ABSTRACT
Canonical Notch signaling has diverse functions during nervous system development and is critical for neural progenitor self-renewal, timing of differentiation and specification of various cell fates. A key feature of Notch-mediated self-renewal is its fluctuating activity within the neural progenitor cell population and the oscillatory expression pattern of the Notch effector Hes1 and its target genes. A negative feedback loop between Hes1 and neurogenic microRNA miR-9 was found to be part of this oscillatory clock. In a recent study we discovered that miR-9 expression is further modulated by direct binding of the Notch intracellular domain/RBPj transcriptional complex to the miR-9_2 promoter. In turn, miR-9 not only targets Hes1 but also Notch2 to attenuate Notch signaling and promote neuronal differentiation. Here, we discuss how the two interwoven feedback loops may provide an additional fail-save mechanism to control proliferation and differentiation within the neural progenitor cell population. Furthermore, we explore potential implications of miR-9-mediated regulation of Notch/Hes1 signaling with regard to neural progenitor homeostasis, patterning, timing of differentiation and tumor formation.
KEYWORDS: differentiation, hes, miR-9, neural stem cells, Notch, oscillation, proliferation
Introduction
How neural stem cells balance proliferation and differentiation is not only a key question in developmental neurobiology, but also highly relevant to our understanding of brain tumor formation and central nervous system (CNS) malformations. One very prominent player for maintaining neural stem cells is the Notch signaling pathway (reviewed by ref. 1). Notch signaling requires direct cell-cell contact, whereby the Notch receptor on one cell is activated by a Notch ligand (Delta/Delta-like (Dll) or Jagged) on a neighboring cell. The Notch receptor is a transmembrane receptor, of which several isoforms exist. The human genome contains 4 Notch receptors – Notch1 to 4. Binding of the ligand triggers a series of proteolytic cleavage steps, which ultimately results in the release of the intracellular domain of Notch (NICD) by the γ-secretase complex (see ref. 2 for detailed information). NICD translocates to the nucleus where it interacts with RBPj and MAML to form a transcriptional complex, which recognizes RBPj motifs in promoter regions activating gene transcription. In the embryonic nervous system, Notch activates the Hes (Hes1 and Hes5) and related Hey genes, which encode inhibitory basic helix-loop-helix (bHLH) proteins. Hes and Hey proteins maintain neural stem cells (NSCs) in an undifferentiated state by suppressing the expression of so-called pro-neural bHLH genes, including Neurogenins (Neurog1, Neurog2) and Ascl1 (reviewed by refs. 3, 4). Notch signaling acts in a context- and dose-dependent manner, and its effects within the cell are tightly controlled by intrinsic feedback loops and epigenetic factors. The expression of Notch pathway components is further regulated at the post-transcriptional level by the action of non-coding RNAs, including microRNAs (reviewed by ref. 5).
In particular, brain-enriched miRNA-9 has been shown to regulate the expression of Notch target genes in several organisms (reviewed by ref. 6). In Drosophila sensory organ precursor cells, miR-9 represses expression of the Notch downstream effector and pro-neural gene senseless, thereby controlling the number of neuronal precursor cells generated.7 In the developing brain of zebrafish and frog, miR-9 regulates the expression of Hes orthologs to drive neuronal differentiation of progenitor cells.8,9
In mouse neural progenitor cells, miR-9 and Hes1 form a double-negative feedback loop, with miR-9 targeting Hes1, and Hes1 repressing the expression of pri-miR-9.10 This auto-regulatory loop between miR-9 and Hes1 is believed to contribute to the ultradian Hes1 oscillations, which are important for neural progenitor proliferation.10
Until recently, studies into the interaction between miR-9 and Notch in human neural progenitor cells have been largely precluded by the limited access to human CNS tissue. With the availability of human pluripotent stem cells (PSCs) this restriction has been eliminated. Using long-term self-renewing neural stem cells (lt-NES cells11,12) derived from human PSCs we have recently identified another regulatory loop between miR-9 and Notch signaling besides the miR-9-Hes1 interaction.13 We found that miR-9 targets Hes1 as well as Notch2 to attenuate Notch activity in human neural stem cells. Reciprocally, miR-9 expression itself depends on active Notch signaling and is induced by binding of the NICD/RBPj complex to the pri-miR-9 locus. In the following, we revisit these findings and discuss what is known about the combinatorial interplay of Notch, Hes1 and miR-9 and its implications on neural progenitor maintenance, identity, differentiation and tumor formation. Finally, we delineate how the interaction of miR-9 with Notch/Hes relates to the vast regulatory network spanning around miR-9.
Mutual interaction between Notch signaling and miR-9 to regulate human neural stem cells
We first assessed expression of miR-9 during spontaneous differentiation of a specific population of human pluripotent stem cell-derived NSCs, i.e. long-term self-renewing neuroepithelial stem cells (lt-NES cells11,12). Expression of both miR-9 (generated from the 5′ arm of the mature miR-9 duplex) and miR-9* (generated from the 3′ arm) increase during lt-NES cell differentiation.14 In agreement with earlier studies, we observed that overexpression of miR-9/9* induces lt-NES cells to shift from proliferation to neuronal differentiation.15 We further found that miR-9/9* targets both Hes1 and Notch2 to decrease Notch activity in human neural stem cells.13 MicroRNA-9/9*-induced differentiation could be reversed by simultaneous overexpression of the active Notch intracellular domain (NICD). In turn, inhibition of miR-9/9* led to a reduction in lt-NES cell differentiation, which could be rescued using the γ-secretase inhibitor DAPT to block Notch signaling.13 The biologically active, mature miRNA is generated via two processing steps from primary (pri-) miRNA transcripts.16 Interestingly, inhibition of Notch signaling decreased both mature miR-9 and miR-9* as well as pri-miR-9_2 levels, while overexpression of NICD had the opposite effect.13 Following up on this, we found that miR-9/9* expression itself is triggered by binding of the NICD/RBPj complex to the miR-9_2 locus creating a relay-like mechanism fine-tuning Notch activity (Fig. 1) and thus human neural stem cell maintenance.13
Figure 1.
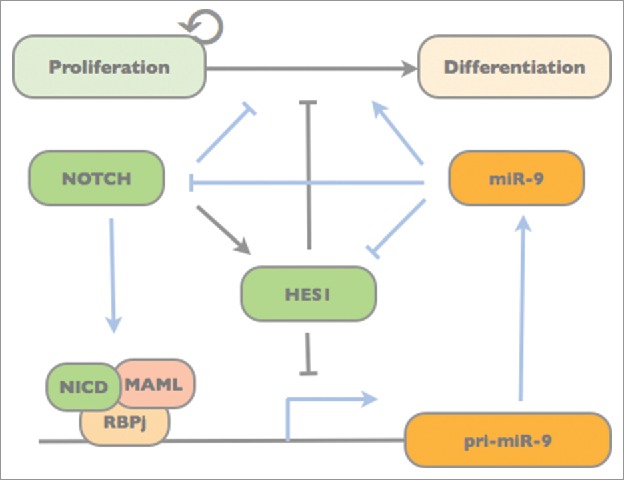
Model of the Notch/Hes1-miR-9 network and its impact on neural progenitor maintenance and differentiation. Blue lines indicate regulatory interactions demonstrated in Roese-Koerner et al. 2016.13 While Notch drives miR-9 expression,13 miR-9 dampens Notch activity by targeting Hes1 and Notch2.10,13 In turn, Hes1, which is also activated by Notch, inhibits miR-9 expression.10
The fact that miR-9 expression depends on active Notch signaling was previously suggested by other studies: Blockage of Notch activity in the developing zebrafish brain reduced expression of pri-miR-9, which according to sequence prediction might also carry potential target sites for the Notch transcriptional complex in its promoter.17 DAPT treatment also reduced the expression of miR-9 and miR-9* in cultured mouse retina.18 Mice lacking the Presenilin1 gene, which codes for the catalytic component of the γ-secretase complex, also exhibit decreased miR-9 expression levels.19 While we assessed the impact and expression of miR-9 and miR-9* together, most of the studies discussed above focused on miR-9 as the preferentially expressed miRNA. Nevertheless, together with our findings on a direct interaction of the NICD/RBPj with the miR-9 locus, these data show that miR-9 expression in the CNS is regulated by Notch signaling in various organisms. The strength of this regulation may, however, be context-dependent and further modulated by the interconnection of Notch and Hes, which was found to negatively affect miR-9 expression (Fig. 1).10
MicroRNA-9 contributes to and dampens Hes oscillation in neural progenitor cells
Interestingly, proliferative neural progenitor cells exhibit oscillatory expression of Hes1 with a period of 2–3 hours, while sustained Hes1 expression retards cell cycling.20 It is assumed that this oscillatory pattern of Hes1 and the out-of phase oscillation of its target genes (Ascl1, Neurog2 and Dll1) is crucial for maintaining the neural progenitor population (Fig. 2).20 Oscillation of the Notch ligand Dll1, which is induced by Neurog2 and repressed by Hes1, might lead to mutual activation of Notch signaling between neighboring cells, thus maintain stemness within the cell population.4,21 Inhibition of Notch signaling in neural progenitor cells, instead, results in a downregulation of Hes1 concomitantly with a sustained upregulation of pro-neural bHLH genes, which drive the cells toward differentiation. In this context, it is very important to understand how the oscillatory pattern of Hes1 is established and maintained within the neural progenitor population and how it is toned down to enable neuronal differentiation.
Figure 2.
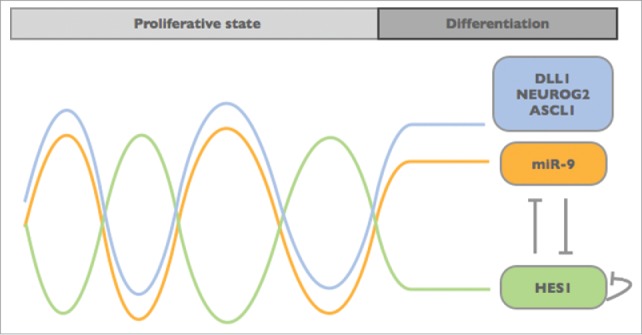
Model of the Hes1-miR-9 oscillation. The negative feedback between miR-9 and HES1 is considered to create an oscillatory pattern resembling the expression of DLL1 and bHLH factors like NEUROG2 and ASCL1. This out-of-phase oscillation may work like an internal clock timing neural progenitor proliferation and differentiation.10,20,23
The expression of Hes1 is induced by Notch signaling, which itself fluctuates within the progenitor population. Hes1 autorepression (Hes1 represses its own transcription) and instability of Hes1 mRNA are considered as the main driving force of the oscillatory pattern (reviewed by ref. 22). It was recently shown that miR-9 binds to and induces degradation of Hes1 mRNA.10 By doing so, miR-9 may contribute to the oscillatory pattern of Hes1. In fact, both miR-9 overexpression as well as its inhibition resulted in an dampening of Hes1 oscillations.10 Vice versa, Hes1 represses the transcription of miR-9, which results in an out-of-phase oscillation of miR-9. Thus, miR-9 and Hes1 seem to form a double-negative feedback loop, which is important for limiting Hes1 oscillations: Over time mature miR-9 accumulates (due to its longer half-life compared with the rather instable Hes1 mRNA and protein) until it reaches a certain threshold at which Hes1 oscillations are dampened, allowing the cells to proceed toward differentiation (Fig. 2).10
Mathematical modeling suggests that the Hes1-miR-9 double-negative feedback loop ensures that neural progenitors can acquire different levels of oscillatory Hes1 expression simply by tuning of miR-9 levels.23 Neural progenitor cells may express high Hes1 levels, which are associated with quiescence, or low Hes1 levels to proceed toward differentiation. Given that miR-9 accumulates over time to induce dampening of Hes1 oscillation, miR-9 might serve as an internal mechanism to measure the time until differentiation.23 Once the cells have embarked for differentiation they do not only down-regulate Notch activity but also the responsiveness of the Hes1 promoter toward NICD/RPBj transcriptional complex.24-26 In this context, the transcriptional repressor MyT1 has recently been found to negatively regulate Hes1 expression and to counteract Notch-mediated activation of the Hes1 locus.27 The expression of MyT1 itself is negatively coupled to Notch activity via its transcriptional activator Ascl1.27 Taken together, these findings suggest that miR-9 contributes to the initial dampening of the Hes1 oscillations at the onset of differentiation, while at later stages of differentiation other factors such as MyT1 join in to directly interfere with Hes1 transcription.
The involvement of Notch signaling in miR-9 expression regulation, upstream of Hes1, adds another layer of complexity to the Hes1-miR-9 interaction. Due to the entanglement of Notch and Hes1, it is quite challenging to decipher and predict the individual impact of Notch and Hes1 on miR-9 expression. This is even further complicated by the fact that miR-9 might target also other components of the transduction cascade, such as Notch ligands, Presenilin and the NICD-cofactor MAML as suggested by target prediction analysis.13 As Notch signaling is highly orchestrated, it might be difficult to separate the contribution of individual miR-9 targets with regard to self-renewal and differentiation of neural progenitor cells. The use of specific target protectors blocking miR-9 binding sites in mRNAs may help to further elucidate the relevance of miR-9 mediated repression for Notch regulation.
MicroRNA-9 and its interaction with Notch/Hes regulate fate specification of neural progenitor cells
Emerging evidence suggests that Notch signaling not only functions to maintain neural progenitor cells but that it also contributes to progenitor cell heterogeneity by regulating fate specification, the frequency of progenitor cell proliferation and their neurogenic versus gliogenic differentiation propensity (reviewed by refs. 1, 28). These processes were also found to be regulated by a cohort of brain-enriched miRNAs, including miR-9 (reviewed by ref. 29).
In developing zebrafish, miR-9 was shown to be involved in timing the transition from proliferation to differentiation. Ablation of miR-9 in zebrafish neural progenitor cells delayed their exit from proliferation and increased the generation of late-born neuronal subtypes.17 Since neural progenitor cells change their competence to generate different neural progeny over time, miR-9 may indirectly contribute to neuronal diversity. In this context, miR-9-mediated targeting of Hes genes was, again, identified as one of the underlying mechanisms. MicroRNA-9 was also demonstrated to influence temporal fate specification during neurogenesis in other model systems (reviewed in ref. 30). For instance, miR-9 – together with let-7 and miR-125 – promotes the transition of murine retinal progenitor cells from the early to the late-born progenitor state by targeting Protogenin and Lin28b.31 Depletion of the miR-9–2 and miR-9–3 loci in mice resulted in altered development of cortical layers, with the production of Cajal-Retzius cells and other early-born cortical cell types being most prominently affected.32,33 Thus, miR-9 may be involved in timing cortical development. Likewise, Notch signaling has also been shown to be important for temporal fate specification of cortical neural progenitors34,35 (see also ref. 36 for a review on this topic). Hence, it would be interesting to assess whether there is a direct interaction of miR-9 and Notch with respect to temporal fate patterning in the cortex.
Neural progenitor cells have to integrate different patterning signals to adopt different spatial identities along the anterior-posterior and dorso-ventral axes of the developing brain (reviewed by ref. 37). Interestingly, miR-9 and its target hairy1 (which belongs to the Hes family) have been shown to act in a region-specific manner in Xenopus neural progenitor cells.8 Although miR-9 inhibition resulted in an impaired neuronal differentiation across the whole Xenopus brain, its net effect on neural progenitor behavior differed along the anterior-posterior axis. In the hindbrain, miR-9 loss-of-function led to an enhanced proliferation, while in the forebrain an increased cell death was observed as the main effect, masking the proliferation effect. Bonev at al. argued, that rather than by miR-9 itself, this divergence seems to be mainly generated at the level of the hairy1 target genes, which appear to be different in hindbrain and forebrain.8
MicroRNA-9-mediated targeting of Hes genes also contributes to the refinement of boundary zones, which separate the different compartments of the brain. These boundary regions often act as important organizing centers for neural patterning and subtype specification. To maintain this function and the residing progenitor pool, boundary regions are characterized by a low neurogenic activity and persistent high Hes1 expression.20,38 MicroRNA-9 shows the opposite expression pattern and is absent from the boundary cells, but highly expressed in the surrounding neurogenic regions of zebrafish and mice.9,17,32 This mutual exclusive expression pattern was suggested to be established by the auto-regulation of miR-9 and Hes1. For instance it has been shown that in the zebrafish embryo miR-9 delimits the spatial extent of the midbrain-hindbrain boundary by clearance of the HES orthologs her5 and her9 in the adjacent tissue.9 Thus, both Hes1 and miR-9 are importantly involved in CNS patterning.
Notch and miR-9 may also have important roles in guiding the generation of astrocytes and oligodendrocytes at later stages of development. The transition from neurogenesis to gliogenesis has been shown to be regulated by Notch signaling. Notch promotes astrocytic differentiation of neural progenitor cells by directly inducing the expression of astroglial genes such as FABP7/BLBP and GFAP, and by cooperating with the transcription factor NFIA and the JAK/STAT pathway (reviewed by ref. 1). In contrast, miR-9 inhibits astroglial differentiation by targeting components of the JAK/STAT pathway.39,40 Zhao et al. found that miR-9 expression is directly induced by the pro-neural bHLH factor Neurog1, which also inhibits astroglial differentiation.40 Interestingly, Neurog1 itself is negatively regulated by Notch signaling.41 Thus, Neurog1-mediated induction of miR-9 might represent an additional loop within the Notch/Hes1/miR-9 auto-regulatory circuitry. However, it still remains to be determined whether there is a direct interaction of Notch and miR-9 with regard to astroglial cell differentiation. While Notch signaling also induces oligodendroglial progenitor cell specification, it has a negative effect on terminal differentiation and maturation of oligodendrocytes (reviewed by ref. 42). Interestingly, in this context, miR-9 seems to have a similar function to Notch and was found to inhibit differentiation of murine oligodendrocyte progenitor cells.43
MicroRNA-9 and Notch in the context of neural stem cell homeostasis and tumor formation
Changes in the balance of NSC proliferation and differentiation may have fatal effects: On the one hand, premature exhaustion of the progenitor pool due to low proliferation rates might manifest in microcephaly and the loss of late-born neural cells. On the other hand, uncontrolled proliferation or reactivation of stemness properties might promote tumor formation. Notch and miR-9 appear to be crucially involved in regulating neural progenitor cell expansion and tumorigenesis.
The impact of Notch on progenitor cell proliferation seems to depend on the level of its activation (reviewed by ref. 28). For instance, treating mouse neural progenitor cells with low doses of the constitutive active N1ICD promoted proliferation, while high levels of N1ICD led to growth arrest.44 Likewise, depending on their expression levels and dynamics, Hes genes can either drive proliferation or induce quiescence.38 Allowing cells to enter quiescence is very important for long-term maintenance of the adult neural progenitor pools that reside in the hippocampal dentate gyrus and in the subventricular zone in mammals (reviewed by ref. 45). Very recently, it was shown that miR-9 potentiated Notch signaling to maintain quiescence in adult zebrafish NSCs.46 In the context of this process, miR-9 is actively shuttled into the nucleus, a phenomenon mediated by binding of the miR-9-Ago complex to the shuttle protein TNRC6.46 The function of these nuclear Ago-miR-9 complexes stands in apparent contrast to the function of cytoplasmatic miR-9 in embryonic zebrafish NSCs, where miR-9 is expressed in cycling NSCs and serves to promote neuronal differentiation, an effect at least partially mediated by interference with Notch signaling.17
Considering its dose-dependent function, it is not surprising that Notch signaling is heavily regulated (reviewed by ref. 5). Interestingly, a primate-specific mechanism to titrate Notch with regard to neural progenitor proliferation has been identified.47 Rani et al. reported on the long non-coding RNA LncND, which is expressed in radial glial cells in the developing human cortex.47 LncND functions as an endogenous sponge for miR-143, which targets Notch mRNAs. In mouse radial glial cells, miR-143 also attenuates Notch signaling.47 However, the additional layer of regulation through LncND is missing, which might be key to the differences in the size of the cortical progenitor population in mice and humans. In fact, LncND gain-of-function in mouse radial glial cells led to an expansion of the neural progenitor population.47
A recent publication suggested a role for miR-9 in regulating postnatal cerebellar growth.48 This study originated from the observation that neonatal loss of N-Myc leads to reduced cerebellar mass due to a decrease in granule neuron progenitor proliferation. Follow-up experiments revealed that N-Myc is a negative regulator of miR-9, and that overexpression of miR-9 inhibits proliferation of granule neuron progenitors.48 Again, it would be interesting to assess whether miR-9 and Notch interact with each other with regard to cortical and cerebellar progenitor cell expansion.
Considering their key roles in regulating neural progenitor proliferation and differentiation, it is tempting to ascribe Notch and miR-9 tumor-promoting and tumor-suppressive effects, respectively. However, the role of both Notch and miR-9 in brain tumors is surprisingly complex. While Notch1 has been shown to be critical for proliferation and survival of glioma cells,49 it was also proposed that Notch cooperates with p53 to keep NSCs in a state of quiescence preventing them from entering a state of fast division.50 In this context, Notch signaling has a tumor-suppressive function, and its inactivation accelerates the growth of mouse gliomas. This is in line with the finding that high Notch activity – measured by HES5 levels – correlates with better survival of patients with specific subtypes of glioma.50 Expression of miR-9 is increased in various types of brain tumors (e.g. refs. 51-53), and many studies have shown that miR-9 regulates tumor cell proliferation (e.g., refs. 53-55) and migration (e.g., refs. 51, 53, 56). However, whether miR-9 affects these two processes positively or negatively depends on the type of brain tumor analyzed. Given these controversial results, more work is needed to understand the network spanning around miR-9 and Notch and to shed light on its role in brain tumorigenesis
Beyond Notch: Interaction of miR-9 with various transcriptional regulators
MicroRNAs often interact with transcriptional regulators forming feedforward or feedback loops (for review see refs. 57,58). Within these circuits miRNAs can either reinforce gene expressional programs or attenuate disturbing transcripts. By doing so, miRNAs contribute to the robustness of gene regulatory networks consolidating cell fate. However, miRNAs can also act as switches to induce cell fate changes. The double-negative feedback loop between miR-9 and Hes1 may represent both, depending on the state of progenitor cells. As long as miR-9 levels are rather low, the out-of-phase oscillation of Hes1 and miR-9 seems to contribute to a proliferative state. However, once a certain miR-9 threshold is reached, it allows switching toward neuronal differentiation.10,23 Besides Notch and Hes, miR-9 interacts with many other factors critical for neural progenitor stemness and differentiation. These factors include, for instance, the transcriptional regulators TLX and REST, both of which interfere with the expression of pro-neural genes including miR-9 itself (Fig. 3).59,60 Thus, both miR-9 and TLX as well as miR-9 and REST form double-negative feedback loops ensuring that miR-9 is only expressed in neural progenitor cells. Once miR-9 expression is induced, it reinforces its own expression by targeting its negative transcriptional regulators. Another interesting double-negative feedback loop, which in fact restricts miR-9 expression to neural cells, comprises Mef2C and HDAC4. In non-neuronal cells cooperative binding of transcription factor Mef2C and HDAC4 inhibits miR-9 expression.61 During neuronal differentiation, Mef2C is relieved from its partner HDAC4, turning it into an activator of the miR-9 locus. Here, again, miR-9 reinforces its own expression by targeting HDAC4 (Fig. 3).61
Figure 3.
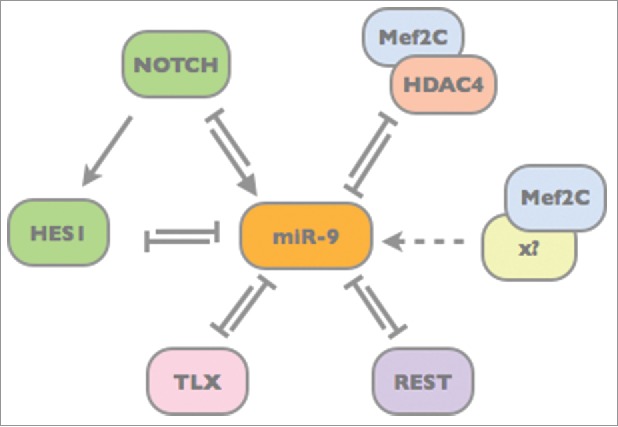
Model of the transcription factor network spanning around miR-9. MicroRNA-9 forms double-negative feedback loops with the transcription factors TLX, REST, HES1 and Mef2C/HDAC4.10,59-61 In turn, miR-9 expression is driven by Notch13 and – once it is freed from HDAC4 binding – by Mef2C.61 The action of these transcriptional regulators converges to ensure timely miR-9 expression in neural progenitor cells.
These complex interactions seem to tie a safety net through which miR-9 is able to buffer changes and pace differentiation and fate commitment of neural progenitors. Results from mathematical modeling suggest that the overall low abundance and stochastic distribution of miR-9 and Hes1 in individual cells might in fact increase robustness of the system toward intrinsic and extrinsic noise by opening up a window for feedback control on whether the correct number of differentiating cells have been generated.62 Interestingly, in drosophila, too, miR-9 was recently found to buffer transcriptional noise and genomic diversity by targeting senseless.63 Along the same line, miR-9 was found to reduce transcriptional noise in mouse by targeting the RNA-stability regulator TTP.64 In non-neuronal cells, TTP prevents the accumulation of neuronal mRNAs, while downregulation of TTP during neuronal differentiation leads to coordinated upregulation of pro-neuronal genes.64 MicroRNA-9* induces neuronal gene expression by targeting BAF53a, an important component of the Swi/Snf chromatin remodeling complex.65 MicroRNA-9 might even affect the miRNA repertoire itself by targeting the RNA-binding proteins Lin28a and Lin28b, which block the processing of a specific miRNA subset favoring differentiation.31,66 In turn, Lin28a binds to and destabilizes pre-miR-9, thereby forming yet another feedback loop to control miR-9 production and neuronal differentiation.67
The broad regulatory potential of miR-9 is also impressively reflected in its ability to induce neuronal reprogramming of fibroblasts upon ectopic expression together with miR-124 and miR-9*.68 Expression of miR-9 itself is induced by BRN2, which is one of the so-called BAM factors (Brn2, Ascl1, Mytl1) that were initially found to be sufficient for converting somatic cells into induced neuron (iNs).69 Follow-up studies showed that while miR-9 is not necessary for the early neural induction phase, it is required to obtain fully mature iNs from human fibroblasts.70,71 The function of miR-9 during iN maturation was linked to a repression of the mRNA splicing regulator PTBP2 by miR-9.71 However, considering the broad target repertoire of miR-9, which includes several key neuronal fate regulators, it is conceivable that other target genes contribute to the consolidation of neuronal differentiation in this conversion paradigm.
Conclusions and future directions
Both Notch/Hes signaling and miR-9 have profound effects on fate specification and neural progenitor cell heterogeneity. For some aspects of neural development, such as balancing proliferation and differentiation, temporal fate specification, and (non-neurogenic) boundary formation a direct interaction of miR-9 and Notch/Hes has been shown. However, it still remains to be determined whether miR-9 and Notch/Hes are directly linked with each other with regard to gliogenesis and tumorigenesis. So far, most of the studies have connected the function of miR-9 to the Hes family as important target genes. It might be interesting to explore the role of miR-9-mediated regulation of Notch signaling upstream of Hes. In this context, one could use specific target protectors to abolish the impact of miR-9 on selective target genes. Moreover, the ambivalent role of miR-9 between noise reduction and switching neural progenitor fate is still to be understood. Simultaneous mapping of miR-9, Notch and Hes1 expression using non-invasive high-resolution imaging and single cell genomics (as described in refs. 58,72) might help to explore their role in CNS development. It would be interesting to assess whether miR-9 and Hes1 are expressed in the same salt and pepper like fashion as Hes1 and Ascl1 (reviewed by ref. 4) and, if so, at which developmental stages and in which progenitor populations oscillatory expression occurs. Further knowledge on miR-9 regulation could be gained using reporter systems to detect miR-9 promoter activity and by targeted deletion of transcription factor binding sites. Deeper insight into the dynamics of the Notch/Hes1 miR-9 network during neural progenitor cell proliferation and differentiation might have important implications for understanding not only normal neural development but also the pathogenic principles underlying developmental disorders and brain tumor formation.
Disclosure of potential conflicts of interest
O.B. is co-founder of and has stock in LIFE & BRAIN GmbH. The other authors have no financial interests to disclose.
Acknowledgments
We thank Nils Christian Braun for critical reading of the manuscript.
Funding
Work in the laboratory of Oliver Brüstle was supported by the EU (HEALTH-F4–2013–602278-NeuroStemCellRepair; FP7-HEALTH-2010–266753-SCR&Tox, COLIPA, 115582-EBiSC; Horizon2020 grant 667301-COSYN; IMI2–2015–05–06 grant 115975–2-ADAPTED); the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept (grant 01ZX1314A-IntegraMent), the North Rhine Westphalian Program „LifeSciences.NRW,” European Regional Development Fund (grant EFRE-0800408-NeuRoWeg); the Ministry of Innovation, Science and Research of the State of North Rhine Westphalia (project StemCellFactory II; #005–1403–0102), the Stem Cell Network North Rhine Westphalia (start-up financing for interdisciplinary and multi-site projects-323–40000513), BONFOR and the Hertie Foundation.
References
- [1].Pierfelice T, Alberi L, Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron 2011; 69:840-55; PMID:21382546; https://doi.org/ 10.1016/j.neuron.2011.02.031 [DOI] [PubMed] [Google Scholar]
- [2].Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development 2011; 138:3593-612; PMID:21828089; https://doi.org/ 10.1242/dev.063610 [DOI] [PubMed] [Google Scholar]
- [3].Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci 2006; 7:93-102; PMID:16429119; https://doi.org/ 10.1038/nrn1847 [DOI] [PubMed] [Google Scholar]
- [4].Shimojo H, Ohtsuka T, Kageyama R. Dynamic expression of Notch signaling genes in neural stem/progenitor cells. Front Neurosci 2011; 5:78; PMID:21716644; https://doi.org/ 10.3389/fnins.2011.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schwanbeck R. The role of epigenetic mechanisms in Notch signaling during development. J Cell Physiol 2015; 230:969-81; PMID:25336183; https://doi.org/ 10.1002/jcp.24851 [DOI] [PubMed] [Google Scholar]
- [6].Coolen M, Katz S, Bally-Cuif L. miR-9: a versatile regulator of neurogenesis. Front Cell Neurosci 2013; 7:220; PMID:24312010; https://doi.org/ 10.3389/fncel.2013.00220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes & Development 2006; 20:2793-805; https://doi.org/ 10.1101/gad.1466306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bonev B, Pisco A, Papalopulu N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev Cell 2011; 20:19-32; PMID:21238922; https://doi.org/ 10.1016/j.devcel.2010.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Leucht C, Stigloher C, Wizenmann A, Klafke R, Folchert A, Bally-Cuif L. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci 2008; 11:641-8; PMID:18454145; https://doi.org/ 10.1038/nn.2115 [DOI] [PubMed] [Google Scholar]
- [10].Bonev B, Stanley P, Papalopulu N. MicroRNA-9 modulates Hes1 ultradian oscillations by forming a double-negative feedback loop. Cell Rep 2012; 2:10-18; PMID:22840391; https://doi.org/ 10.1016/j.celrep.2012.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Koch P, Opitz T, Steinbeck JA, Ladewig J, Brüstle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc Natl Acad Sci U S A 2009; 106:3225-30; PMID:19218428; https://doi.org/ 10.1073/pnas.0808387106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Falk A, Koch P, Kesavan J, Takashima Y, Ladewig J, Alexander M, Wiskow O, Tailor J, Trotter M, Pollard S, et al.. Capture of neuroepithelial-like stem cells from pluripotent stem cells provides a versatile system for in vitro production of human neurons. PLoS One 2012; 7:e29597; PMID:22272239; https://doi.org/ 10.1371/journal.pone.0029597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Roese-Koerner B, Stappert L, Berger T, Braun NC, Veltel M, Jungverdorben J, Evert BO, Peitz M, Borghese L, Brüstle O. Reciprocal regulation between bifunctional miR-9/9* and its transcriptional modulator Notch in human neural stem cell self-renewal and differentiation. Stem Cell Rep 2016; 7:207-19; PMID:27426040; https://doi.org/ 10.1016/j.stemcr.2016.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stappert L, Borghese L, Roese-Koerner B, Weinhold S, Koch P, Terstegge S, Uhrberg M, Wernet P, Brüstle O. MicroRNA-based promotion of human neuronal differentiation and subtype specification. PLoS One 2013; 8:e59011; PMID:23527072; https://doi.org/ 10.1371/journal.pone.0059011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Roese-Koerner B, Stappert L, Koch P, Brüstle O, Borghese L. Pluripotent stem cell-derived somatic stem cells as tool to study the role of microRNAs in early human neural development. Curr Mol Med 2013; 13:707-22; https://doi.org/ 10.2174/1566524011313050003 [DOI] [PubMed] [Google Scholar]
- [16].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; https://doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- [17].Coolen M, Thieffry D, Drivenes Ø, Becker TS, Bally-Cuif L. miR-9 controls the timing of neurogenesis through the direct inhibition of antagonistic factors. Dev Cell 2012; 22:1052-64; PMID:22595676; https://doi.org/ 10.1016/j.devcel.2012.03.003 [DOI] [PubMed] [Google Scholar]
- [18].Georgi SA, Reh TA. Dicer is required for the transition from early to late progenitor state in the developing mouse retina. J Neurosci 2010; 30:4048-61; PMID:20237275; https://doi.org/ 10.1523/JNEUROSCI.4982-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 2003; 9:1274-81; PMID:13130141; https://doi.org/ 10.1261/rna.5980303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shimojo H, Ohtsuka T, Kageyama R. Oscillations in Notch signaling regulate maintenance of neural progenitors. Neuron 2008; 58:52-64; PMID:18400163; https://doi.org/ 10.1016/j.neuron.2008.02.014 [DOI] [PubMed] [Google Scholar]
- [21].Shimojo H, Isomura A, Ohtsuka T, Kori H, Miyachi H, Kageyama R. Oscillatory control of Delta-like1 in cell interactions regulates dynamic gene expression and tissue morphogenesis. Genes Dev 2016; 30:102-16; https://doi.org/ 10.1101/gad.270785.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kageyama R, Shimojo H, Imayoshi I. Dynamic expression and roles of Hes factors in neural development. Cell Tissue Res 2014; 359:125-33; PMID:24850276; https://doi.org/ 10.1007/s00441-014-1888-7 [DOI] [PubMed] [Google Scholar]
- [23].Goodfellow M, Phillips NE, Manning C, Galla T, Papalopulu N. microRNA input into a neural ultradian oscillator controls emergence and timing of alternative cell states. Nat Comms 2014; 5:3399; https://doi.org/ 10.1038/ncomms4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mizutani KI, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 2007; 449:351-5; PMID:17721509; https://doi.org/ 10.1038/nature06090 [DOI] [PubMed] [Google Scholar]
- [25].Kawaguchi A, Ikawa T, Kasukawa T, Ueda HR, Kurimoto K, Saitou M, Matsuzaki F. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development 2008; 135:3113-24; PMID:18725516; https://doi.org/ 10.1242/dev.022616 [DOI] [PubMed] [Google Scholar]
- [26].Nelson BR, Hodge RD, Bedogni F, Hevner RF. Dynamic interactions between intermediate neurogenic progenitors and radial glia in embryonic mouse neocortex: Potential role in Dll1-Notch Signaling. J Neurosci 2013; 33:9122-39; PMID:23699523; https://doi.org/ 10.1523/JNEUROSCI.0791-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vasconcelos FF, Sessa A, Laranjeira C, Raposo AA, Teixeira V, Hagey DW, Tomaz DM, Muhr J, Broccoli V, Castro DS. MyT1 counteracts the neural progenitor program to promote vertebrate neurogenesis. Cell Rep 2016; 17:469-83; PMID:27705795; https://doi.org/ 10.1016/j.celrep.2016.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Giachino C, Taylor V. Notching up neural stem cell homogeneity in homeostasis and disease. Front Neurosci. 2014; 8:32; PMID:24611040; https://doi.org/ 10.3389/fnins.2014.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bian S, Xu TL, Sun T. Tuning the cell fate of neurons and glia by microRNAs. Curr Opin Neurobiol 2013; 23:928-34; PMID:23978589; https://doi.org/ 10.1016/j.conb.2013.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Stappert L, Roese-Koerner B, Brüstle O. The role of microRNAs in human neural stem cells, neuronal differentiation and subtype specification. Cell Tissue Res 2014; 359:47-64; PMID:25172833; https://doi.org/ 10.1007/s00441-014-1981-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].La Torre A, Georgi S, Reh TA. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc Natl Acad Sci U S A 2013; 110:E2362-70; PMID:23754433; https://doi.org/ 10.1073/pnas.1301837110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shibata M, Kurokawa D, Nakao H, Ohmura T, Aizawa S. MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J Neurosci 2008; 28:10415-21; PMID:18842901; https://doi.org/ 10.1523/JNEUROSCI.3219-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci 2011; 31:3407-22; PMID:21368052; https://doi.org/ 10.1523/JNEUROSCI.5085-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wu ZQ, Li D, Huang Y, Chen XP, Huang W, Liu CF, Zhao HQ, Xu RX, Cheng M, Schachner M, et al.. Caspr controls the temporal specification of neural progenitor cells through Notch signaling in the developing mouse cerebral cortex. Cerebral Cortex 2016; 27:1369-85; PMID:26740489; https://doi.org/ 10.1093/cercor/bhv318 [DOI] [PubMed] [Google Scholar]
- [35].Mitzutani KI, Saito T. Progenitors resume generating neurons after temporary inhibition of neurogenesis by Notch activation in the mammalian cerebral cortex. Development 2005; 132:1295-304; PMID:15750183; https://doi.org/ 10.1242/dev.01693 [DOI] [PubMed] [Google Scholar]
- [36].Corbin JG, Gaiano N, Juliano SL, Poluch S, Stancik E, Haydar TF. Regulation of neural progenitor cell development in the nervous system. J Neurochem 2008; 106:2272-87; PMID:18819190; https://doi.org/ 10.1111/j.1471-4159.2008.05522.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Le Dréau G, Martí E. Dorsal-ventral patterning of the neural tube: A tale of three signals. Devel Neurobio 2012; 72:1471-81; https://doi.org/ 10.1002/dneu.22015 [DOI] [PubMed] [Google Scholar]
- [38].Baek JH, Hatakeyama J, Sakamoto S, Ohtsuka T, Kageyama R. Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system. Development 2006; 133:2467-76; PMID:16728479; https://doi.org/ 10.1242/dev.02403 [DOI] [PubMed] [Google Scholar]
- [39].Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 2006; 24:857-64; PMID:16357340; https://doi.org/ 10.1634/stemcells.2005-0441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhao J, Lin Q, Kim KJ, Dardashti FD, Kim J, He F, Sun Y. Ngn1 inhibits astrogliogenesis throughinduction of miR-9 during neuronal fate specification. Elife 2015; 4:e06885; https://doi.org/ 10.7554/eLife.06885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jeon SJ, Fujioka M, Kim SC, Edge AS. Notch signaling alters sensory or neuronal cell fate specification of inner ear stem cells. J Neurosci 2011; 31:8351-8; PMID:21653840; https://doi.org/ 10.1523/JNEUROSCI.6366-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].He L, Lu QR. Coordinated control of oligodendrocyte development by extrinsic and intrinsic signaling cues. Neurosci Bull 2013; 29:129-43; PMID:23494530; https://doi.org/ 10.1007/s12264-013-1318-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Buller B, Chopp M, Ueno Y, Zhang L, Zhang RL, Morris D, Zhang Y, Zhang ZG. Regulation of serum response factor by miRNA-200 and miRNA-9 modulates oligodendrocyte progenitor cell differentiation. Glia 2012; 60:1906-14; PMID:22907787; https://doi.org/ 10.1002/glia.22406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Guentchev M, McKay RD. Notch controls proliferation and differentiation of stem cells in a dose-dependent manner. Euro J Neurosci 2006; 23:2289-96; PMID:16706837; https://doi.org/ 10.1111/j.1460-9568.2006.04766.x [DOI] [PubMed] [Google Scholar]
- [45].Alvarez-Buylla A, Kohwi M, Nguyen TM, Merkle FT. The heterogeneity of adult neural stem cells and the emerging complexity of their niche. Cold Spring Harb Symp Quant Biol 2008; 73:357-65; PMID:19022766; https://doi.org/ 10.1101/sqb.2008.73.019 [DOI] [PubMed] [Google Scholar]
- [46].Katz S, Cussigh D, Urbán N, Blomfield I, Guillemot F, Bally-Cuif L, Coolen M. A nuclear role for miR-9 and Argonaute proteins in balancing quiescent and activated neural stem cell states. Cell Rep 2016; 17:1383-98; PMID:27783951; https://doi.org/ 10.1016/j.celrep.2016.09.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Rani N, Nowakowski TJ, Zhou H, Godshalk SE, Lisi V, Kriegstein AR, Kosik KS. A Primate lncRNA mediates Notch signaling during neuronal development by sequestering miRNA. Neuron 2016; 90:1174-88; PMID:27263970; https://doi.org/ 10.1016/j.neuron.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ma M, Wu W, Li Q, Li J, Sheng Z, Shi J, Zhang M, Yang H, Wang Z, Sun R, et al.. N-myc is a key switch regulating the proliferation cycle of postnatal cerebellar granule cell progenitors. Sci Rep 2015; 5:12740; PMID:26238256; https://doi.org/ 10.1038/srep12740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Purow BW, Haque RM, Noel MW, Su Q, Burdick MJ, Lee J, Sundaresan T, Pastorino S, Park JK, Mikolaenko I, et al.. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res 2005; 65:2353-63; PMID:15781650; https://doi.org/ 10.1158/0008-5472.CAN-04-1890 [DOI] [PubMed] [Google Scholar]
- [50].Giachino C, Boulay J-L, Ivanek R, Alvarado A, Tostado C, Lugert S, Tchorz J, Coban M, Mariani L, Bettler B, et al.. A tumor suppressor function for Notch signaling in forebrain tumor subtypes. Cancer Cell 2015; 28:730-42; PMID:26669487; https://doi.org/ 10.1016/j.ccell.2015.10.008 [DOI] [PubMed] [Google Scholar]
- [51].Nass D, Rosenwald S, Meiri E, Gilad S, Tabibian-Keissar H, Schlosberg A, Kuker H, Sion-Vardy N, Tobar A, Kharenko O, et al.. MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol 2009; 19:375-83; PMID:18624795; https://doi.org/ 10.1111/j.1750-3639.2008.00184.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Malzkorn B, Wolter M, Liesenberg F, Grzendowski M, Stühler K, Meyer HE, Reifenberger G. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol 2010; 20:539-50; PMID:19775293; https://doi.org/ 10.1111/j.1750-3639.2009.00328.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Tan X, Wang S, Yang B, Zhu L, Yin B, Chao T, Zhao J, Yuan J, Qiang B, Peng X. The CREB-miR-9 negative feedback minicircuitry coordinates the migration and proliferation of glioma cells. PLoS One 2012; 7:e49570; PMID:23185366; https://doi.org/ 10.1371/journal.pone.0049570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ferretti E, De Smaele E, Miele E, Laneve P, Po A, Pelloni M, Paganelli A, Di Marcotullio L, Caffarelli E, Screpanti I, et al.. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J 2008; 27:2616-27; PMID:18756266; https://doi.org/ 10.1038/emboj.2008.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Laneve P, Di Marcotullio L, Gioia U, Fiori ME, Ferretti E, Gulino A, Bozzoni I, Caffarelli E. The interplay between microRNAs and the neurotrophin receptor tropomyosin-related kinase C controls proliferation of human neuroblastoma cells. Proc Natl Acad Sci U S A 2007; 104:7957-62; PMID:17483472; https://doi.org/ 10.1073/pnas.0700071104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, Gao FB. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 2010; 6:323-35; PMID:20362537; https://doi.org/ 10.1016/j.stem.2010.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Peláez N, Carthew RW. Biological robustness and the role of microRNAs: A network perspective. Current Topics in Developmental Biology 2012:237-55; PMID:22365741; https://doi.org/ 10.1016/B978-0-12-387038-4.00009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell 2012; 149:515-24; PMID:22541426; https://doi.org/ 10.1016/j.cell.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol 2009; 16:365-71; PMID:19330006; https://doi.org/ 10.1038/nsmb.1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Laneve P, Gioia U, Andriotto A, Moretti F, Bozzoni I, Caffarelli E. A minicircuitry involving REST and CREB controls miR-9-2 expression during human neuronal differentiation. Nucleic Acids Res 2010; 38:6895-905; PMID:20624818; https://doi.org/ 10.1093/nar/gkq604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Davila JL, Goff LA, Ricupero CL, Camarillo C, Oni EN, Swerdel MR, Toro-Ramos AJ, Li J, Hart RP. A positive feedback mechanism that regulates expression of miR-9 during neurogenesis. PLoS One 2014; 9:e94348; PMID:24714615; https://doi.org/ 10.1371/journal.pone.0094348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Phillips NE, Manning CS, Pettini T, Biga V, Marinopoulou E, Stanley P, Boyd J, Bagnall J, Paszek P, Spiller DG, et al.. Stochasticity in the miR-9/Hes1 oscillatory network can account for clonal heterogeneity in the timing of differentiation. Elife 2016; 5:e16118; https://doi.org/ 10.7554/eLife.16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Cassidy JJ, Jha AR, Posadas DM, Giri R, Venken KJT, Ji J, Jiang H, Bellen HJ, White KP, Carthew RW. miR-9a minimizes the phenotypic impact of genomic diversity by buffering a transcription factor. Cell 2013; 155:1556-67; PMID:24360277; https://doi.org/ 10.1016/j.cell.2013.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Dai W, Li W, Hoque M, Li Z, Tian B, Makeyev EV. A post-transcriptional mechanism pacing expression of neural genes with precursor cell differentiation status. Nat Comms 2015; 6:7576; https://doi.org/ 10.1038/ncomms8576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 2009; 460:642-6 PMID: 19561591; https://doi.org/ 10.1038/nature08139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Eda A, Tamura Y, Yoshida M, Hohjoh H. Systematic gene regulation involving miRNAs during neuronal differentiation of mouse P19 embryonic carcinoma cell. Biochem Biophys Res Commun 2009; 388:648-53; PMID:19679099; https://doi.org/ 10.1016/j.bbrc.2009.08.040 [DOI] [PubMed] [Google Scholar]
- [67].Nowak JS, Choudhury NR, de Lima Alves F, Rappsilber J, Michlewski G. Lin28a regulates neuronal differentiation and controls miR-9 production. Nat Comms 2014; 5:3687; https://doi.org/ 10.1038/ncomms4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 2011; 476:228-31; PMID:21753754; https://doi.org/ 10.1038/nature10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010; 463:1035-41; PMID:20107439; https://doi.org/ 10.1038/nature08797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al.. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 2013; 152:82-96; PMID:23313552; https://doi.org/ 10.1016/j.cell.2012.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Xue Y, Qian H, Hu J, Zhou B, Zhou Y, Hu X, Karakhanyan A, Pang Z, Fu XD. Sequential regulatory loops as key gatekeepers for neuronal reprogramming in human cells. Nat Neurosci 2016; 19:807-15; PMID:27110916; https://doi.org/ 10.1038/nn.4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Liu Z, Keller PJ. Emerging Imaging and Genomic Tools for Developmental Systems Biology. Dev Cell 2016; 36:597-610; PMID:27003934; https://doi.org/ 10.1016/j.devcel.2016.02.016 [DOI] [PubMed] [Google Scholar]


