (11) algorithm by applying an oligo (12) in the R statistical software program in Bioconductor (http://www.bioconductor.org/). Background correction, normalization and a calculating expression were included in the process of preprocessing. A total of 18,977 gene expression values were obtained.
DEG analysis
The limma package (13) in Bioconductor was used to analyze DEGs in peri-implantitis samples compared with controls. In the process of the analysis, the P-values of DEGs were calculated using a t-test in the limma package. |log2FC|≥1 and P<0.05 were used as a cut-off criteria.
Gene Ontology (GO) and pathway enrichment analyses
GO is a tool that is used for gene annotation by collecting defined, structured, controlled vocabulary, which includes three main categories: Molecular function (MF), biological process (BP) and cellular component (CC) (14). Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database used for associating related gene sets with their pathways (15). Moreover, DAVID, an integrated data-mining environment, is used to analyze gene lists (16).
GO annotation and KEGG pathway enrichment analyses were conducted for upregulated and downregulated genes by DAVID. Moreover, EASE ≤0.05 and gene counts ≥ were set as the threshold value.
PPI network analysis
The Search Tool for the Retrieval of Interacting Genes (STRING) (17) database provides information regarding the predicted and experimental interactions of proteins. The prediction method of this database came from neighborhood, gene fusion, co-occurrence, co-expression experiments, databases and text mining. Moreover, the interactions of protein pairs in the database are presented with a combined score. In the present study, DEGs were mapped into PPIs and a combined score of >0.9 was used as the cut-off value. In addition, PPI networks were constructed using Cytoscape software version 3.2.1 (18).
Topological properties of the PPI network, including degree (19), subgraph (20) and betweenness (21) centralities were determined using the R software package igraph (22), in order to analyze key genes in the network. A degree was used for describing the importance of protein nodes in the network. Subgraph centrality based on combining network topology and information of protein complexes was used to measure the importance of nodes in the network. The higher the degree and subgraph values were, the more important the nodes were in the network. Moreover, the betweenness centrality is an index describing the global topological properties of the network and could be used to describe how the nodes affect the connectivity between two nodes. Betweenness was defined as the ratio of the number of every path passed per node and the number of the shortest paths. The higher the betweenness values are, the greater the impact of the node in the network is. In addition, R software package igraph version 1.0.1 (22) was used for these three methods.
Module analysis
Network module was one of the characteristics of the protein network and may contain specific biological significance. The Cytoscape software package Molecular Complex Detection (MCODE) (23) was used to analyze the most notable clustering module. Next, the KEGG pathway enriched by DEGs in different modules was analyzed using DAVID online tool. EASE ≤0.05 and count ≥2 were set as the cutoff values.
Results
Data processing and DEG analysis
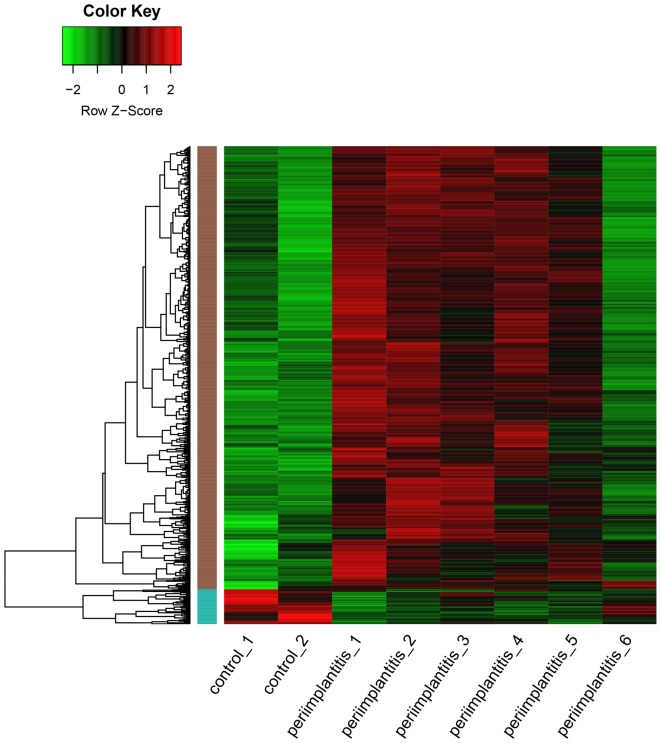
As shown in Fig. 1, a total of 819 DEGs including 759 upregulated and 60 downregulated genes, were identified in the peri-implantitis samples compared with the control ones. As a result, the number of upregulated genes was found to be significantly higher than that of the downregulated ones.
Figure 1.
Heat map of differentially expressed genes. Green represents a lower expression level, red represents higher expression levels and black represents that there is no differential expression amongst the genes.
GO and pathway enrichment analyses
GO and KEGG pathway analyses were performed for upregulated and downregulated DEGs, respectively. The GO terms of upregulated DEGs were mainly associated with the proteasomal protein catabolic process, endoplasmic reticulum, melanosome and structural constituent of the cytoskeleton (Table IA). The downregulated DEGs were mainly enriched in the epidermis and ectoderm development, and the extracellular region part and cadmium ion binding (Table IB).
Table I.
GO for differentially expressed genes.
| A, Upregulated | |||
|---|---|---|---|
| Terms | Description | Counts | P-value |
| GO-BP | |||
| GO: 0010498 | Proteasomal protein catabolic process | 24 | 5.32×10−11 |
| GO: 0043161 | Proteasomal ubiquitin-dependent protein catabolic process | 24 | 5.32×10−11 |
| GO: 0032269 | Negative regulation of cellular protein metabolic process | 30 | 9.32×10−10 |
| GO-CC | |||
| GO: 0005783 | Endoplasmic reticulum | 115 | 1.37×10−21 |
| GO: 0042470 | Melanosome | 30 | 1.77×10−17 |
| GO: 0048770 | Pigment granule | 30 | 1.77×10−17 |
| GO-MF | |||
| GO: 0005200 | Structural constituent of cytoskeleton | 15 | 2.22×10−6 |
| GO: 0070003 | Threonine-type peptidase activity | 8 | 1.13×10−5 |
| B, Downregulated | |||
| Terms | Description | Counts | P-value |
| GO-BP | |||
| GO:0008544 | Epidermis development | 20 | 2.57×10−10 |
| GO:0007398 | Ectoderm development | 20 | 9.84×10−10 |
| GO:0006954 | Inflammatory response | 19 | 9.40×10−6 |
| GO-CC | |||
| GO:0044421 | Extracellular region part | 35 | 1.05×10−5 |
| GO:0005576 | Extracellular region | 54 | 1.33×10−4 |
| GO:0005615 | Extracellular space | 25 | 2.79×10−4 |
| GO-MF | |||
| GO:0046870 | Cadmium ion binding | 4 | 5.27×10−4 |
| GO:0005507 | Copper ion binding | 6 | 6.22×10−3 |
Terms represent the identification number of GO term; description represents the names of GO term; counts represent the number of genes enriched in GO terms. GO, Gene ontology; BP, biological process; CC: cellular component; MF, molecular function.
Two pathways that were significantly enriched by upregulated DEGs were Alzheimer's disease and the proteasome (Table II). Moreover, an important gene HSP90AA1 was significantly enriched in the antigen processing and presentation pathway and the nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathway, and NFKB1 was enriched in NOD-like receptor signaling pathway in KEGG pathway enrichment analysis for upregulated genes. However, downregulated DEGs did not significantly enriched any pathways.
Table II.
KEGG pathway enrichment analysis for upregulated differentially expressed genes.
| Term | Description | Counts | P-value |
|---|---|---|---|
| hsa05010 | Alzheimer's disease | 25 | 1.69×10−5 |
| hsa03050 | Proteasome | 12 | 5.36×10−5 |
| hsa00190 | Oxidative phosphorylation | 18 | 1.19×10−3 |
| hsa05012 | Parkinson's disease | 17 | 2.65×10−3 |
| hsa05016 | Huntington's disease | 21 | 3.41×10−3 |
| hsa00510 | N-Glycan biosynthesis | 9 | 4.31×10−3 |
| hsa04142 | Lysosome | 15 | 7.11×10−3 |
| hsa00480 | Glutathione metabolism | 9 | 7.27×10−3 |
| hsa04612 | Antigen processing and presentation | 12 | 7.74×10−3 |
| hsa04621 | NOD-like receptor signaling pathway | 10 | 8.63×10−3 |
| hsa00970 | Aminoacyl-tRNA biosynthesis | 7 | 2.89×10−2 |
| hsa00020 | Citrate cycle (TCA cycle) | 6 | 3.07×10−2 |
| hsa01040 | Biosynthesis of unsaturated fatty acids | 5 | 3.51×10−2 |
| hsa05130 | Pathogenic Escherichia coli infection | 8 | 4.45×10−2 |
Term represents the identification number of the KEGG pathway; Description represents the name of the KEGG pathway; Counts represent the number of genes enriched in the KEGG pathway. KEGG, Kyoto Encyclopedia of Genes and Genomes; NOD, nucleotide-binding oligomerization domain-like; TCA, tricarboxylic acid.
PPI network analysis
Based on the STRING database, a total of 413 nodes and 1,114 protein pairs were obtained with a combined score >0.9 (Fig. 2). As shown in Fig. 2, the majority of the nodes in the network were upregulated DEGs in peri-implantitis samples. The top 20 most up- and dowregulated nodes are presented in Table III. A number of protein enzyme families, such as the proteasome subunit, α type 1 (PSMA1), proteasome subunit, beta type 1 (PSMB1) and proteasome subunit, α type 4 (PSMA4) were hub nodes based on the subgraph and degree centralities. Heat shock protein HSP90AA1 (90 kDa α, member 1), Ras-related C3 botulinum toxin substrate 1, NFKB1, Jun proto-oncogene were hub nodes with higher betweenness values.
Figure 2.
Protein-protein interaction network of differentially expressed genes in peri-implantitis samples compared with the control ones. Red nodes, upregulated genes; green nodes, downregulated genes.
Table III.
Nodes with higher values in subgraph centrality, betweenness centrality and degree centrality.
| Nodes | Subgragh | Nodes | Betweenness | Nodes | Degree |
|---|---|---|---|---|---|
| PSMA1 | 2.35×107 | HSP90AA1 | 2.78×104 | HSP90AA1 | 34 |
| PSMB1 | 2.32×107 | RAC1 | 1.77×104 | JUN | 27 |
| PSMA4 | 2.31×107 | NFKB1 | 1.38×104 | RAC1 | 27 |
| PSMB4 | 2.30×107 | JUN | 1.21×104 | PSMA1 | 25 |
| PSMA3 | 2.30×107 | HIF1A | 1.07×104 | PSMA4 | 25 |
| PSMB3 | 2.30×107 | CDH1 | 9.21×103 | PSMB1 | 25 |
| PSMB6 | 2.30×107 | SOS1 | 8.93×103 | PSMC2 | 25 |
| PSMC2 | 2.16×107 | CANX | 8.55×103 | PSMB4 | 24 |
| PSMD8 | 2.12×107 | HDAC1 | 8.35×103 | PSMA3 | 24 |
| PSMD5 | 2.08×107 | ATP5B | 8.24×103 | PSMB3 | 24 |
| PSME4 | 2.08×107 | GTF2B | 8.17×103 | PSMB6 | 24 |
| PSMD10 | 2.04×107 | HSPA5 | 7.84×103 | PSMD10 | 24 |
| PSME3 | 1.96×107 | STAT3 | 7.46×103 | PSMD8 | 23 |
| UBE2D1 | 1.54×107 | VCP | 7.32×103 | SEC61A1 | 23 |
| CDC16 | 1.36×107 | RPS5 | 7.14×103 | RPN2 | 23 |
| ANAPC5 | 1.36×107 | NME1 | 6.31×103 | NFKB1 | 23 |
| BUB3 | 1.35×107 | POLR2B | 5.90×103 | PSMD5 | 22 |
| SEC61A1 | 1.23×107 | P4HB | 5.90×103 | PSME4 | 22 |
| SKP1 | 1.16×107 | TXN | 5.67×103 | UBE2D1 | 22 |
| HSPB1 | 1.05×107 | RPN2 | 5.67×103 | RPN1 | 22 |
PSM, proteasome subunit.
Module analysis
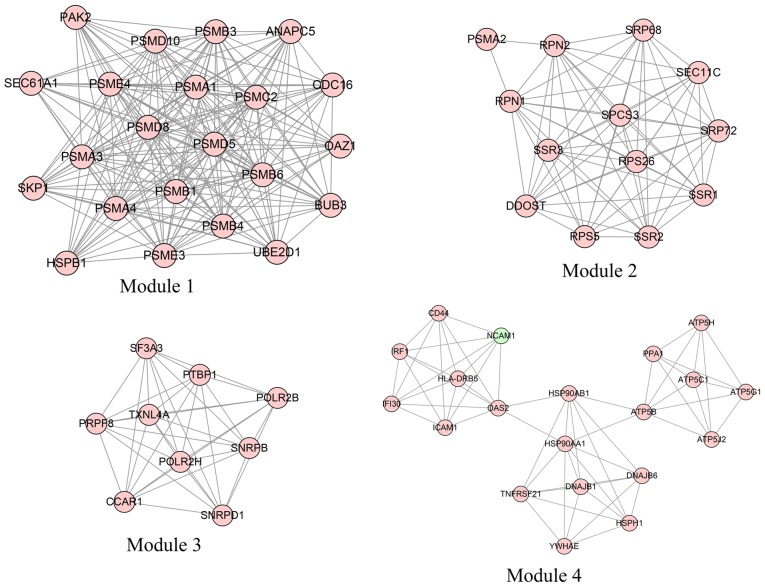
In total, four modules (modules 1, 2, 3 and 4) with score >6 were detected by MCODE (Fig. 3). As shown in Table IV, PSMA1, PSMA4 and PSMB1 were hub nodes with higher node degrees in module 1, and ribophorin 1 (RPN1), ribophorin 2 (RPN2) and dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit (DDOST) were hub nodes in module 2. The hub nodes with higher node degrees in module 3 were pre-mRNA processing factor 8 (PRPF8), small nuclear ribonucleoprotein D1 polypeptide (SNRPD1; 16 kDa) and small nuclear ribonucleoprotein polypeptides B and B1 (SNRPB), and the hub nodes in module 4 were HSP90AA1, ATP synthase, H+ transporting, mitochondrial Fo complex, Subunit F2 (ATP5J2) and ATP synthase, H+ transporting, mitochondrial F1 complex and beta polypeptide (ATP5B). Furthermore, the proteasome pathway was identified as the most significant pathway in module 1.
Figure 3.
Four significant modules identified from the protein-protein interaction network using the molecular complex detection method with a score of >6.0. Module 1: MCODE score=19.238; Module 2: MCODE score=11.333; Module 3: MCODE score=9 and Module 4: MCODE score=6.316.
Table IV.
KEGG pathway enriched by differentially expressed genes in different modules (P<0.05).
| Term | Description | P-value | Genes |
|---|---|---|---|
| Module1 | |||
| hsa03050 | Proteasome | 6.77×10−17 | PSMB4, PSMA1, PSMB6, PSMB1, PSMC2, PSMA4, PSMB3, PSMA3, PSME3, PSME4, PSMD8 |
| hsa04110 | Cell cycle | 9.03×10−3 | ANAPC5, SKP1, CDC16, BUB3 |
| hsa04120 | Ubiquitin mediated proteolysis | 1.16×10−2 | ANAPC5, SKP1, CDC16, UBE2D1 |
| Module2 | |||
| hsa00510 | N-Glycan biosynthesis | 1.63×10−3 | RPN1, RPN2, DDOST |
| hsa03060 | Protein export | 1.10×10−2 | SRP68, SRP72 |
| Module3 | |||
| hsa03040 | Spliceosome | 5.19×10−6 | PRPF8, SNRPD1, SNRPB, TXNL4A, SF3A3 |
| hsa03020 | RNA polymerase | 3.26×10−2 | POLR2H, POLR2B |
| Module4 | |||
| hsa00190 | Oxidative phosphorylation | 1.68×10−5 | ATP5J2, ATP5B, ATP5C1, ATP5G1, ATP5H, PPA1 |
| hsa04612 | Antigen processing and presentation | 1.34×10−3 | HSP90AB1, HSP90AA1, IFI30, HLA-DRB5 |
| hsa05012 | Parkinson's disease | 4.63×10−3 | ATP5B, ATP5C1, ATP5G1, ATP5H |
| hsa05010 | Alzheimer's disease | 9.07×10−3 | ATP5B, ATP5C1, ATP5G1, ATP5H |
| hsa05016 | Huntington's disease | 1.19×10−2 | ATP5B, ATP5C1, ATP5G1, ATP5H |
| hsa04514 | Cell adhesion molecules | 5.00×10−2 | NCAM1, ICAM1, HLA-DRB5 |
Term represents the identification number of the KEGG pathway; description represents the name of the KEGG pathway. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
In the present study, the gene expression patterns obtained from the GEO database revealed a total of 819 genes, including 759 upregulated and 60 downregulated genes, that were differently expressed in peri-implantitis samples compared with controls. The results of the present study demonstrated that HSP90AA1, which had the highest degrees in the PPI network, was significantly identified in module 4. In addition, NFKB1 were also hub nodes with higher betweenness values in the PPI network. Moreover, the proteasome pathway was the most significant pathway in module 1, and may be key mechanisms associated with peri-implantitis progression.
HSP90, a member of the heat shock family of proteins, is essential in determining cell cycle control and survival, hormone and a number of signaling pathways (24). In addition, released HSP90, which functions as a danger signal, can elicit secretion of inflammatory cytokines (25). A cell-impermeable HSP90 inhibitor can prevent inflammatory responses, indicating that extracellular HSP90 is involved in mediating and initiating sterile inflammatory responses (26). Moreover, previous data indicate that HSP90-targeted agents may be helpful for the treatment of inflammatory disease, and that an HSP90 inhibitor can affect multiple signaling processes associated with inflammation (27). Furthermore, inhibition of HSP90 is able to reduce innate immunity responses and diminishes proinflammatory mediator production in immune-stimulated macrophages (28,29). In addition, analysis of inflammatory mediators in crevicular fluid can be used to distinguish peri-implantitis from normal tissue (30).
Systemic markers of inflammation are increased in patients with peri-implantitis (30) and high levels of inflammatory cytokines, such as IL-1β, are associated with signs of early peri-implantitis development (31–33). Furthermore, it has been suggested that fibroblasts express HSP90 that also participates in the pathogenesis of peri-implantitis (7,34). Moreover, HSPs together with vascular and inflammatory biomarkers may be useful as biomarkers of peri-implantitis development (35,36). In the present study, HSP90AA1, a hub node with higher node degrees in module 4, was enriched in antigen processing and the presentation pathway, as well as the NOD-like receptor-signaling pathway. Therefore, the results of the present study are in line with results from former previous studies and indicate that HSP90AA1 may be directly or indirectly important in peri-implantitis development.
In the present study, NFKB1 was shown to also have hub nodes with higher betweenness values in the PPI network. The NFKB1 gene is known to encode the NF-KB p105/p50 isoforms (37). The central pathological pattern of peri-implantitis is inflammatory osteoclastogenesis, which is mediated by proinflammatory mediators and performed by the regulators of osteoclastogenesis, including NF-B, sRANKL and OPG (38,39). A prior study indicated that the NF-B concentration increased 1.5–4-fold in patients with peri-implantitis compared to those with mucositis (6). Moreover, recent studies demonstrated that high levels of NF-B were associated with peri-implantitis (40,41). In addition, NF-B is upregulated in inflammatory bowel disease and is a transcription regulator of the immune response (42). Zou et al (43) reported that NFKB1 could regulate the transcription of genes in the immune response, and was also a key part in coordinating the immune system. Numerous studies have indicated that the NFKB1-94ins/del ATTG promoter polymorphism is associated with inflammatory disease (37,42,44,45). Furthermore, as mentioned in the aforementioned paragraph, inflammation is associated with the pathogenesis of peri-implantitis. Therefore, the results of the present study are in accordance with those from previous studies and suggest that NFKB1 may be a key gene associated with peri-implantitis development. Notably in the current study, HSP90AA1 and NFKB1 were enriched in the NOD-like receptor-signaling pathway. Although the important roles of the NOD-like receptor-signaling pathway have not been fully discussed, it is speculated that HSP90AA1 and NFKB1 may be involved in peri-implantitis progression via the NOD-like receptor-signaling pathway.
In addition to the two aforementioned genes, the proteasome pathway containing the proteasome subunits (PSM) A1, PSMA3, PSMA4, PSMB1, PSMB3, PSMB4, PSMB6, PSMC2, PSMD8, PSME3 and PSME4, were found to be enriched in the most significant pathway in module 1 and to have the highest P-value. Elliott et al (46) indicated that key proteins modulated by the proteasome were involved in controlling inflammatory processes. A prior study demonstrated that the activation of the ubiquitin-proteasome by macrophages is involved in an NF-B-dependent increase in inflammation (47). Moreover, previous studies have indicated that the proteasomal pathway as a targeting goal may be a potential way to treat autoimmune and inflammatory diseases (48–50). In addition, Wang and Maldonado (51) demonstrated that proteasome inhibitors could serve as novel drugs by regulating proinflammatory protein catabolism. Furthermore, as stated above, inflammation has been associated with the pathogenesis of peri-implantitis. Finally, the PSMA1, PSMA3, PSMA4, PSMB1, PSMB3, PSMB4, PSMB6, PSMC2, PSMD8, PSME3 and PSME4 proteins are essential subunits that contribute to the assembly of the proteasome complex (52–55). Therefore, the proteasome and its subunit genes may be important in peri-implantitis progression.
In conclusion, HSP90AA1, NFKB1 and the NOD-like receptor signaling pathway, as well as the proteasome pathway and its subunits genes may be important in the development of peri-implantitis. However, there were certain limitations in the present study, such as no experimental verification and the relatively small sample size used. Therefore, further research investigating the potential mechanisms involved in peri-implantitis are required.
References
- 1.Schminke B, Vom Orde F, Gruber R, Schliephake H, Bürgers R, Miosge N. The pathology of bone tissue during peri-implantitis. J Dent Res. 2015;94:354–361. doi: 10.1177/0022034514559128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mombelli A. Microbiology and antimicrobial therapy of peri-implantitis. Periodontol 2000. 2002;28:177–189. doi: 10.1034/j.1600-0757.2002.280107.x. [DOI] [PubMed] [Google Scholar]
- 3.Berglundh T, Persson L, Klinge B. A systematic review of the incidence of biological and technical complications in implant dentistry reported in prospective longitudinal studies of at least 5 years. J Clin Periodontol. 2002;29:197–212. doi: 10.1034/j.1600-051X.29.s3.12.x. (Suppl 3) 232–233. [DOI] [PubMed] [Google Scholar]
- 4.Becker ST, Beck-Broichsitter BE, Graetz C, Dörfer CE, Wiltfang J, Häsler R. Peri-implantitis versus periodontitis: Functional differences indicated by transcriptome profiling. Clin Implant Dent Relat Res. 2014;16:401–411. doi: 10.1111/cid.12001. [DOI] [PubMed] [Google Scholar]
- 5.Theocharis AD, Seidel C, Borset M, Dobra K, Baykov V, Labropoulou V, Kanakis I, Dalas E, Karamanos NK, Sundan A, Hjerpe A. Serglycin constitutively secreted by myeloma plasma cells is a potent inhibitor of bone mineralization in vitro. J Biol Chem. 2006;281:35116–35128. doi: 10.1074/jbc.M601061200. [DOI] [PubMed] [Google Scholar]
- 6.Rakic M, Struillou X, Petkovic-Curcin A, Matic S, Canullo L, Sanz M, Vojvodic D. Estimation of bone loss biomarkers as a diagnostic tool for peri-implantitis. J Periodontol. 2014;85:1566–1574. doi: 10.1902/jop.2014.140069. [DOI] [PubMed] [Google Scholar]
- 7.Bordin S, Flemmig TF, Verardi S. Role of fibroblast populations in peri-implantitis. Int J Oral Maxillofac Implants. 2009;24:197–204. [PubMed] [Google Scholar]
- 8.Irshad M, Scheres N, Anssari Moin D, Crielaard W, Loos BG, Wismeijer D, Laine ML. Cytokine and matrix metalloproteinase expression in fibroblasts from peri-implantitis lesions in response to viable Porphyromonas gingivalis. J Periodontal Res. 2013;48:647–656. doi: 10.1111/jre.12051. [DOI] [PubMed] [Google Scholar]
- 9.Casado PL, Canullo L, de Almeida Filardy A, Granjeiro JM, Barboza EP, Leite Duarte ME. Interleukins 1β and 10 expressions in the periimplant crevicular fluid from patients with untreated periimplant disease. Implant Dent. 2013;22:143–150. doi: 10.1097/ID.0b013e3182818792. [DOI] [PubMed] [Google Scholar]
- 10.Park SY, Kim KH, Gwak EH, Rhee SH, Lee JC, Shin SY, Koo KT, Lee YM, Seol YJ. Ex vivo bone morphogenetic protein 2 gene delivery using periodontal ligament stem cells for enhanced re-osseointegration in the regenerative treatment of peri-implantitis. J Biomed Mater Res A. 2015;103:38–47. doi: 10.1002/jbm.a.35145. [DOI] [PubMed] [Google Scholar]
- 11.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 12.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altermann E, Klaenhammer TR. PathwayVoyager: pathway mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. BMC Genomics. 2005;6:60. doi: 10.1186/1471-2164-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 20.Estrada E, Rodríguez-Velázquez JA. Subgraph centrality in complex networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;71:056103. doi: 10.1103/PhysRevE.71.056103. [DOI] [PubMed] [Google Scholar]
- 21.Goh KI, Oh E, Kahng B, Kim D. Betweenness centrality correlation in social networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;67:017101. doi: 10.1103/PhysRevE.67.017101. [DOI] [PubMed] [Google Scholar]
- 22.Csardi G, Nepusz T. The igraph software package for complex network research. Inter Journal Complex Systems. 2006;1695:1–9. [Google Scholar]
- 23.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bucci M, Roviezzo F, Cicala C, Sessa WC, Cirino G. Geldanamycin, an inhibitor of heat shock protein 90 (Hsp90) mediated signal transduction has anti-inflammatory effects and interacts with glucocorticoid receptor in vivo. Br J Pharmacol. 2000;131:13–16. doi: 10.1038/sj.bjp.0703549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallin RP, Lundqvist A, Moré SH, von Bonin A, Kiessling R, Ljunggren HG. Heat-shock proteins as activators of the innate immune system. Trends Immunol. 2002;23:130–135. doi: 10.1016/S1471-4906(01)02168-8. [DOI] [PubMed] [Google Scholar]
- 26.Qin S, Ni M, Wang X, Maurier-Mahé F, Shurland DL, Rodrigues GA. Inhibition of RPE cell sterile inflammatory responses and endotoxin-induced uveitis by a cell-impermeable HSP90 inhibitor. Exp Eye Res. 2011;93:889–897. doi: 10.1016/j.exer.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Rice JW, Veal JM, Fadden RP, Barabasz AF, Partridge JM, Barta TE, Dubois LG, Huang KH, Mabbett SR, Silinski MA, et al. Small molecule inhibitors of Hsp90 potently affect inflammatory disease pathways and exhibit activity in models of rheumatoid arthritis. Arthritis Rheum. 2008;58:3765–3775. doi: 10.1002/art.24047. [DOI] [PubMed] [Google Scholar]
- 28.De Nardo D, Masendycz P, Ho S, Cross M, Fleetwood AJ, Reynolds EC, Hamilton JA, Scholz GM. A central role for the Hsp90· Cdc37 molecular chaperone module in interleukin-1 receptor-associated-kinase-dependent signaling by Toll-like receptors. J Biol Chem. 2005;280:9813–9822. doi: 10.1074/jbc.M409745200. [DOI] [PubMed] [Google Scholar]
- 29.Shimp SK, III, Parson CD, Regna NL, Thomas AN, Chafin CB, Reilly CM, Rylander Nichole M. HSP90 inhibition by 17-DMAG reduces inflammation in J774 macrophages through suppression of Akt and nuclear factor-κB pathways. Inflamm Res. 2012;61:521–533. doi: 10.1007/s00011-012-0442-x. [DOI] [PubMed] [Google Scholar]
- 30.Hultin M, Gustafsson A, Hallström H, Johansson LÅ, Ekfeldt A, Klinge B. Microbiological findings and host response in patients with peri-implantitis. Clin Oral Implants Res. 2002;13:349–358. doi: 10.1034/j.1600-0501.2002.130402.x. [DOI] [PubMed] [Google Scholar]
- 31.Tokoro Y, Yamamoto T, Hara K. IL-1 beta mRNA as the predominant inflammatory cytokine transcript: Correlation with inflammatory cell infiltration into human gingiva. J Oral Pathol Med. 1996;25:225–231. doi: 10.1111/j.1600-0714.1996.tb01376.x. [DOI] [PubMed] [Google Scholar]
- 32.Panagakos FS, Aboyoussef H, Dondero R, Jandinski JJ. Detection and measurement of inflammatory cytokines in implant crevicular fluid: A pilot study. Int J Oral Maxillofac Implants. 1996;11:794–799. [PubMed] [Google Scholar]
- 33.Aboyoussef H, Carter C, Jandinski JJ, Panagakos FS. Detection of prostaglandin E2 and matrix metalloproteinases in implant crevicular fluid. Int J Oral Maxillofac Implants. 1998;13:689–696. [PubMed] [Google Scholar]
- 34.Lee SS, Jang JH, Kim KS, Yoo YJ, Kim YS, Lee SK. Failure of bone regeneration after demineralized bone matrix allograft in human maxillary sinus floor elevation. Basic Appl Pathol. 2009;2:125–130. doi: 10.1111/j.1755-9294.2009.01055.x. [DOI] [Google Scholar]
- 35.Bullon P, Fioroni M, Goteri G, Rubini C, Battino M. Immunohistochemical analysis of soft tissues in implants with healthy and peri-implantitis condition, and aggressive periodontitis. Clin Oral Implants Res. 2004;15:553–559. doi: 10.1111/j.1600-0501.2004.01072.x. [DOI] [PubMed] [Google Scholar]
- 36.Borsani E, Salgarello S, Stacchiotti A, Mensi M, Boninsegna R, Ricci F, Zanotti L, Rezzani R, Sapelli P, Bianchi R, Rodella LF. Altered immunolocalization of heat-shock proteins in human peri-implant gingiva. Acta Histochem. 2007;109:221–227. doi: 10.1016/j.acthis.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Borm ME, van Bodegraven AA, Mulder CJ, Kraal G, Bouma G. A NFKB1 promoter polymorphism is involved in susceptibility to ulcerative colitis. Int J Immunogenet. 2005;32:401–405. doi: 10.1111/j.1744-313X.2005.00546.x. [DOI] [PubMed] [Google Scholar]
- 38.Cochran DL. Inflammation and bone loss in periodontal disease. J Periodontol. 2008;79:1569–1576. doi: 10.1902/jop.2008.080233. (8 Suppl) [DOI] [PubMed] [Google Scholar]
- 39.Theoleyre S, Wittrant Y, Tat SK, Fortun Y, Redini F, Heymann D. The molecular triad OPG/RANK/RANKL: Involvement in the orchestration of pathophysiological bone remodeling. Cytokine Growth Factor Rev. 2004;15:457–475. doi: 10.1016/j.cytogfr.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Rakić M, Nikolić-Jakoba N, Struillout X, Petković-Curcin A, Stamatović N, Matić S, Janković S, Aleksić Z, Vasilić D, Leković V, Vojvodić D. Receptor activator of nuclear factor kappa B (RANK) as a determinant of peri-implantitis. Vojnosanit Pregl. 2013;70:346–351. doi: 10.2298/VSP1304346R. [DOI] [PubMed] [Google Scholar]
- 41.Rakic M, Lekovic V, Nikolic-Jakoba N, Vojvodic D, Petkovic-Curcin A, Sanz M. Bone loss biomarkers associated with peri-implantitis. A cross-sectional study. Clin Oral Implants Res. 2013;24:1110–1116. doi: 10.1111/j.1600-0501.2012.02518.x. [DOI] [PubMed] [Google Scholar]
- 42.Karban AS, Okazaki T, Panhuysen CI, Gallegos T, Potter JJ, Bailey-Wilson JE, Silverberg MS, Duerr RH, Cho JH, Gregersen PK, et al. Functional annotation of a novel NFKB1 promoter polymorphism that increases risk for ulcerative colitis. Hum Mol Genet. 2004;13:35–45. doi: 10.1093/hmg/ddh008. [DOI] [PubMed] [Google Scholar]
- 43.Zou YF, Wang F, Feng XL, Tao JH, Zhu JM, Pan FM, Su H. Association of NFKB1-94ins/delATTG promoter polymorphism with susceptibility to autoimmune and inflammatory diseases: A meta-analysis. Tissue antigens. 2011;77:9–17. doi: 10.1111/j.1399-0039.2010.01559.x. [DOI] [PubMed] [Google Scholar]
- 44.Li H, Gao L, Shen Z, Li CY, Li K, Li M, Lv YJ, Li CX, Gao TW, Liu YF. Association study of NFKB1 and SUMO4 polymorphisms in Chinese patients with psoriasis vulgaris. Arch Dermatol Res. 2008;300:425–433. doi: 10.1007/s00403-008-0843-4. [DOI] [PubMed] [Google Scholar]
- 45.Kosoy R, Concannon P. Functional variants in SUMO4, TAB2, and NFkappaB and the risk of type 1 diabetes. Genes Immun. 2005;6:231–235. doi: 10.1038/sj.gene.6364174. [DOI] [PubMed] [Google Scholar]
- 46.Elliott PJ, Zollner TM, Boehncke WH. Proteasome inhibition: A new anti-inflammatory strategy. J Mol Med (Berl) 2003;81:235–245. doi: 10.1007/s00109-003-0422-2. [DOI] [PubMed] [Google Scholar]
- 47.Marfella R, D'Amico M, Di Filippo C, Baldi A, Siniscalchi M, Sasso FC, Portoghese M, Carbonara O, Crescenzi B, Sangiuolo P, et al. Increased activity of the ubiquitin-proteasome system in patients with symptomatic carotid disease is associated with enhanced inflammation and may destabilize the atherosclerotic plaque: Effects of rosiglitazone treatment. J Am Coll Cardiol. 2006;47:2444–2455. doi: 10.1016/j.jacc.2006.01.073. [DOI] [PubMed] [Google Scholar]
- 48.Goldberg AL, Rock K. Not just research tools-proteasome inhibitors offer therapeutic promise. Nat Med. 2002;8:338–340. doi: 10.1038/nm0602-639b. [DOI] [PubMed] [Google Scholar]
- 49.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 50.Ciechanover A. The ubiquitin proteolytic system and pathogenesis of human diseases: A novel platform for mechanism-based drug targeting. Biochem Soc Trans. 2003;31:474–481. doi: 10.1042/bst0310474. [DOI] [PubMed] [Google Scholar]
- 51.Wang J, Maldonado MA. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell Mol Immunol. 2006;3:255–261. [PubMed] [Google Scholar]
- 52.Nothwang HG, Tamura T, Tanaka K, Ichihara A. Sequence analyses and inter-species comparisons of three novel human proteasomal subunits, HsN3, HsC7-I and HsC10-II, confine potential proteolytic active-site residues. Biochim Biophys Acta. 1994;1219:361–368. doi: 10.1016/0167-4781(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 53.Gallastegui N, Groll M. The 26S proteasome: Assembly and function of a destructive machine. Trends Biochem Sci. 2010;35:634–642. doi: 10.1016/j.tibs.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 54.Le Tallec B, Barrault MB, Courbeyrette R, Guérois R, Marsolier-Kergoat MC, Peyroche A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol Cell. 2007;27:660–674. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 55.Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]